Identification of Emerging Hazards in Mussels by the Galician Emerging Food Safety Risks Network (RISEGAL). A First Approach
Abstract
1. Introduction
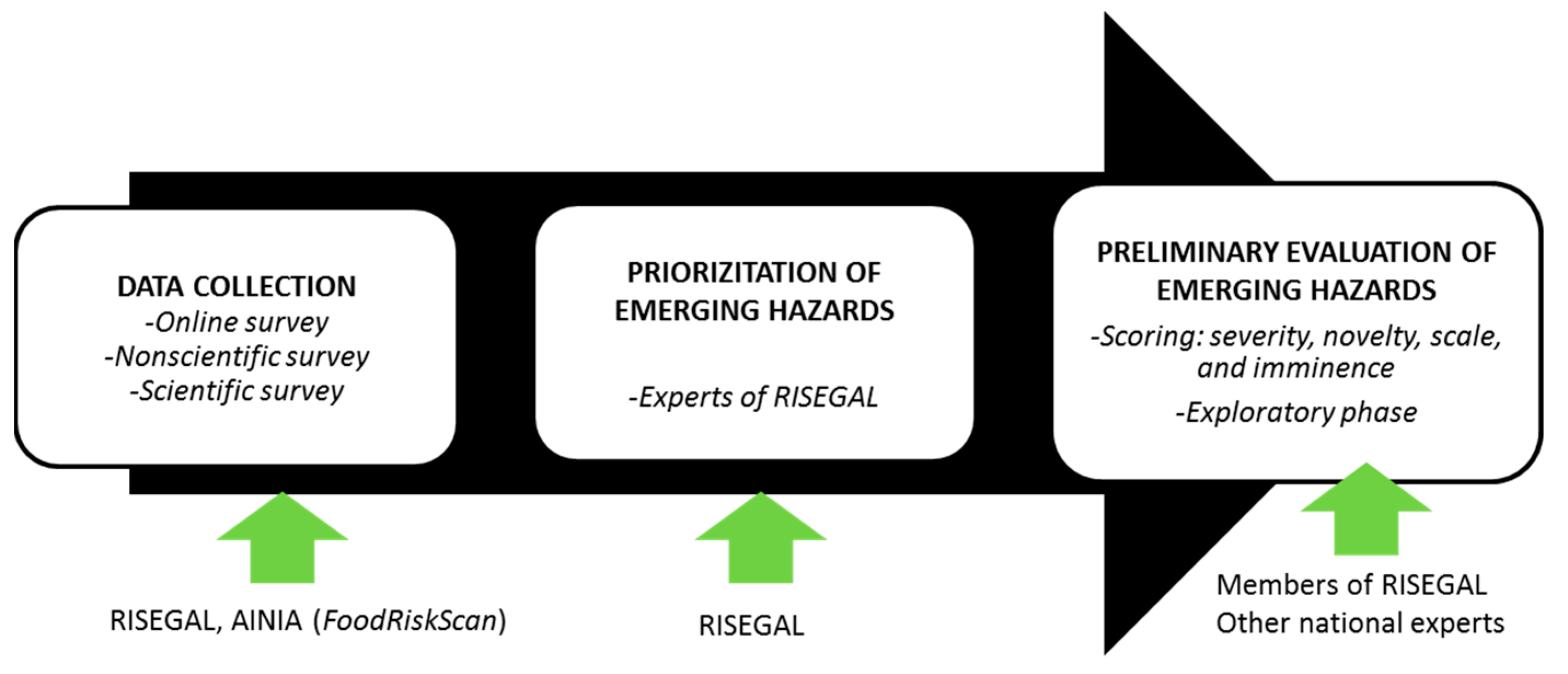
2. Materials and Methods
2.1. Online Survey
2.2. Nonscientific Survey
2.3. Scientific Survey
2.4. Preliminary Assessment by Scoring
2.5. Exploratory Study
2.5.1. Shellfish Sampling
2.5.2. Determination of Vibrio parahaemolyticus and Vibrio vulnificus
2.5.3. Determination of Cultivable Heterotrophic Bacteria
2.5.4. Isolation of Antibiotic-Resistant Bacteria
2.5.5. Identification of Ciprofloxacin-Resistant Bacteria
2.5.6. Detection of Antibiotic Resistance Genes
2.5.7. Determination of Hepatitis E Virus HEV in Mussels
2.5.8. Determination of Antibiotic Residues in Mussels
2.5.9. Determination of Tetrodotoxin
N2a Assay
LC–MS/MS Analysis
Extraction and SPE-ENVI-Carb Clean-Up
HILIC LC–MS/MS Analysis
3. Results and Discussion
3.1. Identification of Food Safety Emerging Problems in Bivalve by RISEGAL
3.2. Data Collection
3.2.1. Online Survey
3.2.2. Nonscientific Literature Survey
3.2.3. Scientific Literature Survey
3.3. Prioritization of Emerging Hazards
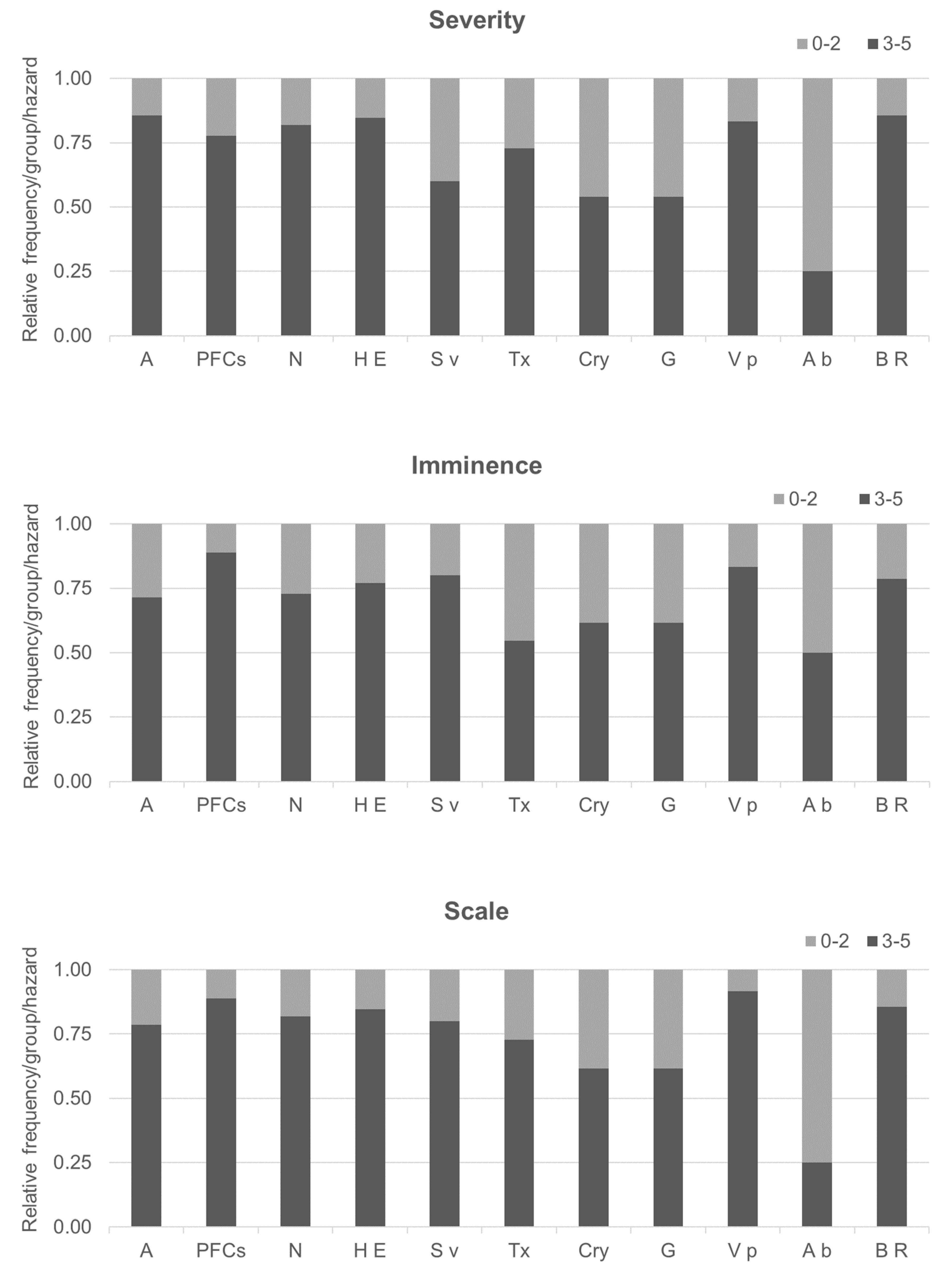
3.4. Preliminary Evaluation of Emerging Hazards
3.4.1. Scoring
3.4.2. Exploratory Study
- Hepatitis E virus
- Antibiotic-resistant bacteria (ARB)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority. Development and implementation of a system for the early identification of emerging risks in food and feed. EFSA J. 2010, 8. [Google Scholar] [CrossRef]
- Agencia Gallega de Innovación Xunta de Galicia. RIS3T—Smart Specialisation Strategy in Galicia 2014–2020. Available online: http://gain.xunta.gal/artigos/74/investigacion+referencia+competitiva?locale=es_ES (accessed on 10 August 2020).
- Labarta, U.; Fernández-Reiriz, M.J. The Galician mussel industry: Innovation and changes in the last forty years. Ocean Coast. Manag. 2019, 167, 208–218. [Google Scholar] [CrossRef]
- Sauvé, G. (Ed.) Molluscan Shellfish Safety; Springer: Dordrecht, The Netherlands, 2014; ISBN 978-94-007-6587-0. [Google Scholar]
- Børresen, T. (Ed.) Improving Seafood Products for the Consumer; Elsevier: Boca Raton, FL, USA, 2008; ISBN 978-1-84569-019-9. [Google Scholar]
- WHO. Food Safety—Climate Change and the Role of WHO. Available online: http://www.who.int/foodsafety/publications/all/climate_change/en/ (accessed on 6 August 2020).
- Strubbia, S.; Schaeffer, J.; Besnard, A.; Wacrenier, C.; Le Mennec, C.; Garry, P.; Desdouits, M.; Le Guyader, F.S. Metagenomic to evaluate norovirus genomic diversity in oysters: Impact on hexamer selection and targeted capture-based enrichment. Int. J. Food Microbiol. 2020, 323, 108588. [Google Scholar] [CrossRef]
- Emerging Risks Identification on Food and Feed—EFSA. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/5359 (accessed on 5 August 2020).
- Cavallo, R.A.; Acquaviva, M.I.; Stabili, L. Culturable heterotrophic bacteria in seawater and Mytilus galloprovincialis from a Mediterranean area (Northern Ionian Sea—Italy). Environ. Monit. Assess. 2009, 149, 465–475. [Google Scholar] [CrossRef]
- Varela, A.R.; Macedo, G.N.; Nunes, O.C.; Manaia, C.M. Genetic characterization of fluoroquinolone resistant Escherichia coli from urban streams and municipal and hospital effluents. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Bernárdez, M.; Rodríguez-Herrera, J.J.; Comesaña, Á.S.; Cabo, M.L. Identification and metagenetic characterisation of Listeria monocytogenes-harbouring communities present in food-related industrial environments. Food Control 2019, 95, 6–17. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Rivadulla, E.; Varela, M.F.; Mesquita, J.R.; Nascimento, M.S.J.; Romalde, J.L. Detection of hepatitis E virus in shellfish harvesting areas from Galicia (Northwestern Spain). Viruses 2019, 11, 618. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Chiesa, L.M.; Nobile, M.; Malandra, R.; Panseri, S.; Arioli, F. Occurrence of antibiotics in mussels and clams from various FAO areas. Food Chem. 2018, 240, 16–23. [Google Scholar] [CrossRef]
- Manger, R.L.; Leja, L.S.; Lee, S.Y.; Hungerford, J.M.; Wekell, M.M. Tetrazolium-Based cell bioassay for neurotoxins active on voltage-sensitive sodium channels: Semiautomated assay for Saxitoxins, Brevetoxins, and Ciguatoxins. Anal. Biochem. 1993, 214, 190–194. [Google Scholar] [CrossRef]
- Manger, R.L.; Leja, L.S.; Lee, S.Y.; Hungerford, J.M.; Hokama, Y.; Dickey, R.W.; Granade, H.R.; Lewis, R.; Yasumoto, T.; Wekell, M.M. Detection of sodium channel toxins: Directed cytotoxicity assays of purified ciguatoxins, brevetoxins, saxitoxins, and seafood extracts. J. AOAC Int. 1995, 78, 521–527. [Google Scholar] [CrossRef]
- Leão, J.M.; Lozano-Leon, A.; Giráldez, J.; Vilariño, Ó.; Gago-Martínez, A. Preliminary results on the evaluation of the occurrence of tetrodotoxin associated to marine Vibrio spp. in bivalves from the Galician Rias (Northwest of Spain). Mar. Drugs 2018, 16, 81. [Google Scholar] [CrossRef]
- Turner, A.D.; McNabb, P.S.; Harwood, D.T.; Selwood, A.I.; Boundy, M.J. Single-Laboratory validation of a multitoxin ultra-performance LC-hydrophilic interaction LC-MS/MS method for quantitation of paralytic shellfish toxins in Bivalve shellfish. J. AOAC Int. 2015, 98, 609–621. [Google Scholar] [CrossRef] [PubMed]
- European Union Reference Laboratory for Marine Biotoxins. Determination of tetrodotoxin by HILIC-MS/MS. In Eurlmb TTX Sop; European Union Reference Laboratory for Marine Biotoxins: Vigo, Spain, 2015; Volume 25. [Google Scholar]
- Boundy, M.J.; Selwood, A.I.; Harwood, D.T.; McNabb, P.S.; Turner, A.D. Development of a sensitive and selective liquid chromatography-mass spectrometry method for high throughput analysis of paralytic shellfish toxins using graphitised carbon solid phase extraction. J. Chromatogr. A 2015, 1387, 1–12. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). EFSA’s activities on emerging risks in 2014. EFSA Support. Publ. 2015, 12, 838E. [Google Scholar] [CrossRef]
- Fernández-Sanjuan, M.; Faria, M.; Lacorte, S.; Barata, C. Bioaccumulation and effects of perfluorinated compounds (PFCs) in zebra mussels (Dreissena polymorpha). Environ. Sci. Pollut. Res. Int. 2013, 20, 2661–2669. [Google Scholar] [CrossRef]
- Fatta-Kassinos, D.; Meric, S.; Nikolaou, A. Pharmaceutical residues in environmental waters and wastewater: Current state of knowledge and future research. Anal. Bioanal. Chem. 2011, 399, 251–275. [Google Scholar] [CrossRef]
- Tacconelli, E.; Pezzani, M.D. Public health burden of antimicrobial resistance in Europe. Lancet Infect. Dis. 2019, 19, 4–6. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Andremont, A.; Bengtsson-Palme, J.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Fick, J.; Flach, C.-F.; Gaze, W.H.; Kuroda, M.; et al. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018, 117, 132–138. [Google Scholar] [CrossRef]
- Manaia, C.M. Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-Direct proportionality between abundance and risk. Trends Microbiol. 2017, 25, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Grande, C.; Reid, P.C.; Hélaouët, P.; Edwards, M.; Höfle, M.G.; Brettar, I.; Colwell, R.R.; Pruzzo, C. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. USA 2016, 113, E5062–E5071. [Google Scholar] [CrossRef] [PubMed]
- Pratheepa, V.; Alex, A.; Silva, M.; Vasconcelos, V. Bacterial diversity and tetrodotoxin analysis in the viscera of the gastropods from Portuguese coast. Toxicon 2016, 119, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P. The current host range of hepatitis E viruses. Viruses 2019, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.H.; Emerson, S.U. Hepatitis E: An emerging awareness of an old disease. J. Hepatol. 2008, 48, 494–503. [Google Scholar] [CrossRef]
- Grodzki, M.; Schaeffer, J.; Piquet, J.-C.; Saux, J.-C.L.; Chevé, J.; Ollivier, J.; Pendu, J.L.; Guyader, F.S.L. Bioaccumulation efficiency, tissue distribution, and environmental occurrence of hepatitis E virus in Bivalve shellfish from France. Appl. Environ. Microbiol. 2014, 80, 4269–4276. [Google Scholar] [CrossRef]
- Romalde, J.L.; Rivadulla, E.; Varela, M.F.; Barja, J.L. An overview of 20 years of studies on the prevalence of human enteric viruses in shellfish from Galicia, Spain. J. Appl. Microbiol. 2018, 124, 943–957. [Google Scholar] [CrossRef]
- Mesquita, J.R.; Oliveira, D.; Rivadulla, E.; Abreu-Silva, J.; Varela, M.F.; Romalde, J.L.; Nascimento, M.S.J. Hepatitis E virus genotype 3 in mussels (Mytilus galloprovinciallis), Spain. Food Microbiol. 2016, 58, 13–15. [Google Scholar] [CrossRef]
- Manso, C.F.; Romalde, J.L. Detection and characterization of hepatitis A virus and norovirus in mussels from Galicia (NW Spain). Food Environ. Virol. 2013, 5, 110–118. [Google Scholar] [CrossRef]
- Krog, J.S.; Larsen, L.E.; Schultz, A.C. Enteric porcine viruses in farmed shellfish in Denmark. Int. J. Food Microbiol. 2014, 186, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Muscillo, M. GIV noroviruses and other enteric viruses in bivalves: A preliminary study. New Microbiol. 2012, 35, 27–34. [Google Scholar]
- Fusco, G.; Anastasio, A.; Kingsley, D.H.; Amoroso, M.G.; Pepe, T.; Fratamico, P.M.; Cioffi, B.; Rossi, R.; La Rosa, G.; Boccia, F. Detection of hepatitis A virus and other enteric viruses in shellfish collected in the Gulf of Naples, Italy. Int. J. Environ. Res. Public Health 2019, 16, 2588. [Google Scholar] [CrossRef]
- Suffredini, E.; Lanni, L.; Arcangeli, G.; Pepe, T.; Mazzette, R.; Ciccaglioni, G.; Croci, L. Qualitative and quantitative assessment of viral contamination in bivalve molluscs harvested in Italy. Int. J. Food Microbiol. 2014, 184, 21–26. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, Z.; Crossan, C.; Craft, J.; Scobie, L. First report of the presence of hepatitis E virus in Scottish-Harvested shellfish purchased at retail level. Food Environ. Virol. 2018, 10, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, Y.; Oshiro, Y.; Hasegawa, N.; Fukuda, K.; Abei, M.; Nishi, M.; Okamoto, H.; Ohkohchi, N. Clinical features of hepatitis E virus infection in Ibaraki, Japan: Autochthonous hepatitis E and acute-on-chronic liver failure. Tohoku J. Exp. Med. 2015, 235, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, Y.; Isoda, N.; Sato, Y.; Iwaki, T.; Ono, K.; Ido, K.; Sugano, K.; Takahashi, M.; Nishizawa, T.; Okamoto, H. Infection of a Japanese patient by genotype 4 hepatitis E virus while traveling in Vietnam. J. Clin. Microbiol. 2004, 42, 3883–3885. [Google Scholar] [CrossRef]
- Said, B.; Ijaz, S.; Kafatos, G.; Booth, L.; Thomas, H.L.; Walsh, A.; Ramsay, M.; Morgan, D. Hepatitis E outbreak on cruise ship. Emerg. Infect. Dis. 2009, 15, 1738–1744. [Google Scholar] [CrossRef]
- Marti, E.; Huerta, B.; Rodríguez-Mozaz, S.; Barceló, D.; Jofre, J.; Balcázar, J.L. Characterization of ciprofloxacin-resistant isolates from a wastewater treatment plant and its receiving river. Water Res. 2014, 61, 67–76. [Google Scholar] [CrossRef]
- Marti, E.; Balcázar, J.L. Aeromonas rivipollensis sp. nov., a novel species isolated from aquatic samples. J. Basic Microbiol. 2015, 55, 1435–1439. [Google Scholar] [CrossRef]
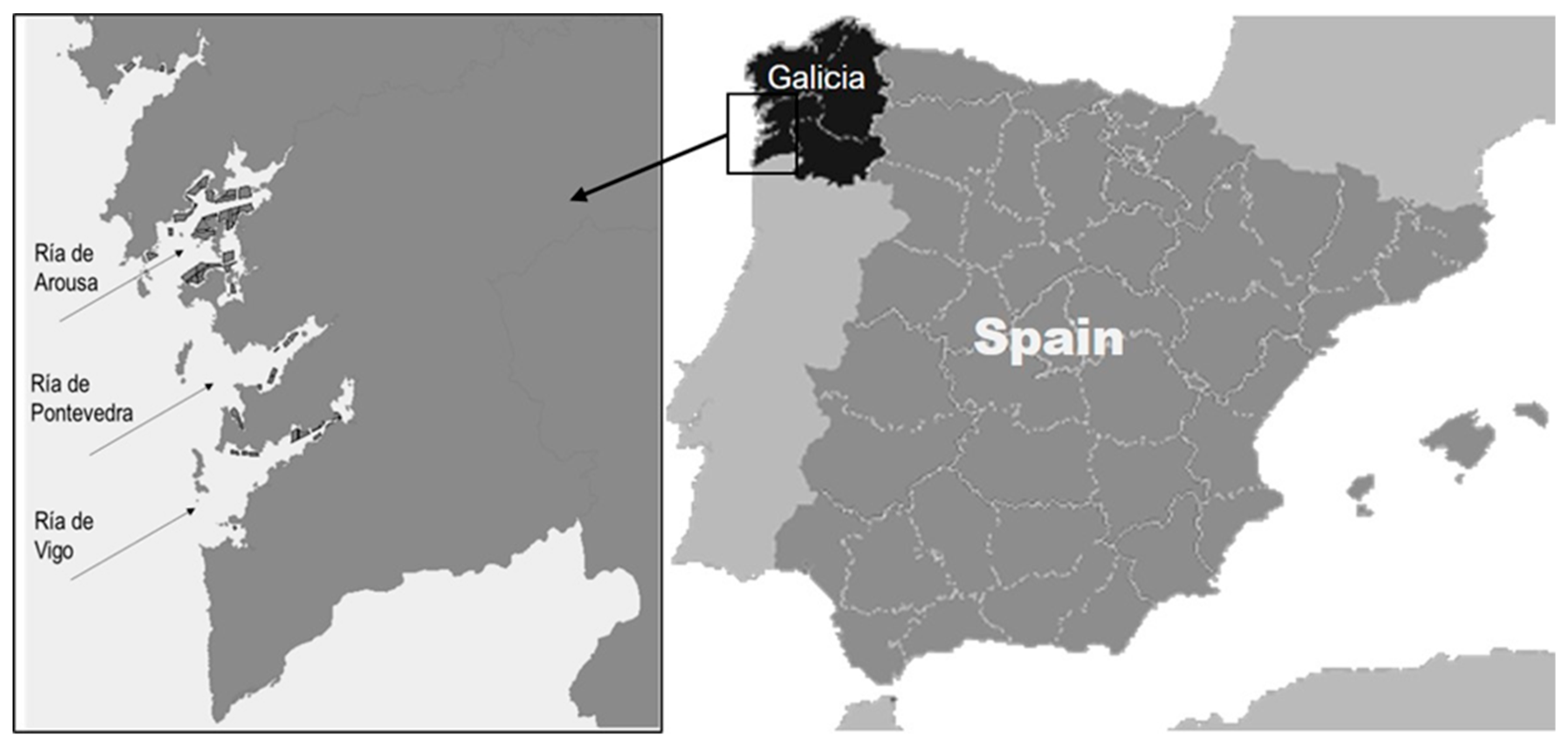
| Month | Site 1 | Site 2 | Site 3 |
|---|---|---|---|
| Ría de Arousa | Ría de Vigo | Ría de Pontevedra | |
| May | ** | ||
| June | ** | * | * |
| July | ** | * | * |
| Q.1. Could you identify any “emerging problem” in food safety that could affect the production and commercialization chain of bivalve mollusks? Yes/no. |
| Q.2. Brief description of the identified emerging problem. Note: the answer should describe briefly the emerging problem about which she/he is thinking. |
| Q.3. In your opinion, which group of bivalve mollusks could be mostly affected by the identified emerging problem? Clam/mussel/oyster/other. |
| Q.4. What is the time scale for the identified emerging problem to occur? short term (<2 years), medium term (2–10 years), and long term (>10 years). |
| Q.5. Could you identify the step of the value chain most affected by the problem? Production/transformation/distribution. |
| Q.6. To which sector do you belong? Food industry worker/sanitary inspector or researcher/citizen. |
| HAZARD | AGENT | NEW | CLASSIFICATION | RELATED CAUSE | VULNERABLE GROUPS | TEMPORAL SCALE | PRESENCE IN GALICIA |
|---|---|---|---|---|---|---|---|
| Drug residues in bivalvos | Antimicrobials | Yes | Increased susceptibility | Changes in consumption habits | Immunocompromised | 1–3 years | Likely |
| Organic pollutant residues in bivalves | Perfluorinated compounds | Yes | Increased exposure | Direct human intervention | Childhood | 3–10 years | Likely |
| Nanoparticles residues in bivalves. Transport capacity of other pollutants | Nanoparticles | Yes | Increased exposure | Changes in consumption habits | Childhood | 1–3 years | Likely |
| Hepatitis E virus in bivalves | Hepatitis E virus | Yes | New scenarios | Control not required Scarce data | Any group | 1–3 years | Likely |
| Sapovirus in Galician and import bivalves | Sapovirus | Yes | Increased exposure | Control not required Scarce data | Any group | 1–3 years | Likely |
| Tetrodotoxin in European coast | Tetrodotoxin | Yes | New scenarios | Environmental factors | Any group | 3–10 years | Insufficient data |
| Cryptosporidium in bivalves | Cryptosporidium | No | Incresased exposure | Control not required Scarce data | Any group | * | Insufficient data |
| Giardia in bivalves | Giardia | No | Undefined | Control not required Scarce data | Any group | * | Insufficient data |
| Vibrio parahaemolyticus in bivalves | Vibrio parahaemolyticus | No | New scenarios | Environmental factors | Any group | 1–3 years | Likely |
| Arcobacter spp. in bivalves molluscs cultivated in Galicia | Arcobacter spp. (A. butzuli) | No | Increased exposure | Environmental factors | Any group | 3–10 years | Insufficient data |
| Bivalves as reservoirs of antibiotic resistant bacteria and their relationship to the transmission of resistance genes | Antibiotic-resistant bacteria of last resort | No | Increased susceptibility | Direct human intervention | Immunocompromised and old adult | >10 years | Insufficient data |
| Production Zone | May | June | July |
|---|---|---|---|
| Site 1 | 1.09 × 103 | 15 | n.d. |
| Site 2 | 3.01 × 103 | 260 | n.d. |
| ORIGIN | Site 1 | Site 2 | Site 3 | |||
|---|---|---|---|---|---|---|
| DATE | CHB | CRB | CHB | CRB | CHB | CRB |
| May 2019 | 6.80 (0.33) 6.25 (0.22) | * * | * | * | * | * |
| June 2019 | 4.79 (0.07) 5.68 (0.10) | n.d. 125 | 4.45 (0.21) | 100 | * | * |
| July 2019 | 5.39 (0.37) 6.02 (0.11) | <15 n.d. | * | * | 6.74 (0.40) | 500 |
| Production Zone | May | June | July |
|---|---|---|---|
| Site 1 | * | Vagococcus fluvialis | * |
| Site 2 | * | Aeromonas rivipollensis Pediococcus pentosaceus | * |
| Site 3 | * | Escherichia coli |
| Species (phylum) | Class 1 Integron | β-Lactam | β-Lactam | Quinolone | Aminoglycoside | Sulfonamide |
|---|---|---|---|---|---|---|
| IntI1 280 bp | blaTEM 1080 bp | blaCTX-M 540 bp | qnrS 463 bp | aac(6) 482 bp | sul1 789 bp | |
| Vagococcus fluvialis | - | - | - | - | + | - |
| Pediococcus pentosaceus | - | - | - | - | + | - |
| Aeromonas rivipollensis | + | - | + | + | + | + |
| Escherichia coli | + | + | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López Cabo, M.; L. Romalde, J.; Simal-Gandara, J.; Gago Martínez, A.; Giráldez Fernández, J.; Bernárdez Costas, M.; Pascual del Hierro, S.; Pousa Ortega, Á.; Manaia, C.M.; Abreu Silva, J.; et al. Identification of Emerging Hazards in Mussels by the Galician Emerging Food Safety Risks Network (RISEGAL). A First Approach. Foods 2020, 9, 1641. https://doi.org/10.3390/foods9111641
López Cabo M, L. Romalde J, Simal-Gandara J, Gago Martínez A, Giráldez Fernández J, Bernárdez Costas M, Pascual del Hierro S, Pousa Ortega Á, Manaia CM, Abreu Silva J, et al. Identification of Emerging Hazards in Mussels by the Galician Emerging Food Safety Risks Network (RISEGAL). A First Approach. Foods. 2020; 9(11):1641. https://doi.org/10.3390/foods9111641
Chicago/Turabian StyleLópez Cabo, Marta, Jesús L. Romalde, Jesus Simal-Gandara, Ana Gago Martínez, Jorge Giráldez Fernández, Marta Bernárdez Costas, Santiago Pascual del Hierro, Ánxela Pousa Ortega, Célia M. Manaia, Joana Abreu Silva, and et al. 2020. "Identification of Emerging Hazards in Mussels by the Galician Emerging Food Safety Risks Network (RISEGAL). A First Approach" Foods 9, no. 11: 1641. https://doi.org/10.3390/foods9111641
APA StyleLópez Cabo, M., L. Romalde, J., Simal-Gandara, J., Gago Martínez, A., Giráldez Fernández, J., Bernárdez Costas, M., Pascual del Hierro, S., Pousa Ortega, Á., Manaia, C. M., Abreu Silva, J., & Rodríguez Herrera, J. (2020). Identification of Emerging Hazards in Mussels by the Galician Emerging Food Safety Risks Network (RISEGAL). A First Approach. Foods, 9(11), 1641. https://doi.org/10.3390/foods9111641










