Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Culture
2.2. Acid Tolerance and Bile Tolerance
2.3. Antimicrobial Activity
2.4. C. elegans Culture Conditions
2.5. C. elegans Pmk-1-Mediated Screening for Anti-Aging Activity
2.6. C. elegans Lifespan Analysis
2.7. Measurement of Intestinal Colonization of C. elegans
2.8. Whole-Genome Sequencing and Pangenome Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Functional Analysis of Potential Probiotics
3.2. Screening of Probiotics That Have an Anti-Aging Effect on C. elegans
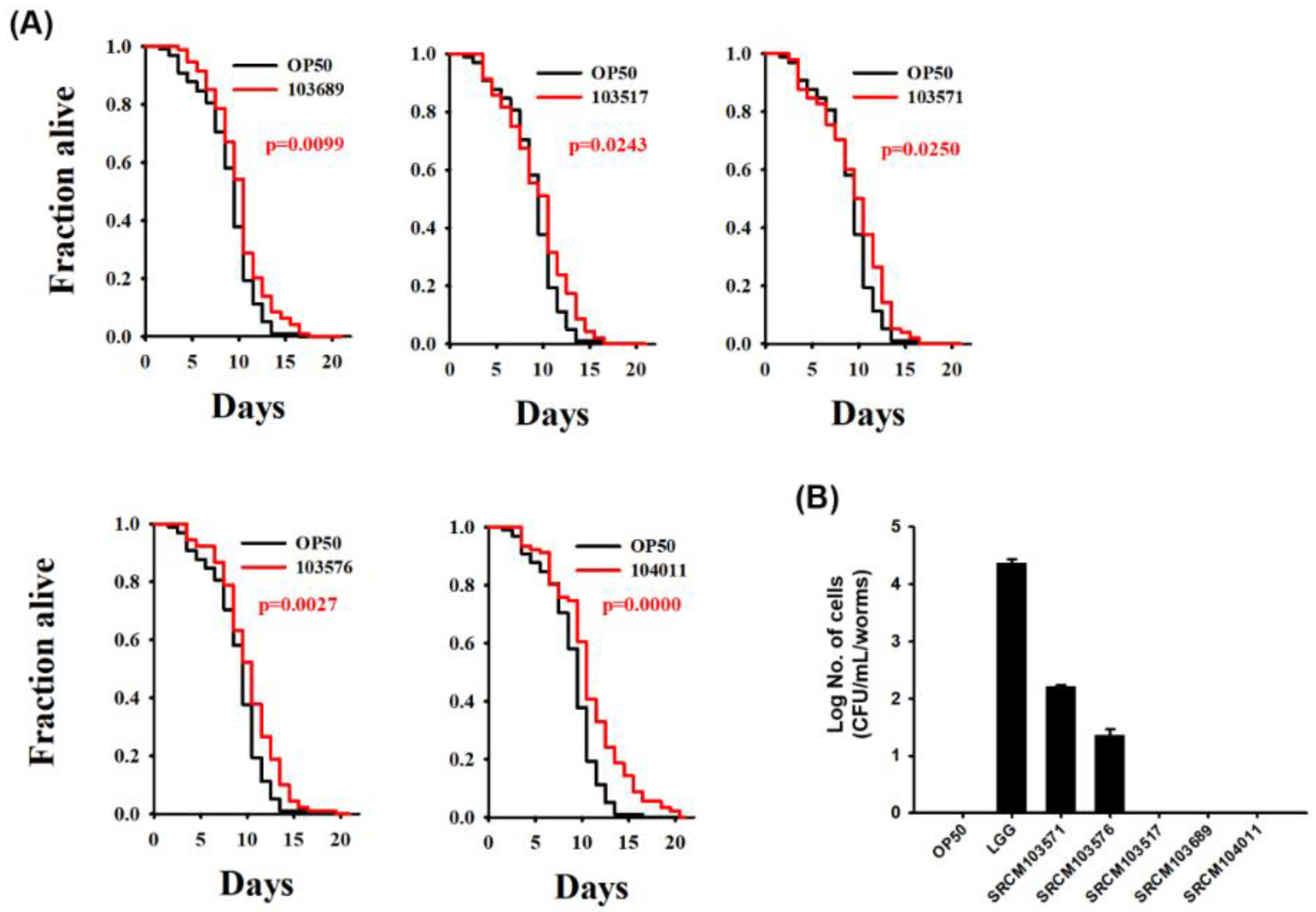
3.3. Bacillus Extends the Lifespan of C. elegans
3.4. Colonization of Bacteria on C. elegans Intestine
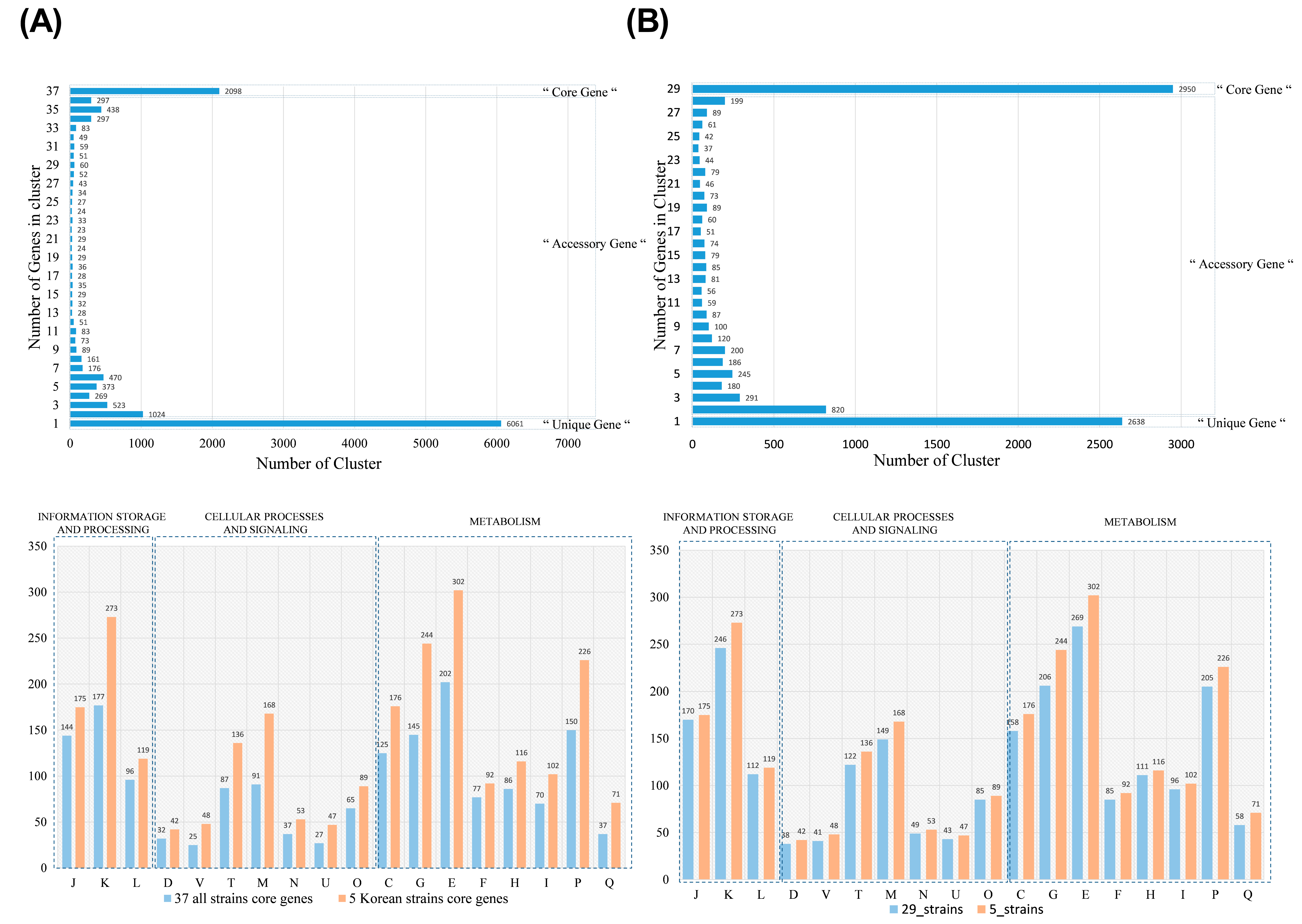
3.5. Whole-Genome Sequencing and Pangenome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Daliri, E.; Lee, B. Current trends and future perspectives on functional foods and nutraceuticals. In Beneficial Microorganisms in Food and Nutraceuticals; Springer: Cham, Switzerland, 2015; pp. 221–244. ISBN 978-3-319-23176-1. [Google Scholar]
- Das, G.; Paramithiotis, S.; Sundaram Sivamaruthi, B.; Wijaya, C.H.; Suharta, S.; Sanlier, N.; Shin, H.-S.; Patra, J.K. Traditional fermented foods with anti-aging effect: A concentric review. Food Res. Int. 2020, 134, 109269. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Hong, V.M.; Yang, J.; Hyun, H.; Im, J.J.; Hwang, J.; Yoon, S.; Kim, J.E. A review of fermented foods with beneficial effects on brain and cognitive function. Prev. Nutr. Food Sci. 2016, 21, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, M.; De Rosa, M.; Stellavato, A.; Lamberti, M.; Marzaioli, I.; Giuliano, M. In vitro evaluation of Lactobacillus plantarum DSMZ 12028 as a probiotic: Emphasis on innate immunity. Int. J. Food Microbiol. 2009, 135, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ji, H. Influence of Probiotics on Dietary Protein Digestion and Utilization in the Gastrointestinal Tract. Available online: https://www.eurekaselect.com/162238/article (accessed on 19 November 2020).
- Fooks, L.J.; Fuller, R.; Gibson, G.R. Prebiotics, probiotics and human gut microbiology. Int. Dairy J. 1999, 9, 53–61. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Kiran, M.; Gharat, P.; Vakharia, M.; Ranganathan, N. Specific probiotics for chronic kidney disease: A review. Indian Pract. 2019, 72, 29–40. [Google Scholar]
- La Fata, G.; Weber, P.; Mohajeri, M.H. Probiotics and the gut immune system: Indirect regulation. Probiotics Antimicrob. Proteins 2018, 10, 11–21. [Google Scholar] [CrossRef]
- Sharif, M.K.; Mahmood, S.; Ahsan, F. Chapter 2—Role of probiotics toward the improvement of gut health with special reference to colorectal cancer. In Diet, Microbiome and Health; Holban, A.M., Grumezescu, A.M., Eds.; Handbook of Food Bioengineering; Academic Press: Faisalabad, Pakistan, 2018; pp. 35–50. ISBN 978-0-12-811440-7. [Google Scholar]
- Chun, B.H.; Kim, K.H.; Jeong, S.E.; Jeon, C.O. The effect of salt concentrations on the fermentation of doenjang, a traditional Korean fermented soybean paste. Food Microbiol. 2020, 86, 103329. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Lee, N.-K.; Han, E.J.; Paik, H.-D. Characterization of subtilin KU43 produced by Bacillus subtilis KU43 isolated from traditional Korean fermented foods. Food Sci. Biotechnol. 2012, 21, 1433–1438. [Google Scholar] [CrossRef]
- Khan, I.; Kang, S.C. Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi—A traditional Korean fermented food. Food Control. 2016, 60, 88–94. [Google Scholar] [CrossRef]
- Duc, L.H.; Hong, H.A.; Barbosa, T.M.; Henriques, A.O.; Cutting, S.M. Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 2004, 70, 2161–2171. [Google Scholar] [CrossRef] [PubMed]
- Mingmongkolchai, S.; Panbangred, W. Bacillus probiotics: An alternative to antibiotics for livestock production. J. Appl. Microbiol. 2018, 124, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- De Boer, A.S.; Diderichsen, B. On the safety of Bacillus subtilis and B. amyloliquefaciens: A review. Appl. Microbiol. Biotechnol. 1991, 36, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Komura, T.; Ikeda, T.; Hoshino, K.; Shibamura, A.; Nishikawa, Y. Caenorhabditis elegans as an alternative model to study senescence of host defense and the prevention by immunonutrition. Adv. Exp. Med. Biol. 2012, 710, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.H.; Yun, B.; Choi, H.J.; Ryu, S.; Lee, W.J.; Oh, M.-H.; Song, M.-H.; Kim, J.N.; Oh, S.; Kim, Y.; et al. Simple evaluation of Listeria monocytogenes pathogenesis using Caenorhabditis elegans animal model. Food Sci. Anim. Resour. 2019, 39, 84–92. [Google Scholar] [CrossRef]
- Liu, C.; Lu, J.; Lu, L.; Liu, Y.; Wang, F.; Xiao, M. Isolation, structural characterization and immunological activity of an exopolysaccharide produced by Bacillus licheniformis 8-37-0-1. Bioresour. Technol. 2010, 101, 5528–5533. [Google Scholar] [CrossRef]
- Oh, A.; Daliri, E.B.; Oh, D.H. Screening for potential probiotic bacteria from Korean fermented soybean paste: In vitro and Caenorhabditis elegans model testing. LWT 2018, 88, 132–138. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Park, M.R.; Ryu, S.; Maburutse, B.E.; Oh, N.S.; Kim, S.H.; Oh, S.; Jeong, S.-Y.; Jeong, D.-Y.; Oh, S.; Kim, Y. Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of Caenorhabditis elegans through a nuclear hormone receptor. Sci. Rep. 2018, 8, 7441. [Google Scholar] [CrossRef]
- Heo, J.; Shin, D.; Chang, S.Y.; Bogere, P.; Park, M.R.; Ryu, S.; Lee, W.J.; Yun, B.; Lee, H.K.; Kim, Y.; et al. Comparative genome analysis and evaluation of probiotic characteristics of Lactobacillus plantarum strain JDFM LP11. Korean J. Food Sci. Anim Resour. 2018, 38, 878–888. [Google Scholar] [CrossRef]
- Fuller, R. (Ed.) Probiotics: The Scientific Basis; Springer Nature: Dordrecht, The Netherlands, 1992; ISBN 978-94-010-5043-2. [Google Scholar]
- Kim, Y.; Cho, J.-Y.; Kuk, J.-H.; Moon, J.-H.; Cho, J.-I.; Kim, Y.-C.; Park, K.-H. Identification and antimicrobial activity of phenylacetic acid produced by Bacillus licheniformis isolated from fermented soybean, Chungkook-Jang. Curr. Microbiol. 2004, 48, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Kim, J.-C.; Kwon, T. Effect of Chongkukjang on blood glucose and lipid profile in neonatal streptozotocin-induced diabetic rats. Food Sci. Biotechnol. 2003, 12, 544–547. [Google Scholar]
- Kim, K.P.; Rhee, C.H.; Park, H.D. Degradation of cholesterol by Bacillus subtilis SFF34 isolated from Korean traditional fermented flatfish. Lett. Appl. Microbiol. 2002, 35, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Syahbanu, F.; Giriwono, P.E.; Tjandrawinata, R.R.; Suhartono, M.T. Molecular analysis of a fibrin-degrading enzyme from Bacillus subtilis K2 isolated from the Indonesian soybean-based fermented food moromi. Mol. Biol. Rep. 2020, 47, 8553–8563. [Google Scholar] [CrossRef] [PubMed]
- Wilks, J.C.; Kitko, R.D.; Cleeton, S.H.; Lee, G.E.; Ugwu, C.S.; Jones, B.D.; BonDurant, S.S.; Slonczewski, J.L. Acid and base stress and transcriptomic responses in Bacillus subtilis. Appl. Environ. Microbiol. 2009, 75, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Ciarciaglini, G.; Hill, P.J.; Davies, K.; McClure, P.J.; Kilsby, D.; Brown, M.H.; Coote, P.J. Germination-induced bioluminescence, a route to determine the inhibitory effect of a combination preservation treatment on bacterial spores. Appl. Environ. Microbiol. 2000, 66, 3735–3742. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Zhang, L.; Liu, W.; Zhang, Y.; Zhang, X.; Sun, T. In vitro assessment of probiotic properties of Bacillus isolated from naturally fermented congee from Inner Mongolia of China. World J. Microbiol. Biotechnol. 2010, 26, 1369–1377. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Kamei, T.; Baek, D.H.; Shin, S.Y.; Sung, M.H.; Soda, K.; Yagi, T.; Misono, H. Isolation of Bacillus subtilis (chungkookjang), a poly-gamma-glutamate producer with high genetic competence. Appl. Microbiol. Biotechnol. 2001, 57, 764–769. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Yang, H.-J.; Jeong, S.-Y.; Jeong, D.-Y. Identification of characterization and statistical optimization of medium constituent for Bacillus subtilis SCJ4 isolated from Korean traditional fermented food. Korean J. Microbiol. 2015, 51, 48–60. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef]
- Foster, K.J.; Cheesman, H.K.; Liu, P.; Peterson, N.D.; Anderson, S.M.; Pukkila-Worley, R. Innate immunity in the C. elegans intestine is programmed by a neuronal regulator of AWC olfactory Neuron Development. Cell Rep. 2020, 31, 107478. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Feinbaum, R.; Alloing, G.; Emerson, F.E.; Garsin, D.A.; Inoue, H.; Tanaka-Hino, M.; Hisamoto, N.; Matsumoto, K.; Tan, M.-W.; et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 2002, 297, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Heo, J.; Park, M.R.; Song, M.-H.; Kim, J.N.; Jo, S.H.; Jeong, D.-Y.; Lee, H.K.; Kim, Y.; Oh, S. Genome characteristics of Lactobacillus fermentum strain JDFM216 for application as probiotic bacteria. J. Microbiol. Biotechnol. 2017, 27, 1266–1271. [Google Scholar] [CrossRef] [PubMed]
- Goldin, B.R. Health benefits of probiotics. Br. J. Nutr. 1998, 80, S203–S207. [Google Scholar] [CrossRef]
- De Groote, M.A.; Frank, D.N.; Dowell, E.; Glode, M.P.; Pace, N.R. Lactobacillus rhamnosus GG bacteremia associated with probiotic use in a child with short gut syndrome. Pediatr. Infect. Dis. J. 2005, 24, 278–280. [Google Scholar] [CrossRef]
- Hawrelak, J.A.; Whitten, D.L.; Myers, S.P. Is Lactobacillus rhamnosus GG effective in preventing the onset of antibiotic-associated diarrhoea: A systematic review. Digestion 2005, 72, 51–56. [Google Scholar] [CrossRef]
- Lee, J.; Choe, J.; Kim, J.; Oh, S.; Park, S.; Kim, S.; Kim, Y. Heat-killed Lactobacillus spp. cells enhance survivals of Caenorhabditis elegans against Salmonella and Yersinia infections. Lett. Appl. Microbiol. 2015, 61, 523–530. [Google Scholar] [CrossRef]
- Kim, Y.; Mylonakis, E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect. Immun. 2012, 80, 2500–2508. [Google Scholar] [CrossRef]
- Iatsenko, I.; Yim, J.J.; Schroeder, F.C.; Sommer, R.J. B. subtilis GS67 protects C. elegans from Gram-positive pathogens via fengycin-mediated microbial antagonism. Curr. Biol. 2014, 24, 2720–2727. [Google Scholar] [CrossRef]
- Roselli, M.; Schifano, E.; Guantario, B.; Zinno, P.; Uccelletti, D.; Devirgiliis, C. Caenorhabditis elegans and probiotics interactions from a prolongevity perspective. Int. J. Mol. Sci. 2019, 20, 5020. [Google Scholar] [CrossRef]
- Komura, T.; Ikeda, T.; Yasui, C.; Saeki, S.; Nishikawa, Y. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 2013, 14, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Pal Bhadra, M.; Bhadra, U. Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species. Biogerontology 2017, 18, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Tissenbaum, H.A. Using C. elegans for aging research. Invertebr. Reprod. Dev. 2015, 59, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Chompoo, J.; Taira, N.; Fukuta, M.; Tawata, S. Significant longevity-extending effects of Alpinia zerumbet leaf extract on the life span of Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 2013, 77, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, L.; Zheng, X.; Fu, T.; Guo, H.; Ren, F. Lactobacillus salivarius strain FDB89 induced longevity in Caenorhabditis elegans by dietary restriction. J. Microbiol. 2013, 51, 183–188. [Google Scholar] [CrossRef]
- Ayala, F.R.; Bauman, C.; Cogliati, S.; Leñini, C.; Bartolini, M.; Grau, R. Microbial flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity. Microb. Cell 2017, 4, 133–136. [Google Scholar] [CrossRef]
- Gasbarrini, G.; Bonvicini, F.; Gramenzi, A. Probiotics history. J. Clin. Gastroenterol. 2016, 50 (Suppl. 2), S116–S119. [Google Scholar] [CrossRef]
- Vasiljevic, T.; Shah, N.P. Probiotics—From Metchnikoff to bioactives. Int. Dairy J. 2008, 18, 714–728. [Google Scholar] [CrossRef]
- Kwon, G.; Lee, J.; Lim, Y.-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016, 6, 31713. [Google Scholar] [CrossRef]
- Troemel, E.R.; Chu, S.W.; Reinke, V.; Lee, S.S.; Ausubel, F.M.; Kim, D.H. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006, 2, e183. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, Y.; Liu, R.; Zheng, X.; Zhang, M.; Guo, H.; Zhang, H.; Ren, F. The Transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci. Rep. 2017, 7, 7408. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Isolauri, E.; Salminen, S.; Sanz, Y. The impact of probiotic on gut health. Curr. Drug Metab. 2009, 10, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Gueimonde, M.; Sanz, Y.; Salminen, S. Adhesion properties and competitive pathogen exclusion ability of bifidobacteria with acquired acid resistance. J. Food Prot. 2006, 69, 1675–1679. [Google Scholar] [CrossRef]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Development of new probiotics by strain combinations: Is it possible to improve the adhesion to intestinal mucus? J. Dairy Sci. 2007, 90, 2710–2716. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Donaldson, G.P.; Mikulski, Z.; Boyajian, S.; Ley, K.; Mazmanian, S.K. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013, 501, 426–429. [Google Scholar] [CrossRef]
- Ahimou, F.; Paquot, M.; Jacques, P.; Thonart, P.; Rouxhet, P.G. Influence of electrical properties on the evaluation of the surface hydrophobicity of Bacillus subtilis. J. Microbiol. Methods 2001, 45, 119–126. [Google Scholar] [CrossRef]
- Khan, M.W.; Zadeh, M.; Bere, P.; Gounaris, E.; Owen, J.; Klaenhammer, T.; Mohamadzadeh, M. Modulating intestinal immune responses by lipoteichoic acid-deficient Lactobacillus acidophilus. Immunotherapy 2012, 4, 151–161. [Google Scholar] [CrossRef]
- Gonzales-Siles, L.; Karlsson, R.; Schmidt, P.; Salvà-Serra, F.; Jaén-Luchoro, D.; Skovbjerg, S.; Moore, E.R.B.; Gomila, M. A pangenome approach for discerning species-unique gene markers for identifications of Streptococcus pneumoniae and Streptococcus pseudopneumoniae. Front. Cell Infect. Microbiol. 2020, 10, 222. [Google Scholar] [CrossRef]
- Balaev, V.V.; Prokofev, I.I.; Gabdoulkhakov, A.G.; Betzel, C.; Lashkov, A.A. Crystal structure of pyrimidine-nucleoside phosphorylase from Bacillus subtilis in complex with imidazole and sulfate. Acta Cryst. F Struct. Biol. Commun. 2018, 74, 193–197. [Google Scholar] [CrossRef]
- Thomas, M.; Wrzosek, L.; Ben-Yahia, L.; Noordine, M.-L.; Gitton, C.; Chevret, D.; Langella, P.; Mayeur, C.; Cherbuy, C.; Rul, F. Carbohydrate metabolism is essential for the colonization of Streptococcus thermophilus in the digestive tract of gnotobiotic rats. PLoS ONE 2011, 6, e28789. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, G.; Chen, W.; Guo, B. Metabolism of fructooligosaccharides in Lactobacillus plantarum ST-III via differential gene transcription and alteration of cell membrane fluidity. Appl. Environ. Microbiol. 2015, 81, 7697–7707. [Google Scholar] [CrossRef] [PubMed]
- Rattanaprasert, M.; van Pijkeren, J.-P.; Ramer-Tait, A.E.; Quintero, M.; Kok, C.R.; Walter, J.; Hutkins, R.W. Genes involved in galactooligosaccharide metabolism in Lactobacillus reuteri and their ecological role in the gastrointestinal tract. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 2004, 25, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Siegers, K.; Heinzmann, S.; Entian, K.-D. Biosynthesis of lantibiotic nisin posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 1996, 271, 12294–12301. [Google Scholar] [CrossRef] [PubMed]
- Velho, R.V.; Basso, A.P.; Segalin, J.; Costa-Medina, L.F.; Brandelli, A. The presence of sboA and spaS genes and antimicrobial peptides subtilosin A and subtilin among Bacillus strains of the Amazon basin. Genet. Mol. Biol. 2013, 36, 101–104. [Google Scholar] [CrossRef]
- Qiao, J.Q.; Tian, D.W.; Huo, R.; Wu, H.J.; Gao, X.W. Functional analysis and application of the cryptic plasmid pBSG3 harboring the RapQ-PhrQ system in Bacillus amyloliquefaciens B3. Plasmid 2011, 65, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, X.; Zhu, Y.; Niu, L.; Teng, M.; Sun, B.; Li, X. Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus. Acta Cryst D 2015, 71, 1768–1776. [Google Scholar] [CrossRef]
- Yan, L.; Liu, G.; Zhao, B.; Pang, B.; Wu, W.; Ai, C.; Zhao, X.; Wang, X.; Jiang, C.; Shao, D.; et al. Novel biomedical functions of surfactin A from Bacillus subtilis in wound healing promotion and scar inhibition. J. Agric. Food Chem. 2020, 68, 6987–6997. [Google Scholar] [CrossRef]
- Wang, S.; Ye, Q.; Wang, K.; Zeng, X.; Huang, S.; Yu, H.; Ge, Q.; Qi, D.; Qiao, S. Enhancement of macrophage function by the antimicrobial peptide sublancin protects mice from methicillin-resistant Staphylococcus aureus. J. Immunol. Res. 2019, 2019, 3979352. [Google Scholar] [CrossRef]
- Di Luccia, B.; D’Apuzzo, E.; Varriale, F.; Baccigalupi, L.; Ricca, E.; Pollice, A. Bacillus megaterium SF185 induces stress pathways and affects the cell cycle distribution of human intestinal epithelial cells. Benef. Microbes 2016, 7, 609–620. [Google Scholar] [CrossRef]
- Park, K.-Y.; Jung, K.-O.; Rhee, S.-H.; Choi, Y.H. Antimutagenic effects of doenjang (Korean fermented soypaste) and its active compounds. Mutat. Res. 2003, 523–524, 43–53. [Google Scholar] [CrossRef]


| Gene | Gene Description | Strains |
|---|---|---|
| deoA | Thymidine phosphorylase | SRCM103571, SRCM103689 |
| haeIIIM_2 | Modification methylase HaeIII | SRCM103571, SRCM103689 |
| vgb | Virginiamycin B lyase | SRCM103517, SRCM103689, SRCM104011 |
| vsr | Very short patch repair protein | SRCM103571, SRCM103689 |
| group_6918 | Response regulator aspartate phosphatase I | SRCM103571, SRCM104011 |
| group_16459 | Chromosomal replication initiator protein DnaA | SRCM103517, SRCM103571 |
| group_16672 | Competence pheromone | SRCM103571, SRCM103689 |
| Gene | Gene Description | Strains |
|---|---|---|
| comX | Competence pheromone | SRCM103571, SRCM103689 |
| tagU_1 | Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagU | SRCM103517, SRCM103571, SRCM103689, SRCM104011 |
| cscB | Sucrose permease | SRCM103571, SRCM103689 |
| lacF_2 | Lactose transport system permease protein LacF | SRCM103571, SRCM103689 |
| lacG | Lactose transport system permease protein LacG | SRCM103571, SRCM103689 |
| maa_1 | Maltose O-acetyltransferase | SRCM103571, SRCM103689 |
| nisB | Nisin biosynthesis protein NisB | SRCM103571, SRCM103689 |
| nisC_2 | Nisin biosynthesis protein NisC | SRCM103571, SRCM103689 |
| rapI | Response regulator aspartate phosphatase I | SRCM103517, SRCM104011 |
| sacA | Sucrose-6-phosphate hydrolase | SRCM103571, SRCM103689 |
| saeR | Response regulator SaeR | SRCM103517, SRCM104011 |
| scrK_2 | Fructokinase | SRCM103517, SRCM104011 |
| spaS | Lantibiotic subtilin | SRCM103571, SRCM103689 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.J.; Shin, D.; Shin, M.; Yun, B.; Kang, M.; Yang, H.-J.; Jeong, D.-Y.; Kim, Y.; Oh, S. Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods. Foods 2020, 9, 1805. https://doi.org/10.3390/foods9121805
Choi HJ, Shin D, Shin M, Yun B, Kang M, Yang H-J, Jeong D-Y, Kim Y, Oh S. Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods. Foods. 2020; 9(12):1805. https://doi.org/10.3390/foods9121805
Chicago/Turabian StyleChoi, Hye Jin, Donghyun Shin, Minhye Shin, Bohyun Yun, Minkyoung Kang, Hee-Jong Yang, Do-Youn Jeong, Younghoon Kim, and Sangnam Oh. 2020. "Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods" Foods 9, no. 12: 1805. https://doi.org/10.3390/foods9121805
APA StyleChoi, H. J., Shin, D., Shin, M., Yun, B., Kang, M., Yang, H.-J., Jeong, D.-Y., Kim, Y., & Oh, S. (2020). Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods. Foods, 9(12), 1805. https://doi.org/10.3390/foods9121805





