Are Dietary Lectins Relevant Allergens in Plant Food Allergy?
Abstract
1. Introduction
2. Allergenic Potential of Lectins from Higher Plants
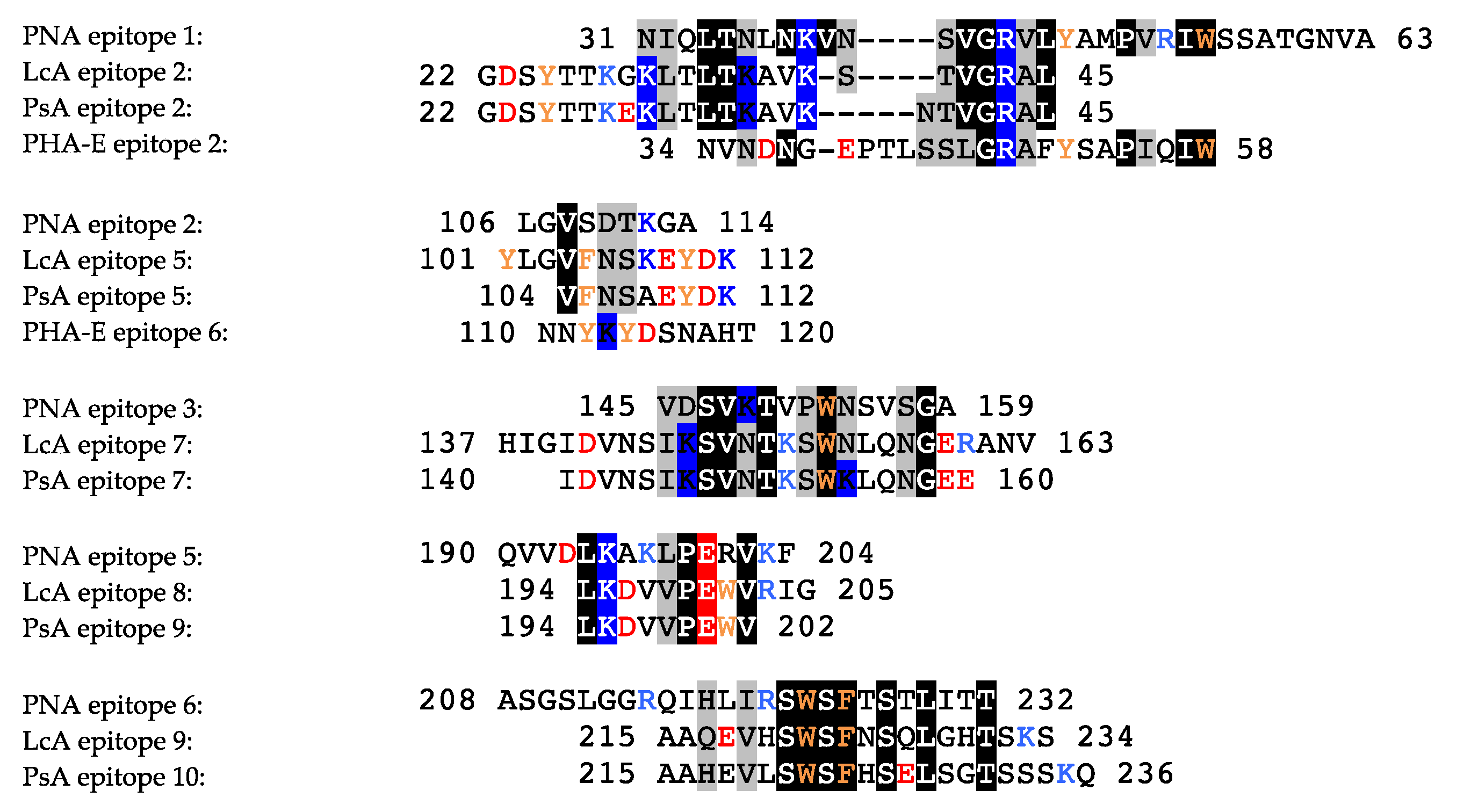
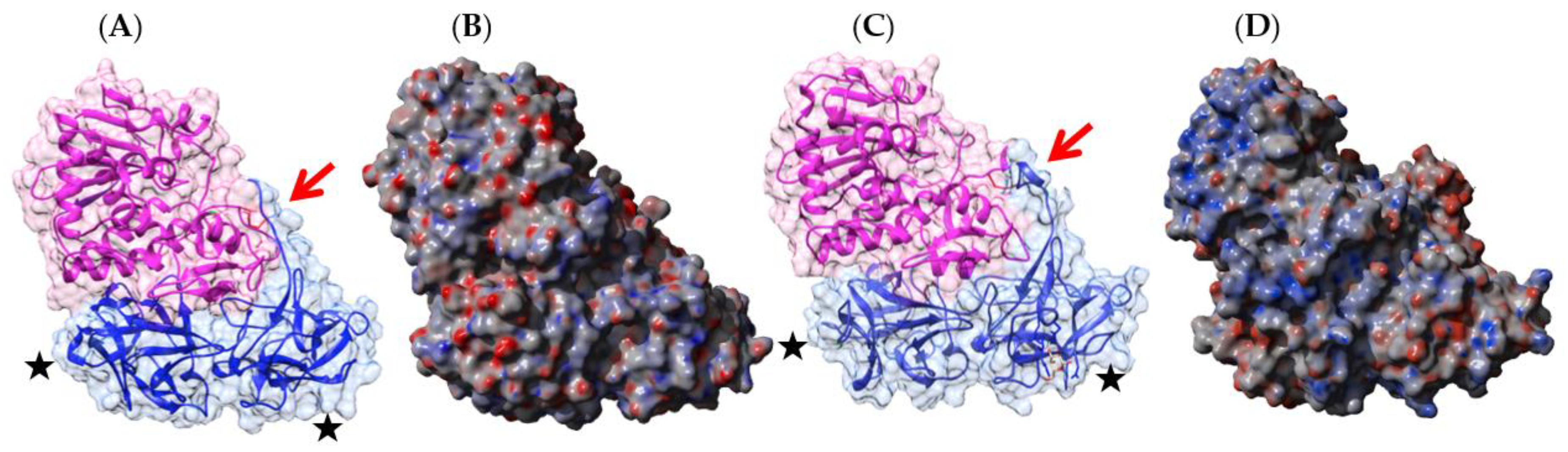
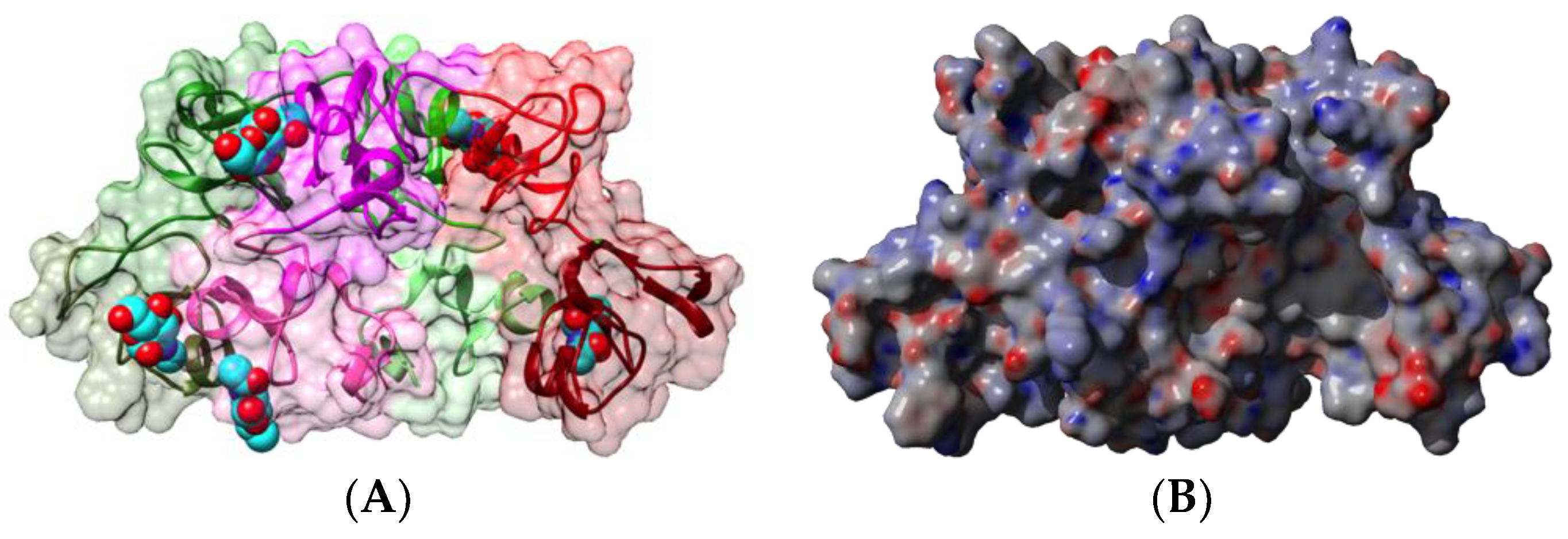
2.1. Legume Lectins
- -
- the homotetrameric single-chain lectins, comprise both man-specific (e.g., Con A from Jackbean, Canavalia ensiformis, and DgL from Dioclea grandiflora) and Gal/GalNAc-specific lectins (e.g., PNA from peanut, Arachis hypogaea, and SBA from soybean, Glycine max).
- -
- the homodimeric two-chain lectins, exclusively comprise man-specific lectins (e.g., LcA from lentil, Lens culinaris, and PsA from garden pea, Pisum sativum).
2.1.1. Homotetrameric Single-Chain Lectins
2.1.2. Homodimeric Two-Chain Lectins
2.2. Type II Ribosome Inactivating Proteins (RIP-II)
2.3. Wheat Germ Agglutinin (WGA)
2.4. Chitinases Containing a Hevein Domain
2.5. Jacalin-Related Lectins
2.6. GNA-Like Lectins
2.7. Nictaba-Related Lectins
3. Discussion
4. Bioinformatics
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACA | Allium cepa (onion) agglutinin/lectin |
| ASA | Allium sativum (garlic) agglutinin/lectin |
| BanLec | Banana (Musa acuminata) lectin |
| CBS | Carbohydrate-binding site |
| DbA | Dolichos biflorus (horse gram) agglutinin/lectin |
| GNA | Galanthus nivalis (snowdrop) agglutinin/lectin |
| IgE | Immunoglobulin E |
| INFγ | Interferon-gamma |
| LcA | Lens culinaris (lentil) agglutinin/lectin |
| moAb | Monoclonal antibody |
| Nictaba | Nicotiana tabacum (tobacco) agglutinin/lectin |
| PHA-E | Erythroagglutinin of Phaseolus (kidney bean) agglutinin/lectin |
| PHA-L | Leucoagglutinin of Phaseolus agglutinin/lectin |
| PNA | Peanut (Arachis hypogaea) agglutinin/lectin (also known as Ara h agglutinin) |
| PsA | Pisum sativum (garden pea) agglutinin/lectin |
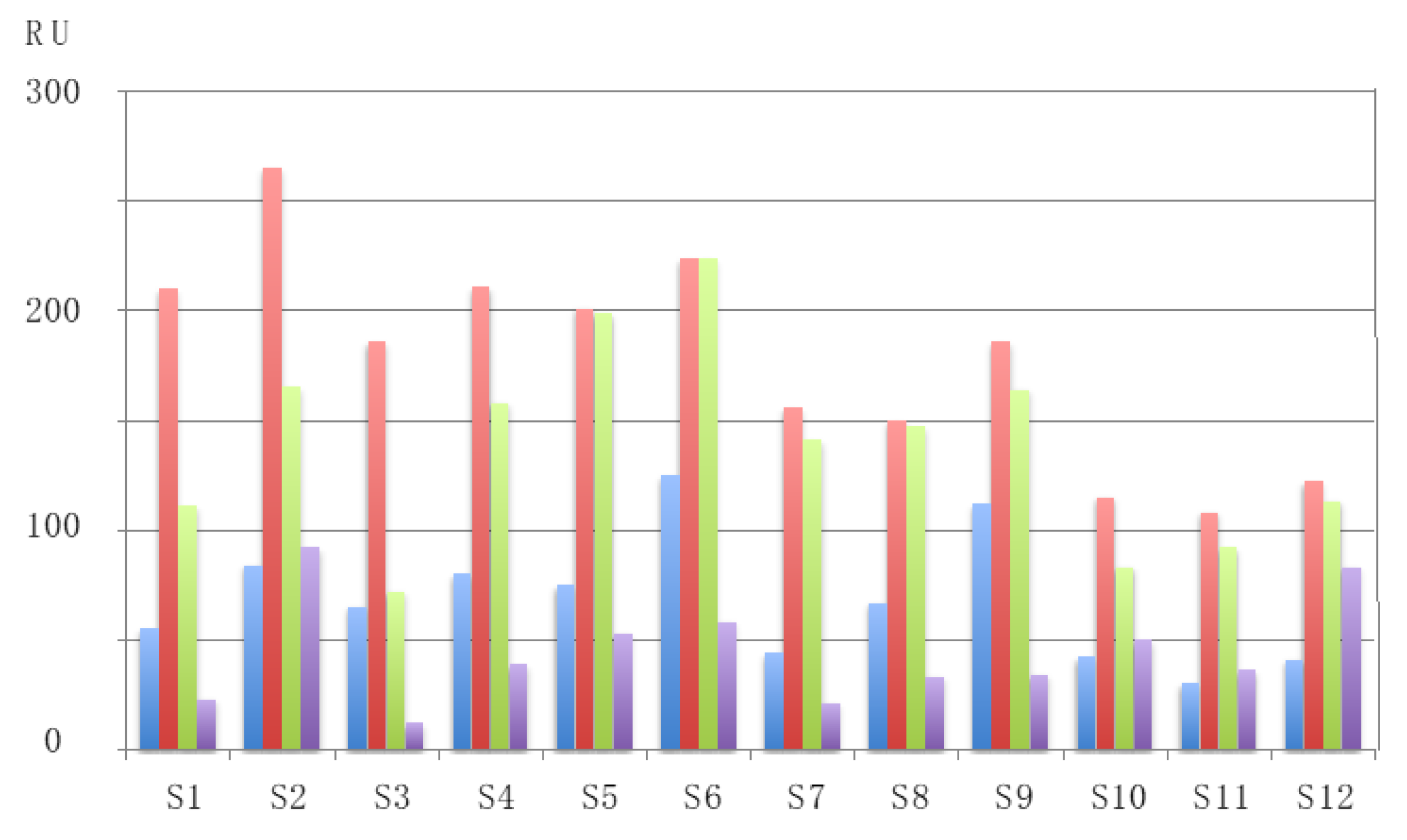
| RU | Resonance unit |
| SBA | Soybean (Glycine max) agglutinin/lectin |
| SNA | Sambucus nigra (black elderberry) agglutinin/lectin |
| SPR | Surface plasmon resonance |
| WGA | Wheat (Triticum aestivum) germ agglutinin/lectin (also known as Tri a 18) |
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Iweala, O.I.; Choudhary, S.K.; Commins, S.P. Food allergy. Curr. Gastroenterol. Rep. 2018, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Sathe, S.K.; Sharma, G.M. Effects of food processing on food allergens. Mol. Nutr. Food Res. 2009, 53, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 2004, 113, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Mills, E.N. Molecular properties of food allergens. J. Allergy Clin. Immunol. 2005, 115, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, A.R.; Scheurer, S.; Vieths, S. Food allergens: Molecular and immunological aspects, allergen databases and cross-reactivity. Chem. Immunol. Allergy 2015, 101, 18–29. [Google Scholar] [PubMed]
- Barre, A.; Sordet, C.; Culerrier, R.; Rancé, F.; Didier, A.; Rougé, P. Vicilin allergens of peanut and tree nuts (walnut, hazelnut and cashew nut) share structurally related IgE-binding epitopes. Mol. Immunol. 2008, 45, 1231–1240. [Google Scholar] [CrossRef]
- Leoni, C.; Volpicella, M.; Dileo, M.C.G.; Gattulli, B.A.R. Chitinases as food allergens. Molecules 2019, 24, 2087. [Google Scholar] [CrossRef]
- Azofra García, J.; Cuesta-Herranz, J.; Perea Lam, N.; Díaz-Perales, A. Anaphylaxis mediated by thaumatin-like proteins. J. Investig. Allergol. Clin. Immunol. 2014, 24, 228–449. [Google Scholar]
- Radauer, C.; Lackner, P.; Breiteneder, H. The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 2008, 8, 286. [Google Scholar] [CrossRef]
- Bienvenu, F.; Barre, A.; Viel, S.; Garnier, L.; Guyon, C.; Favre-Metz, C.; Pauli, G.; Bienvenu, J.; Rougé, P. Are defensins important plant allergens? Rev. Fr. Allergol. 2013, 53, 585–590. [Google Scholar] [CrossRef]
- Pascal, M.; Muñoz-Cano, R.; Reina, Z.; Palacín, A.; Vilella, R.; Picado, C.; Juan, M.; Sánchez-López, J.; Rueda, M.; Salcedo, G.; et al. Lipid transfer protein syndrome: Clinical pattern, cofactor effect and profile of molecular sensitization to plant-foods and pollens. Clin. Exp. Allergy 2012, 42, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Ladics, G.S.; Budziszewski, G.J.; Herman, R.A.; Herouet-Guicheney, C.; Joshi, S.; Lipscomb, E.A.; McClain, S.; Ward, J.M. Measurement of endogenous allergens in genetically modified soybean-short communication. Regul. Toxicol. Pharmacol. 2014, 70, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Peumans, W.J.; Barre, A.; Rougé, P. Plant lectins: A composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit. Rev. Plant Sci. 1998, 17, 575–692. [Google Scholar] [CrossRef]
- Peumans, W.J.; Van Damme, E.J.M.; Barre, A.; Rougé, P. Classification of plant lectins in families of structurally and evolutionary related proteins. Adv. Exp. Med. Biol. 2001, 491, 27–54. [Google Scholar] [PubMed]
- Barre, A.; Bourne, Y.; Van Damme, E.J.M.; Rougé, P. Overview of the structure-function relationships of mannose-specific lectins from plants, algae and fungi. Int. J. Mol. Sci. 2019, 20, 254. [Google Scholar] [CrossRef]
- Rougé, P.; Culerrier, R.; Granier, C.; Rancé, F.; Barre, A. Characterization of IgE-binding epitopes of peanut (Arachis hypogaea) PNA lectin allergen cross-reacting with other structurally related legume lectins. Mol. Immunol. 2010, 47, 2359–2366. [Google Scholar] [CrossRef]
- Fialová, D.; Tichá, M.; Kocourek, J. Studies on phytohemagglutinins XXVI. A new type of a lentil hemagglutinin isolated from Lens esculenta Moench., subsp. Microsperma (Baumg.) barulina. Biochim. Biophys. Acta 1975, 393, 170–181. [Google Scholar] [CrossRef]
- Rougé, P. Biologie des Hémagglutinines chez le Pois (Pisum sativum L.). Ph.D. Thesis, Université Paul Sabatier, Toulouse, France, 27 January 1977. [Google Scholar]
- Trembley, R.; Feng, M.; Menassa, R.; Huner, N.P.A.; Jevnikar, A.M.; Ma, S. High-yield expression of recombinant soybean agglutinin in plants using transient and stable systems. Transgenic Res. 2011, 20, 345–356. [Google Scholar] [CrossRef]
- Lotan, R.; Skutelsky, E.; Danon, D.; Sharon, N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J. Biol. Chem. 1975, 250, 8518–8523. [Google Scholar]
- Etzler, M.E. The Dolichos biflorus lectin family: A model system for studying legume lectin structure and function. In Lectins, Biology-Biochemistry, Clinical Biochemistry; Van Driessche, E., Rougé, P., Beeckmans, S., Bøg-Hansen, T.C., Eds.; Textop: Hellerup, Denmark, 1996; Volume 11, pp. 3–9. [Google Scholar]
- Rigas, D.A.; Osgood, E.E. Purification and properties of the phytohemagglutinin of Phaseolus vulgaris. J. Biol. Chem. 1955, 212, 607–616. [Google Scholar] [CrossRef]
- LeVine, D.; Kaplan, M.J.; Greenaway, P.J. The purification and characterization of wheat germ agglutinin. Biochem. J. 1972, 129, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Van Damme, E.J.M. The role of lectins in the defense of plants. In Lectins, Biology-Biochemistry, Clinical Biochemistry; Van Driessche, E., Franz, H., Beeckmans, S., Pfüller, U., Kallikorm, A., Bøg-Hansen, T.C., Eds.; Textop: Hellerup, Denmark, 1993; Volume 8, pp. 82–91. [Google Scholar]
- Nikolic, J.; Mrkic, I.; Grozdanovic, M.; Popovic, M.; Petersen, A.; Jappe, U.; Gavrovic-Jankulovic, M. Protocol for simultaneous isolation of three important banana allergens. J. Chromatogr. B 2014, 962, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Pomés, A.; Davies, J.M.; Gadermaier, G.; Hilger, C.; Holzhauser, T.; Lidholm, J.; Lopata, A.L.; Mueller, G.A.; Nandy, A.; Radauer, C.; et al. WHO/IUIS allergen nomenclature: Providing a common language. Mol. Immunol. 2018, 100, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, M.; Sumazaki, R.; Isovama, S.; Takita, H. Interaction of lectins with human IgE: IgE-binding property and histamine-releasing activity of twelve plant lectins. Int. Arch. Allergy Immunol. 1992, 98, 18–25. [Google Scholar] [CrossRef]
- Haas, H.; Falcone, F.H.; Schramm, G.; Haisch, K.; Gibbs, B.F.; Klaucke, J.; Pöppelmann, M.; Becker, W.-M.; Gabius, H.-J.; Schlaak, M. Dietary lectins can induce in vitro release of IL-4 and IL-13 from human basophils. Eur. J. Immunol. 1999, 29, 918–927. [Google Scholar] [CrossRef]
- Moreno, A.N.; Jamur, M.C.; Oliver, C.; Roque-Barreira, M.C. Mast cell degranulation induced by lectins: Effect on neutrophil recruitment. Int. Arch. Allergy Immunol. 2003, 132, 221–230. [Google Scholar] [CrossRef]
- Sànchez-Monge, R.; Blanco, C.; Díaz-Perales, A.; Collada, C.; Carrillo, T.; Aragoncillo, C.; Salcedo, G. Isolation and characterization of major banana allergens: Identification as fruit class I chitinases. Clin. Exp. Allergy 1999, 29, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Sutton, R.; Skerritt, J.H.; Baldo, B.A.; Wrigley, C.W. The diversity of allergens involved in baker’s asthma. Clin. Allergy 1984, 14, 93–107. [Google Scholar] [CrossRef]
- Volpicella, M.; Leoni, C.; Fanizza, I.; Distaso, M.; Leoni, G.; Farioli, L.; Naumann, T.; Pastorello, E.; Ceci, L.R. Characterization of maize chitinase-A, a tough allergenic molecule. Allergy 2017, 72, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Hänninen, A.R.; Mikkola, J.H.; Kalkkinen, N.; Turjanmaa, K.; Ylitalo, L.; Reunala, T.; Palosuo, T. Increased allergen production in turnip (Brassica rapa) by treatments activating defense mechanisms. J. Allergy Clin. Immunol. 1999, 104, 194–201. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Biagini, R.E.; Karnani, R.; Hamilton, R.; Murphy, K.; Bernstein, C.; Arif, S.A.; Berendts, B.; Yeang, H.Y. In vivo sensitization to purified Hevea brasiliensis proteins in health care workers sensitized to natural rubber latex. J. Allergy Clin. Immunol. 2003, 111, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Sowka, S.; Hsieh, L.S.; Krebitz, M.; Akasawa, A.; Martin, B.M.; Starrett, D.; Peterbauer, C.K.; Scheiner, O.; Breiteneder, H. Identification and cloning of Pers a 1, a 32-kDa endochitinase and major allergen of avocado, and its expression in yeast Pichia pastoris. J. Biol. Chem. 1998, 273, 28091–28097. [Google Scholar] [CrossRef] [PubMed]
- Bublin, M.; Mari, A.; Ebner, C.; Knulst, A.; Scheiner, O.; Hoffmann-Sommergruber, K.; Breiteneder, H.; Radauer, C. IgE sensitization profiles toward green and gold kiwifruits differ among patients allergic to kiwifruit from 3 European countries. J. Allergy Clin. Immunol. 2004, 114, 1169–1175. [Google Scholar] [CrossRef]
- Chen, Y.T.; Hsu, L.H.; Huang, I.P.; Tsai, T.C.; Lee, G.C.; Shaw, J.F. Gene cloning and characterization of a novel recombinant antifungal chitinase from papaya (Carica papaya). J. Agric. Food Chem. 2007, 55, 714–722. [Google Scholar] [CrossRef]
- Díaz-Perales, A.; Collada, C.; Blanco, C.; Sanchez-Monge, R.; Carrillo, T.; Aragoncilllo, C.; Salcedo, G. Cross-reactions in the latex-fruit syndrome: A relevant role of chitinase but not of complex asparagine-linked glycans. J. Allergy Clin. Immunol. 1999, 104, 681–687. [Google Scholar] [CrossRef]
- Pastorello, E.A.; Farioli, L.; Pravettoni, V.; Ortolani, C.; Fortunato, D.; Giuffrida, M.G.; Perono Garoffo, L.; Calamari, A.M.; Brenna, O.; Conti, A. Identification of grape and wine allergens as an endochitinase 4, a lipid-transfer protein, and a thaumatin. J. Allergy Allergy Clin. Immunol. 2003, 111, 350–359. [Google Scholar] [CrossRef]
- Albanesi, M.; Pasculli, C.; Giliberti, L.; Rossi, M.P.; Di Bona, D.; Caiaffa, M.F.; Macchia, L. Immunological characterization of onion (Allium cepa) allergy. Adv. Dermatol. Allergol. 2019, XXXVI, 98–103. [Google Scholar] [CrossRef]
- Clement, F.; Pramod, S.N.; Venkatesh, Y.P. Identity of the immunomodulatory proteins from garlic (Allium sativum) with the major garlic lectins or agglutinins. Int. Immunopharmacol. 2010, 10, 316–324. [Google Scholar] [CrossRef]
- Burks, A.W.; Cockrell, G.; Connaughton, C.; Guin, J.; Allen, W.; Helm, R.M. Identification of peanut agglutinin and soybean trypsin inhibitor as minor legume allergens. Int. Arch. Allergy Immunol. 1994, 105, 143–149. [Google Scholar] [CrossRef]
- Pramod, S.N.; Krishnakantha, T.P.; Venkatesh, Y.P. Effect of horse gram lectin (Dolichos biflorus agglutinin) on degranulation of mast cells and basophils of atopic subjects: Identification as an allergen. Int. Immunopharmacol. 2006, 6, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.; Xu, C.; Bae, H.; Caperna, T.J.; Garrett, W.M. Proteomic analysis of allergen and antinitritional proteins in wild and cultivated soybean seeds. J. Plant Biochem. Biotechnol. 2006, 15, 103–108. [Google Scholar] [CrossRef]
- García Jiménez, S.; Pastor Vargas, C.; de la Heras, M.; Sanz Matoto, A.; Vivanco, F.; Sastre, J. Allergen characterization of chia seeds (Salvia hispanica), a new allergenic food. J. Investig. Allergol. Clin. Immunol. 2015, 25, 55–56. [Google Scholar]
- Koshte, V.L.; Aalbers, M.; Calkhoven, P.G.; Aalberse, R.C. The potent IgG4-inducing antigen in banana is a mannose-binding lectin, BanLec-I. Int. Arch. Allergy Immunol. 1992, 97, 17–24. [Google Scholar] [CrossRef]
- González-Mancebo, E.; López-Torrejón, G.; González de Olano, D.; Santos, S.; Gandolfo-Cano, M.; Meléndez, A.; Salcedo, G.; Cuesta-Herranz, J.; Vivanco, F.; Pastor-Vargas, C. Identification of potential allergens involved in systemic reactions to melon and watermelon. Ann. Allergy Asthma Immunol. 2010, 104, 271–272. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Rougé, P.; Peumans, W.J. Plant lectins. In Carbohydrate-protein interactions: Plant Lectins; Kamerling, J.P., Boons, G.J., Lee, Y.C., Suzuki, A., Taniguchi, N., Voragen, A.G.I., Eds.; Elsevier: New York, NY, USA, 2007; pp. 564–599. [Google Scholar]
- Vojdani, A. Immune reactivities to peanut proteins, agglutinins, and oleosins. Antern. Ther. Health Med. 2015, 21, 73–79. [Google Scholar]
- White, B.L.; Gökce, E.; Nepomuceno, A.I.; Muddiman, D.C.; Sanders, T.H.; Davis, J.P. Comparative proteomic analysis and IgE binding properties of peanut seed and testa (skin). J. Agric. Food Chem. 2013, 61, 3957–3968. [Google Scholar] [CrossRef] [PubMed]
- Zabel, P.L.; Noujaim, A.A.; Shysh, A.; Bray, J. Radioiodinated peanut lectin: A potential radiopharmaceutical for immunodetection of carcinoma expressing the T antigen. Eur. J. Nucl. Med. 1983, 8, 250–254. [Google Scholar] [CrossRef]
- Goldstein, I.J.; Poretz, R.D. Isolation, physicochemical characterization, and carbohydrate-binding specificity of lectins. In The Lectins, Properties, Functions, and Applications in Biology and Medicine; Liener, I.E., Sharon, N., Goldstein, I.J., Eds.; Academic Press, Inc.: Orlando, FL, USA, 1986; pp. 33–247. [Google Scholar]
- Haas, H.; Herzig, K.H.; André, S.; Galle, J.; Gronow, A.; Gabius, H.-J. Low-dose intragastric administration of Phaseolus vulgaris agglutinin (PHA) does not induce immunoglobulin E (IgE) production in Sprague-Dawley rats. Glycoconj. J. 2001, 18, 273–275. [Google Scholar] [CrossRef]
- Rougé, P.; Culerrier, R.; Thibeau, F.; Didier, A.; Barre, A. A case of severe anaphylaxis to kidney bean: Phaseolin (vicilin) and PHA (lectin) identified as putative allergens. Allergy 2011, 66, 301–302. [Google Scholar] [CrossRef]
- Kasera, R.; Singh, B.P.; Lavasa, S.; Prasad, K.N.; Sahoo, R.C.; Singh, A.B. Kidney bean: A major sensitizer among legumes in asthma and rhinitis patients from India. PLoS ONE 2011, 6, e27193. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Verma, A.K.; Sharma, A.; Kumar, D.; Tripathi, A.; Chaudhari, B.P.; Das, M.; Jain, S.K.; Dwivedi, P.D. Phytohemagglutinins augment red kidney bean (Phaseolus vulgaris L.) induced allergic manifestations. J. Proteom. 2013, 93, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A.; Das, M.; Jain, S.K.; Dwivedi, P.D. Leucoagglutinating phytohemagglutinin: Purification, characterization, proteolytic digestion and assessment for allergenicity potential in BALB/c mice. Immunopharmacol. Immunotoxicol. 2014, 36, 138–144. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhao, J.; Elfalleh, W.; Jemaà, M.; Sun, H.; Sun, X.; Tang, M.; He, Q.; Wu, Z.; Lang, F. In silico identification and in vitro analysis of B- and T-cell epitopes of the black turtle bean (Phaseolus vulgaris L.) lectin. Cell Physiol. Biochem. 2018, 49, 1600–1614. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, S.; Tang, M.; Sun, X.; Zhang, Z.; Ye, Y.; Cao, X.; Sun, H. Low-pH induced structural changes, allergenicity and in vitro digestibility of lectin from black turtle bean (Phaseolus vulgaris L.). Food Chem. 2019, 283, 183–190. [Google Scholar] [CrossRef]
- Lu, M.; Jin, Y.; Cerny, R.; Ballmer-Weber, B.; Goodman, R.E. Combining 2-DE immunoblots and mass spectrometry to identify putative soybean (Glycine max) allergens. Food. Chem. Toxicol. 2018, 116, 207–215. [Google Scholar] [CrossRef]
- Kleine-Tebbe, J.; Kagey-Sobotka, A.; MacGlashan, D.W.; Lichtenstein, L.M.; MacDonald, S.M. Lectins do not distinguish between heterogenous IgE molecules as defined by differential activity of an IgE-dependent histamine releasing factor. J. Allergy Clin. Immunol. 1996, 98, 181–188. [Google Scholar] [CrossRef]
- Risler, J.L.; Delorme, M.O.; Delacroix, H.; Henaut, A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol. 1988, 204, 1019–1029. [Google Scholar] [CrossRef]
- Peumans, W.J.; Hao, Q.; Van Damme, E.J.M. Ribosome-inactivating proteins from plants: More than RNA N-glycosidases? FASEB J. 2001, 15, 493–506. [Google Scholar] [CrossRef]
- Förster-Waldl, E.; Marchetti, M.; Schöll, I.; Focke, M.; Radauer, C.; Kinaciyan, T.; Nentwich, I.; Jäger, S.; Schmid, E.R.; Boltz-Nitulescu, G.; et al. Type I allergy to elderberry (Sambucus nigra) is elicited by a 33.2 kDa allergen with significant homology to ribosomal inactivating proteins. Clin. Exp. Allergy 2003, 33, 1703–1710. [Google Scholar] [CrossRef]
- Szalai, K.; Schöll, I.; Förster-Waldl, E.; Polito, A.; Bolognesi, E.; Untermayr, E.; Riemer, A.B.; Boltz-Nitulescu, G.; Stirpe, F.; Jensen-Jarolim, E. Occupational sensitization to ribosome-inactivating proteins in researchers. Clin. Exp. Allergy 2005, 35, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.; Cabrero, P.; Basterrechea, J.E.; Tejero, J.; Cordoba-Diaz, D.; Girbes, T. Isolation and molecular characterization of two lectins from dwarf elder (Sambucus ebulus L.) blossoms related to the Sam n 1 allergen. Toxins 2013, 5, 1767–1779. [Google Scholar] [CrossRef] [PubMed]
- Luther, P.; Sehrt, I.; Bergmann, K.C.; Reutgen, H. Allergy and lectins: Action between IgE-mediated histamine release and glycoproteins from Viscum album L. (mistletoe). Acta Biol. Med. Ger. 1978, 37, 1623–1628. [Google Scholar] [PubMed]
- Yoon, T.J.; Yoo, Y.C.; Kang, T.B.; Her, E.; Kim, S.H.; Kim, K.; Azuma, I.; Kim, J.B. Cellular and humoral adjuvant activity of lectins isolated from Korean mistletoe (Viscum album colaratum). Int. Immunopharmacol. 2001, 1, 881–889. [Google Scholar] [CrossRef]
- Reyes-López, C.A.; Hernández-Santoyo, A.; Pedraza-Escalona, M.; Mendoza, G.; Hernández-Arana, A.; Rodríguez-Romero, A. Insights into a conformational epitope of Hev b 6.02 (hevein). Biochem. Biophys. Res. Commun. 2004, 314, 123–130. [Google Scholar] [CrossRef]
- Pedraza-Escalona, M.; Becerril-Luján, B.; Agundis, C.; Domínguez-Ramírez, L.; Pereyra, A.; Riaño-Umbarila, L.; Rodríguez-Romero, A. Analysis of B-cell epitopes from the allergen Hev b 6.02 revealed by using blocking antibodies. Mol. Immunol. 2009, 46, 668–676. [Google Scholar] [CrossRef]
- Alenius, H.; Kalkkinen, N.; Lukka, M.; Reunala, T.; Turjanmaa, K.; Mäkinen-Kiljunen, S.; Yip, E.; Palosuo, T. Prohevein from the rubber tree (Hevea brasiliensis) is a major latex allergen. Clin. Exp. Allergy 1995, 25, 659–665. [Google Scholar] [CrossRef]
- Chen, Z.; Posch, A.; Lohaus, C.; Raulf-Heimsoth, M.; Meyer, H.E.; Baur, X. Isolation and identification of hevein as a major IgE-binding polypeptide in Hevea latex. J. Allergy Clin. Immunol. 1997, 99, 402–409. [Google Scholar] [CrossRef]
- Karisola, P.; Kotovuori, A.; Poikonen, S.; Niskanen, E.; Kalkkinen, N.; Turjanmaa, K.; Palosuo, T.; Reunala, T.; Alenius, H.; Kulomaa, M.S. Isolated hevein-like domains, but not 31-kD endochitinases, are responsible for IgE-mediated in vitro and in vivo reactions in latex-fruit syndrome. J. Allergy Clin. Immunol. 2005, 115, 598–605. [Google Scholar] [CrossRef]
- Mikkola, J.H.; Alenius, H.; Kalkkinen, N.; Turjanmaa, K.; Palosuo, T.; Reunala, T. Hevein-like protein domains as a possible cause for allergen cross-reactivity between latex and banana. J. Allergy Clin. Immunol. 1998, 102, 1005–1012. [Google Scholar] [CrossRef]
- Díaz-Perales, A.; Collada, C.; Blanco, C.; Sánchez-Monge, R.; Carrillo, T.; Aragoncillo, C.; Salcedo, G. Class I chitinases with hevein-like domain, but not class II enzymes, are relevant chestnut and avocado allergens. J. Allergy Clin. Immunol. 1998, 102, 127–133. [Google Scholar] [CrossRef]
- Sánchez-Monge, R.; Blanco, C.; López-Torrejón, G.; Cumplido, J.; Recas, M.; Figueroa, J.; Carrillo, T.; Salcedo, G. Differential allergen sensitization patterns in chestnut allergy with or without associated latex-fruit syndrome. J. Allergy Clin. Immunol. 2006, 118, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Posch, A.; Cremer, R.; Raulf-Heimsoth, M.; Baur, X. Identification of hevein (Hev b 6.02) in Hevea latex as a major cross-reacting allergen with avocado fruit in patients with latex allergy. J. Allergy Clin. Immunol. 1998, 102, 476–481. [Google Scholar] [CrossRef]
- Posch, A.; Wheeler, C.H.; Chen, Z.; Flagge, A.; Dunn, M.J.; Papenfuss, F.; Raulf-Heimsoth, M.; Baur, X. Class I endochitinase containing a hevein domain is the causative allergen in latex-associated avocado allergy. Clin. Exp. Allergy 1999, 29, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, P.M.; Sánchez-Monge, R.; Díaz-Perales, A.; Salcedo, G.; Ansótegui, J.; Sanz, M.L. Latex-vegetable syndrome due to custard apple and aubergine, new variations of the hevein symphony. J. Investig. Allergol. Clin. Immunol. 2005, 15, 308–311. [Google Scholar] [PubMed]
- Danhash, N.; Wagemakers, C.A.; van Kan, J.A.; de Wit, P.J. Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol. Biol. 1993, 22, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- O’Riordain, G.; Radauer, C.; Hoffmann-Sommergruber, K.; Adhami, F.; Peterbauer, C.K.; Blanco, C.; Godnic-cvar, J.; Scheiner, O.; Ebner, C.; Breiteneder, H. Cloning and molecular characterization of the Hevea brasiliensis allergen Hev b 11, a class I chitinase. Clin. Exp. Allergy 2002, 32, 455–462. [Google Scholar] [CrossRef]
- Blanco, C.; Díaz-Perales, A.; Collada, C.; Sánchez-Monge, R.; Aragoncillo, C.; Castillo, R.; Ortega, N.; Alvarez, M.; Carrillo, T.; Salcedo, G. Class I chitinases as potential panallergens involved in the latex-fruit syndrome. J. Allergy Clin. Immunol. 1999, 103, 507–513. [Google Scholar] [CrossRef]
- Palosuo, T.; Panzani, R.C.; Singh, A.B.; Ariano, R.; Alenius, H.; Turjanmaa, K. Allergen cross-reactivity between proteins of the latex from Hevea brasiliensis, seeds and pollen of Ricinus communis, and pollen of Mercurialis annua, members of the Euphorbiaceae family. Allergy Asthma Proc. 2002, 23, 141–147. [Google Scholar]
- Wagner, S.; Breiteneder, H. The latex-fruit syndrome. Biochem. Soc. Trans. 2002, 30, 935–940. [Google Scholar] [CrossRef]
- Swanson, M.D.; Boudreaux, D.M.; Salmon, L.; Chugh, J.; Winter, H.C.; Meagher, J.L.; André, S.; Murphy, P.V.; Oscarson, S.; Roy, R.; et al. Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell 2015, 163, 746–758. [Google Scholar] [CrossRef] [PubMed]
- Krithika, N.; Pramod, S.N.; Mahesh, P.A.; Venkatesh, Y.P. Banana lectin (BanLec) induces non-specific activation of basophils and mast cells in atopic subjects. Eur. Ann. Allergy Clin. Immunol. 2018, 50, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Stojanović, M.M.; Zivković, I.P.; Petrusić, V.Z.; Kosec, D.J.; Dimitrijević, L.A.; Gavrović-Jankulović, M.D. In vitro stimulation of Balb/c and C57 BL/6 splenocytes by a recombinantly produced banana lectin isoform results in both proliferation of T cells and an increased secretion of interferon-γ. Int. Immunopharmacol. 2010, 10, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Barbosa--Lorenzi, V.C.; Cecilio, N.T.; de Almeida Buranello, P.A.; Pranchevicius, M.C.; Goldman, M.H.; Pereira-da-Silva, G.; Roque-Barreira, M.C.; Jamur, M.C.; Oliver, C. Recombinant ArtinM activates mast cells. BMC Immunol. 2016, 17, 22. [Google Scholar] [CrossRef]
- Mondal, H.A.; Chakrabort, D.; Majumder, P.; Roy, P.; Roy, A.; Bhattacharya, S.G.; Das, S. Allergenicity assessment of Allium sativum leaf agglutinin, a potential candidate protein for developing sap sucking insect resistant food crops. PLoS ONE 2011, 6, e27716. [Google Scholar] [CrossRef]
- Almogren, A.; Shakoor, Z.; Adam, M.H. Garlic and onion sensitization among Saudi patients screened for food allergy: A hospital based study. Afr. Health Sci. 2013, 13, 689–693. [Google Scholar] [CrossRef]
- Treudler, R.; Reuter, A.; Engin, A.M.; Simon, J.C. A case of anaphylaxis after garlic ingestion: Is alliinase the only culprit allergen? J. Investig. Allergol. Clin. Immunol. 2015, 25, 374–375. [Google Scholar]
- Das, A.; Ghosh, P.; Das, S. Expression of Colocasia esculenta tuber agglutinin in Indian mustard provides resistance against Lipaphis erysimi and the expressed protein is non-allergenic. Plant Cell. Rep. 2018, 37, 849–863. [Google Scholar] [CrossRef]
- Schouppe, D.; Rougé, P.; Lasanajak, Y.; Barre, A.; Smith, D.F.; Proost, P.; Van Damme, E.J.M. Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconj. J. 2010, 27, 613–623. [Google Scholar] [CrossRef][Green Version]
- Van Damme, E.J.M.; Barre, A.; Rougé, P.; Van Leuven, F.; Peumans, W.J. The NeuAc(α2,6)-Gal/GalNAc-binding lectin from elderberry (Sambucus nigra) bark, a type-2 ribosome-inactivating protein with an unusual specificity and structure. Eur. J. Biochem. 1996, 235, 128–137. [Google Scholar] [CrossRef]
- Rougé, P.; Brunet, E.; Borges, J.P.; Jauneau, A.; Saggio, B.; Bourrier, T.; Fancé, F.; Didier, A.; Barre, A. Proteins with cupin motif as major seed allergens. Rev. Fr. Allergol. 2011, 51, 36–40. [Google Scholar] [CrossRef]
- Villanueva, M.A. Elimination of artifacts on native western blots arising from endogenous lectin activity. J. Biochem. Biophys. Methods 2002, 50, 141–149. [Google Scholar] [CrossRef]
- Pramod, S.N.; Venkatesh, Y.P.; Mahesh, P.A. Potato lectin activates basophils and mast cells of atopic subjects by its interaction with core chitobiose of cell-bound non-specific immunoglobulin E. Clin. Exp. Immunol. 2007, 148, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997, 15, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Das, K.; Ravishankar, R.; Suguna, K.; Surolia, A.; Vijayan, M. Conformation, protein-carbohydrate interactions and a novel subunit association in the refined structure of peanut lectin-lactose complex. J. Mol. Biol. 1996, 259, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Hamelryck, T.W.; Loris, R.; Bouckaert, J.; Dao-Thi, M.H.; Strecker, G.; Imberty, A.; Fernandez, E.; Wyns, L.; Etzler, M. Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus. J. Mol. Biol. 1999, 286, 1161–1177. [Google Scholar] [CrossRef]
- Dessen, A.; Gupta, D.; Sabesan, S.; Brewer, C.F.; Sacchettini, J.C. X-ray crystal structure of the soybean agglutinin cross-linked with a biantennary analog of the blood group I carbohydrate antigen. Biochemistry 1995, 34, 4933–4942. [Google Scholar] [CrossRef]
- Casset, F.; Hamelryck, T.; Loris, R.; Brisson, J.R.; Tellier, C.; Dao-Thi, M.H.; Wyns, L.; Poortmans, F.; Pérez, S.; Imberty, A. NMR, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction. J. Biol. Chem. 1995, 270, 25619–25628. [Google Scholar] [CrossRef]
- Rutenber, E.; Katzin, B.J.; Ernst, S.; Collins, E.J.; Mlsna, D.; Ready, M.P.; Robertus, J.D. Crystallographic refinement of ricin to 2.5 Å. Proteins 1991, 10, 240–250. [Google Scholar] [CrossRef]
- Schwefel, D.; Maierhofer, C.; Beck, J.G.; Seeberger, S.; Diederichs, K.; Möller, H.M.; Welte, W.; Wittmann, V. Structural basis of multivalent binding to wheat germ agglutinin. J. Am. Chem. Soc. 2010, 132, 8704–8719. [Google Scholar] [CrossRef]
- Ramachandraiah, G.; Chandra, N.R.; Surolia, A.; Vijayan, M. Re-refinement using reprocessed data to improve the quality of the structure: A case study involving garlic lectin. Acta Crystallogr. D Biol. Crysyallogr. 2002, 58, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parametrizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Buts, L.; Dao-Thi, M.H.; Loris, R.; Wyns, L.; Etzler, M.; Hamelryck, T. Weak protein-protein interactions in lectins: The crystal structure of a vegetative lectin from the legume Dolichos biflorus. J. Mol. Biol. 2001, 309, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Shetty, K.N.; Latha, V.L.; Rao, R.N.; Nadimpalli, S.K.; Suguna, K. Affinity of galactose-specific legume lectin from Dolichos lablab to adenine revealed by X-ray crystallography. IUBMB Life 2013, 65, 633–644. [Google Scholar] [CrossRef]
- Nagae, M.; Soga, K.; Morita-Matsumoto, K.; Hanashima, S.; Ikeda, A.; Yamamoto, K.; Yamaguchi, Y. Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition-mode using a common legume lectin fold. Glycobiology 2014, 24, 368–378. [Google Scholar] [CrossRef]
- Nagae, M.; Kanagawa, M.; Morita-Matsumoto, K.; Hanashima, S.; Kizuka, Y.; Taniguchi, N.; Yamaguchi, Y. Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins. Sci. Rep. 2016, 6, 22973. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Crystallogr. 1993, 126, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
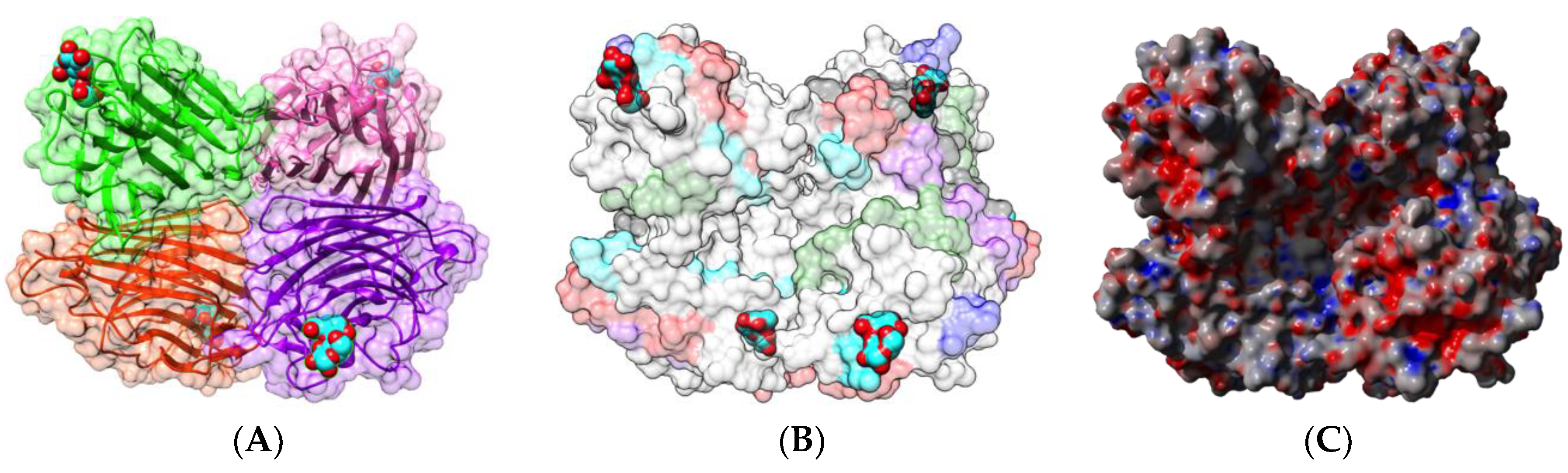
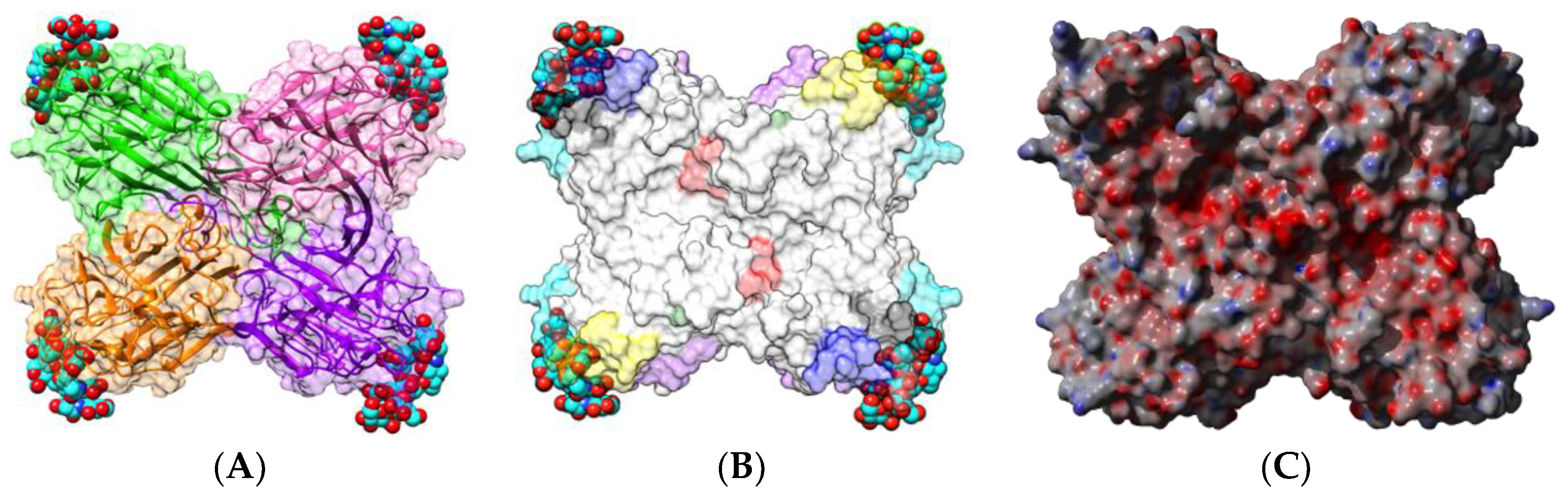
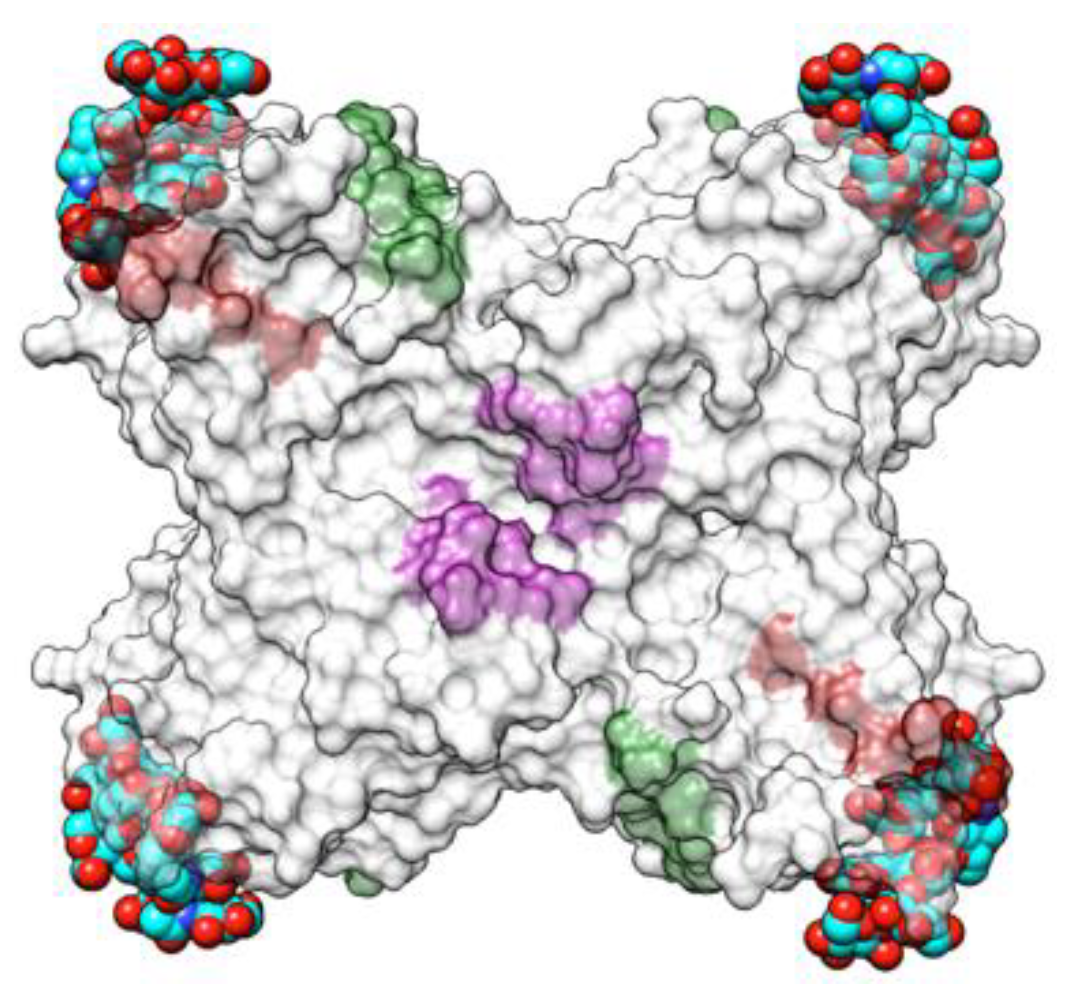
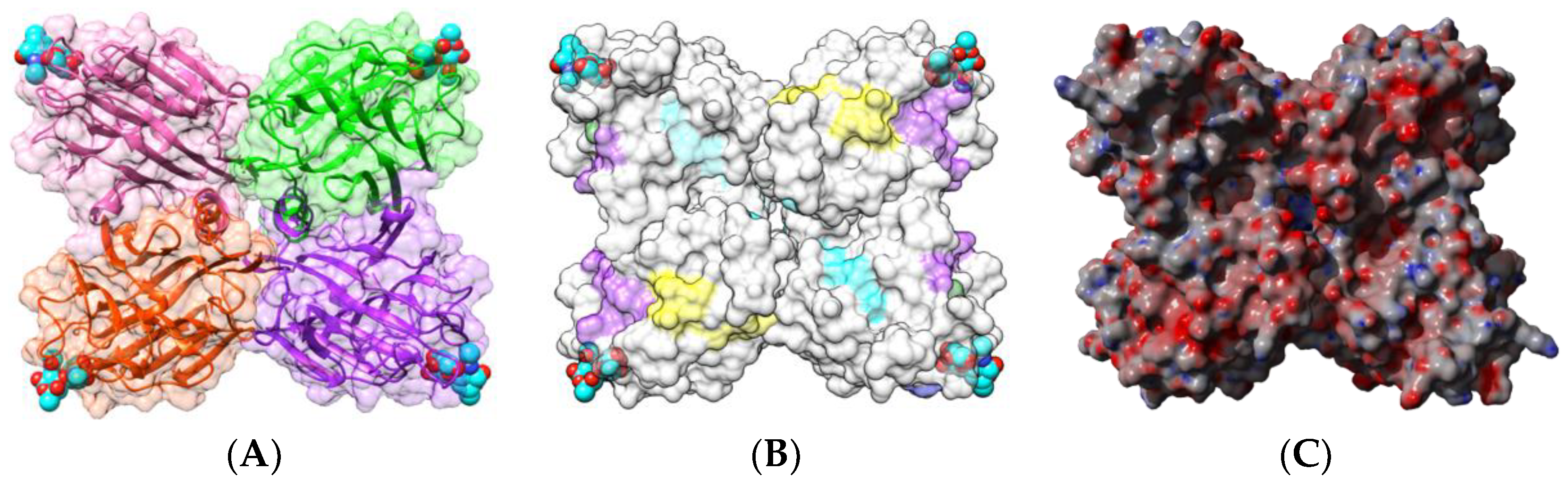
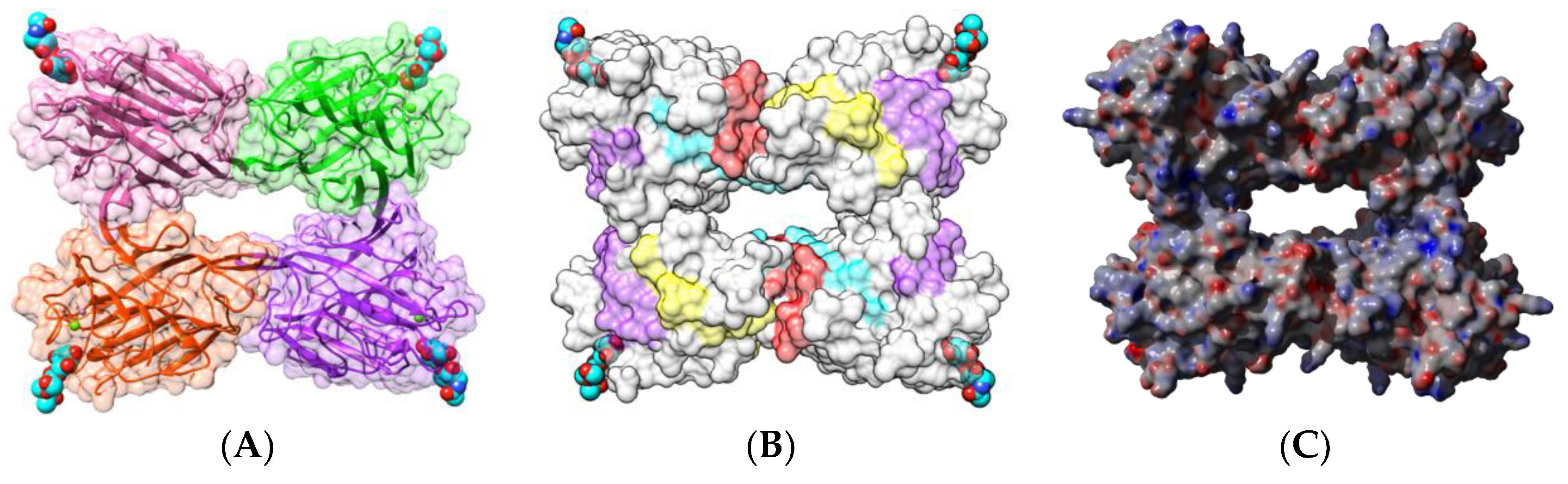
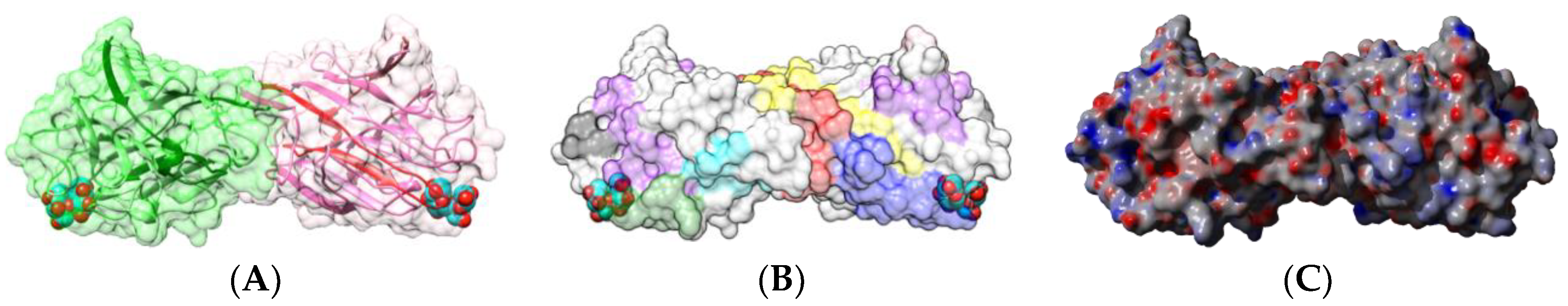
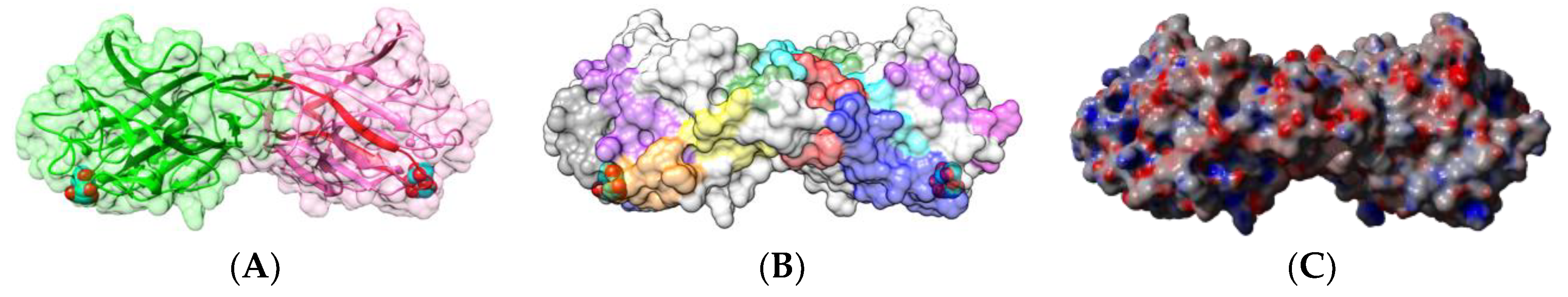
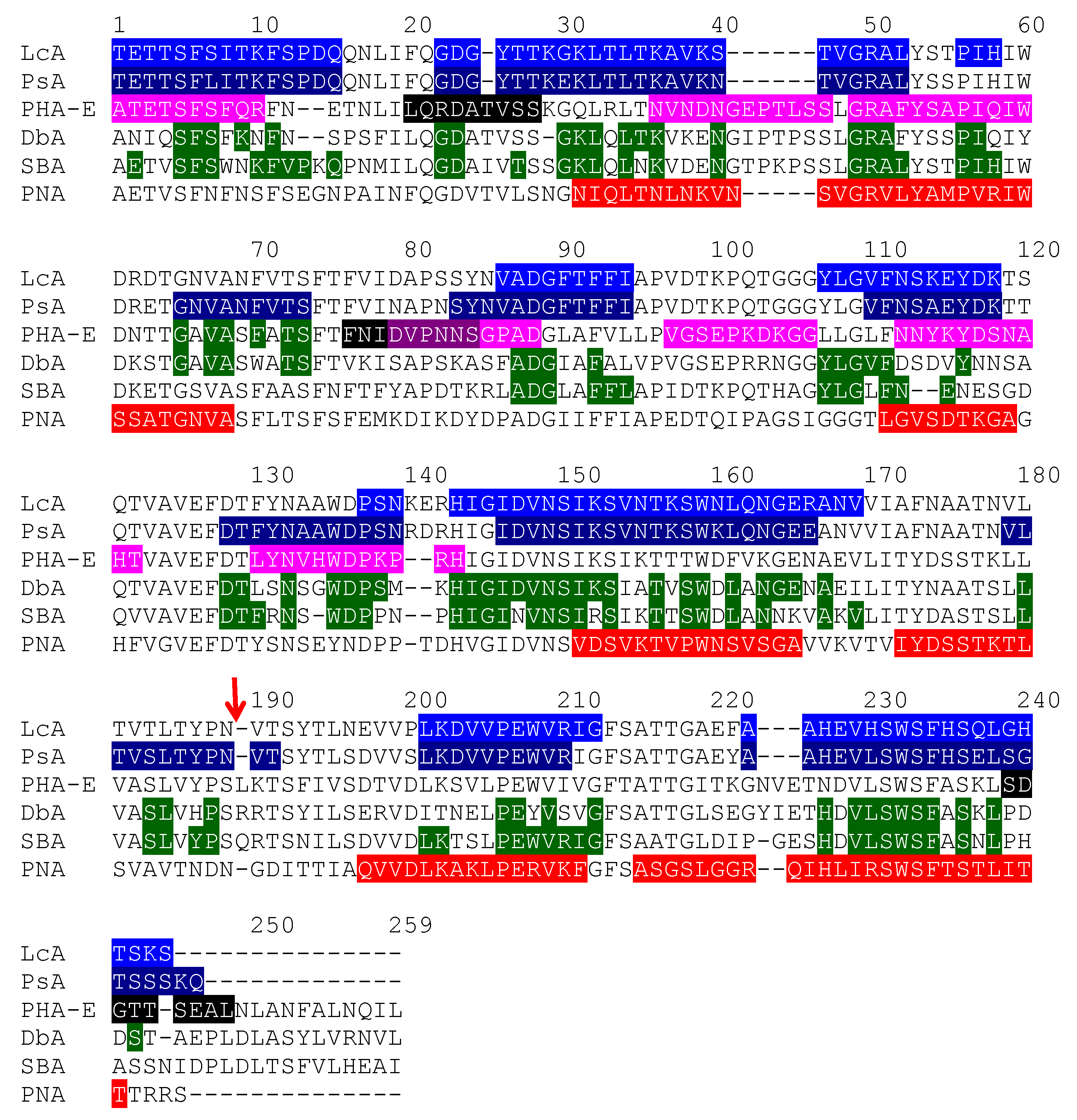

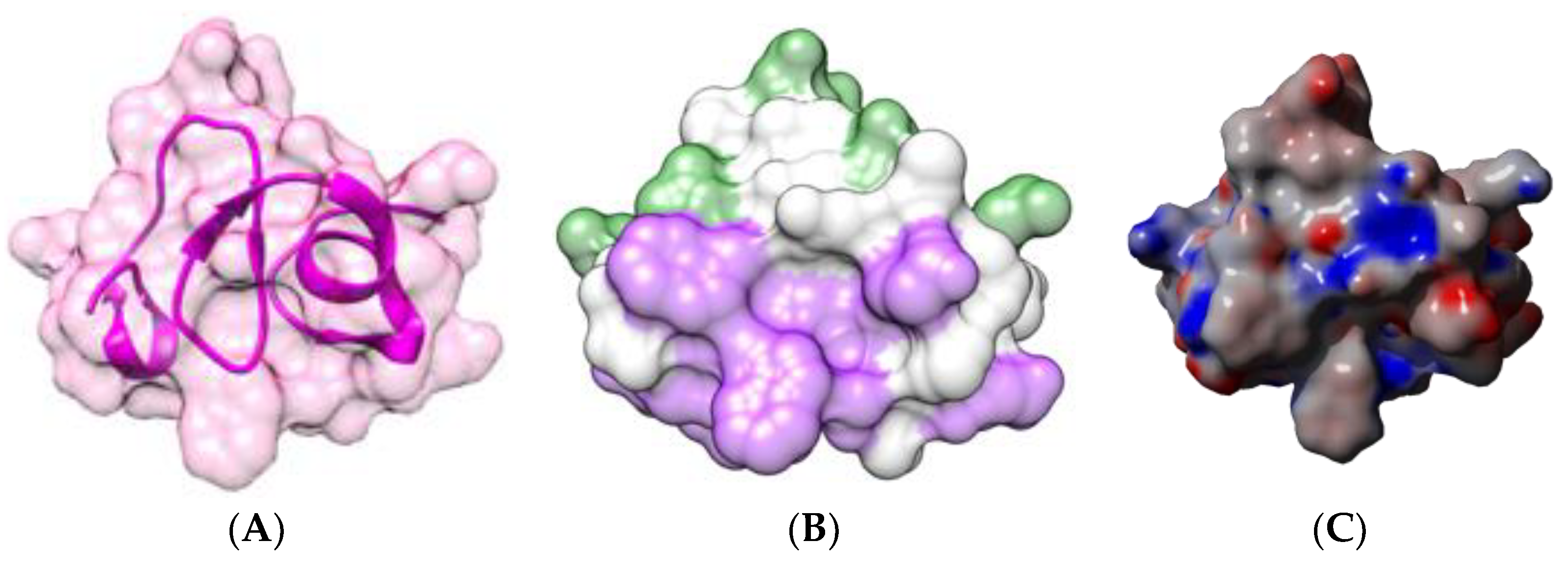
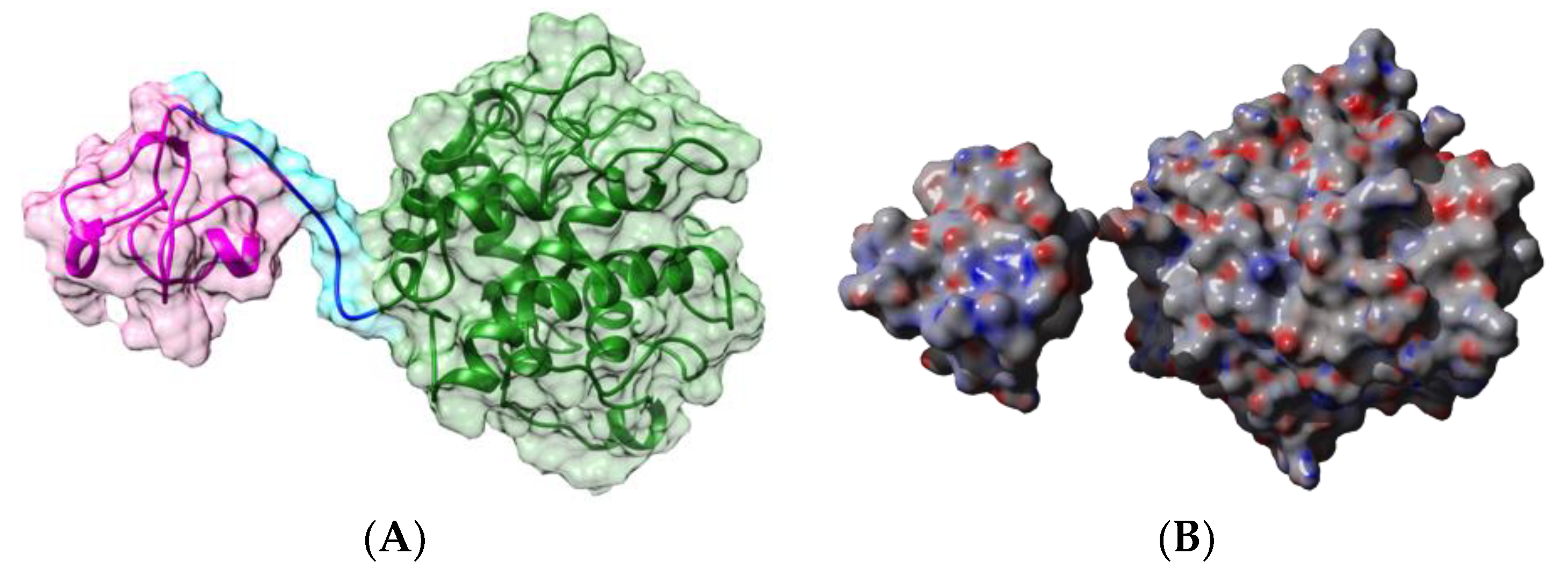
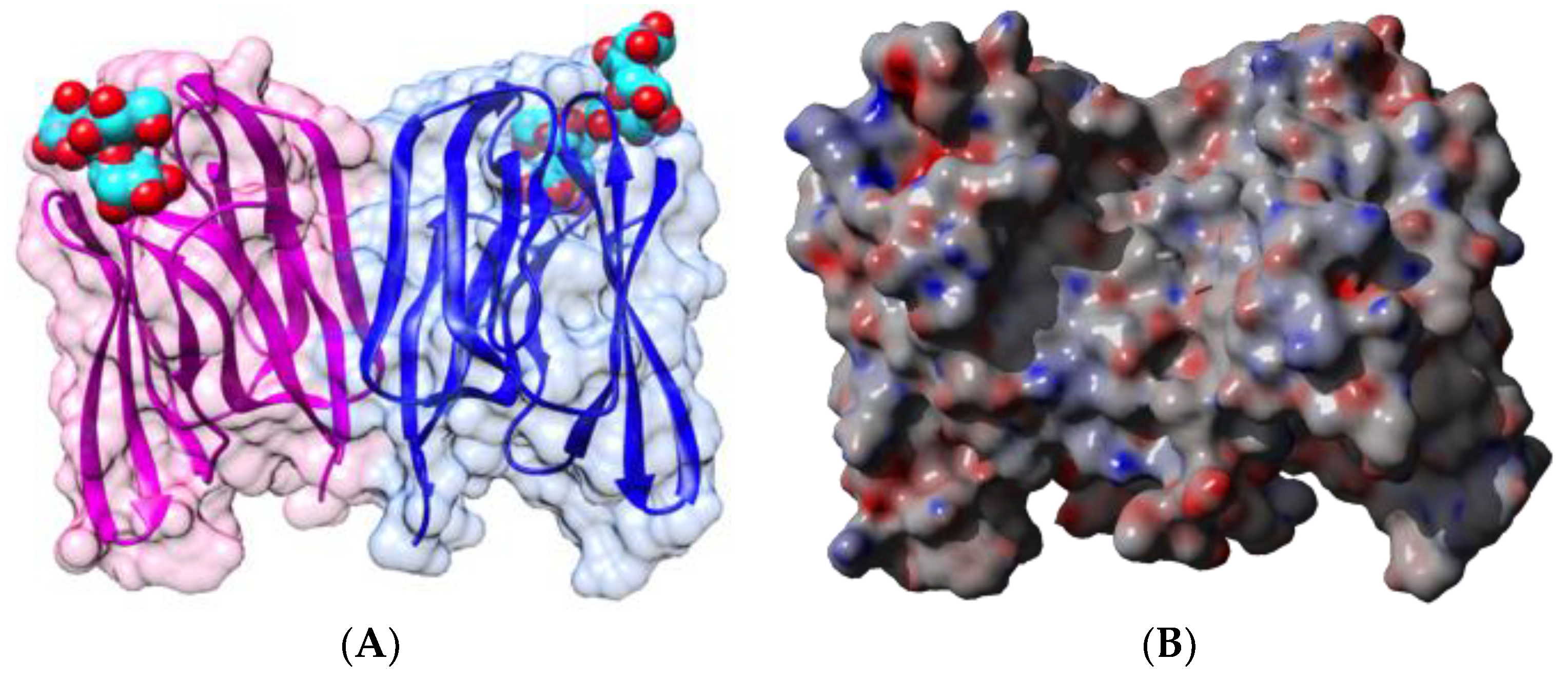
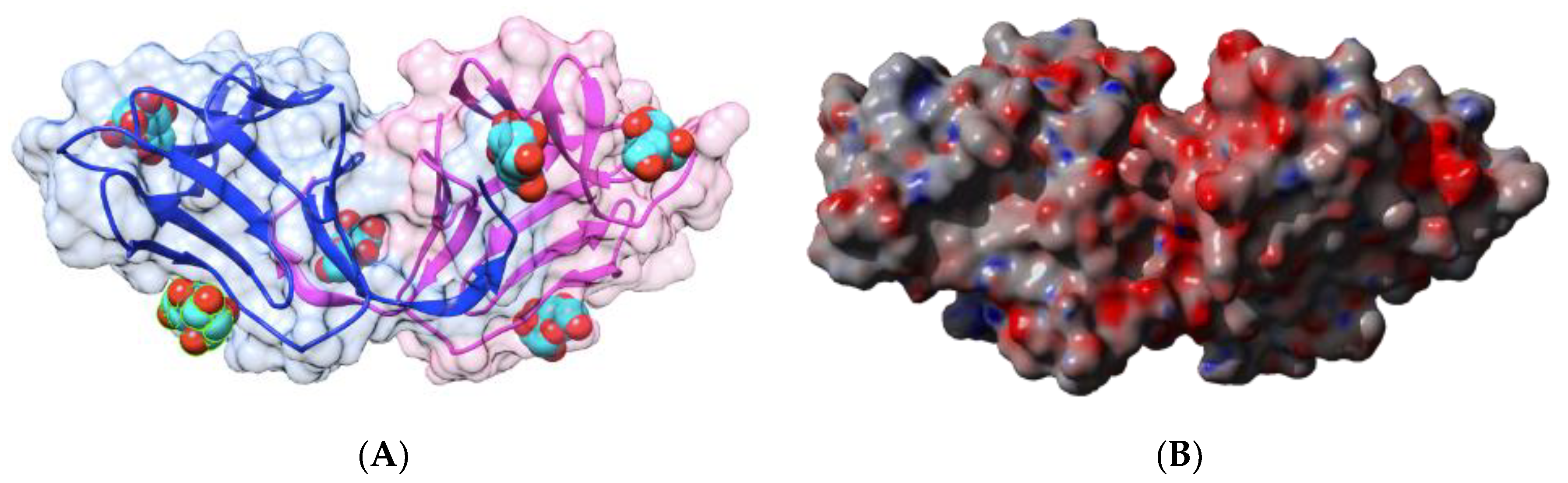
| Edible Plant | Lectin | Lectin Content (% of Total Protein) | Ref. |
|---|---|---|---|
| Lentil (Lens culinaris) | LcA | 2.5% | [18] |
| Pea (Pisum sativum) | PsA | 2.5% | [19] |
| Soybean (Glycine max) | SBA | 2% | [20] |
| Peanut (Arachis hypogaea) | PNA | 1.5% | [21] |
| Horse gram (Dolichos biflorus) | DbA | 10% | [22] |
| Kidney bean (Phaseolus vulgaris) | PHA | 1% | [23] |
| Wheat (Triticum aestivum) | WGA | 3% | [24] |
| Garlic (Allium sativum) | ASA | 10% | [25] |
| Black elderberry (Sambucus nigra) | SNA-I | 3% | [25] |
| Banana (Musa acuminata) | Mus a 2 | 2.3% | [26] |
| Allergen | Protein | Plant Species | IUIS Approved | Food Allergen | Ref. |
|---|---|---|---|---|---|
| Mus a 2 | Class I chitinase | Musa acuminata | + | + | [31] |
| Tri a 18 | Wheat germ agglutinin | Triticum aestivum | + | + | [32] |
| Zea m 8 | Class IV chitinase | Zea mays | + | + | [33] |
| Bra r 2 | Chitin-binding protein | Brassica rapa | + | + | [34] |
| Cas s 5 | Chitinase | Castanea sativa | + | + | * |
| Hev b 6 | Hevein precursor | Hevea brasiliensis | + | Contact | [35] |
| Hev b 11 | Class I chitinase | Hevea brasiliensis | + | Contact | * |
| Pers a 1 | Class I chitinase | Persea americana | + | + | [36] |
| Act c chitinase | Class I chitinase | Actinidia chinensis | + | [37] | |
| Car p chitinase | Class I chitinase | Carica papaya | + | [38] | |
| Lyc es chitinase | Class I chitinase | Lycopersicum esculentum | + | [39] | |
| Ann ch chitinase | Class I chitinase | Annona cherimola | + | [39] | |
| Pas ed chitinase | Class I chitinase | Passiflora edulis | + | [39] | |
| Vit v chitinase | Class IV chitinase | Vitis vinifera | + | [40] | |
| AcA | GNA-like Lectin | Allium cepa | + | [41] | |
| AsA | GNA-like lectin | Allium sativum | + | [42] | |
| Ara h agglutinin | Legume lectin | Arachis hypogaea | + | [43] | |
| DbA | Legume lectin | Dolichos biflorus | + | [44] | |
| SBA | Legume lectin | Glycine max | + | [45] | |
| LcA | Legume lectin | Lens culinaris | + | [29] | |
| PHA-E | Legume lectin | Phaseolus vulgaris | + | [29] | |
| PHA-L | Legume lectin | Phaseolus vulgaris | + | [29] | |
| PsA | Legume lectin | Pisum sativum | + | [29] | |
| Chia lectin | Legume lectin | Salvia hispanica | [29] | ||
| Ricin | RIP-II | Ricinus communis | + | [46] | |
| SNA-I | RIP-II | Sambucus nigra | + | [29] | |
| BanLec | Jacalin-related lectin | Musa acuminata | + | [47] | |
| Cuc m 17 kDa | Nictaba-related lectin | Cucumis melo | + | [48] |
| Lectin Family | Structural Scaffold | Simple Sugar Specificity | Complex Sugar Specificity |
|---|---|---|---|
| Agaricus bisporus lectin family | β-sandwich | Gal/GalNAc | T-antigen |
| Amaranthin family | β-trefoil | Gal/GalNAc | T-antigen |
| Class V chitinase homolog family | TIM-barrel (α8β8) | Man | High-mannose glycans |
| Cyanivirin family | β-barrel | Man | High-mannose glycans |
| Euonymus europaeus lectin family | β-trefoil | Gal, Man | B + H group + high-mannose glycans |
| GNA-like family | β-trefoil | Man | High-mannose glycans |
| Hevein family | hevein domain | GlcNAc | chitin |
| Jacalin-related lectin family | β-barrel | Gal, GalNAc, Man | T-antigen, High-mannose glycans |
| Legume lectin family | β-sandwich | Gal, GalNAc, Man | Complex glycans, High-mannose glycans |
| Lysin motif domain family | αβ | GlcNAc | Chitin, lipochito- oligosaccharides, |
| Nictaba family | β-sandwich | GlcNAc, Man | Chitooligosaccharides, high-mannose glycans, complex glycans |
| Ricin B family * | β-trefoil | Gal | Lactose |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Damme, E.J.M.V.; Simplicien, M.; Benoist, H.; Rougé, P. Are Dietary Lectins Relevant Allergens in Plant Food Allergy? Foods 2020, 9, 1724. https://doi.org/10.3390/foods9121724
Barre A, Damme EJMV, Simplicien M, Benoist H, Rougé P. Are Dietary Lectins Relevant Allergens in Plant Food Allergy? Foods. 2020; 9(12):1724. https://doi.org/10.3390/foods9121724
Chicago/Turabian StyleBarre, Annick, Els J.M. Van Damme, Mathias Simplicien, Hervé Benoist, and Pierre Rougé. 2020. "Are Dietary Lectins Relevant Allergens in Plant Food Allergy?" Foods 9, no. 12: 1724. https://doi.org/10.3390/foods9121724
APA StyleBarre, A., Damme, E. J. M. V., Simplicien, M., Benoist, H., & Rougé, P. (2020). Are Dietary Lectins Relevant Allergens in Plant Food Allergy? Foods, 9(12), 1724. https://doi.org/10.3390/foods9121724







