Isolation of Microalgae from Mediterranean Seawater and Production of Lipids in the Cultivated Species
Abstract
1. Introduction
2. Materials and Methods
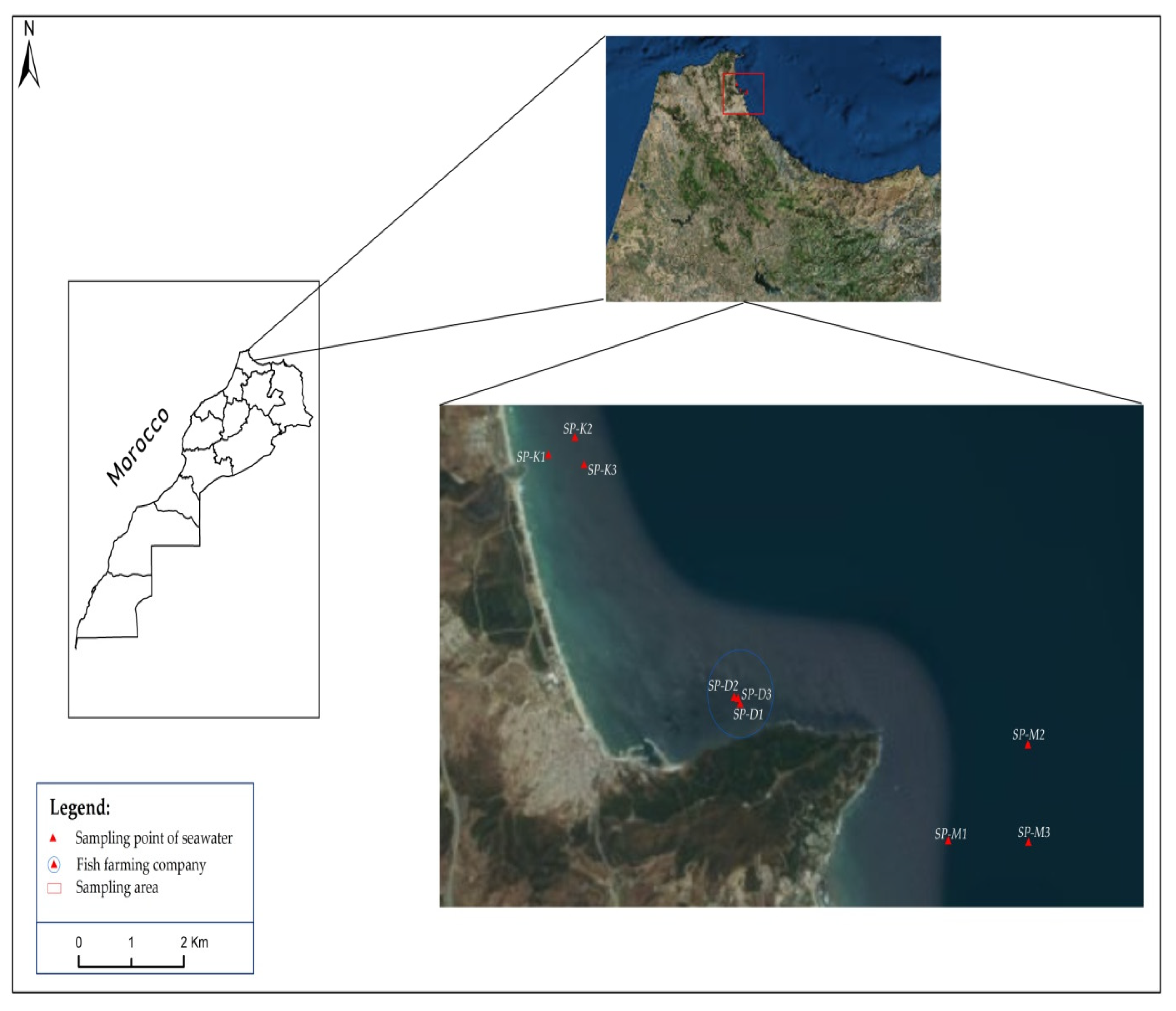
2.1. Sampling and Isolation
2.2. Strain Identification
2.3. Lipid Analysis
Extraction and Measurement of Lipid Contents
2.4. HPLC Analysis
2.5. Statistical Analysis
3. Results and Discussion
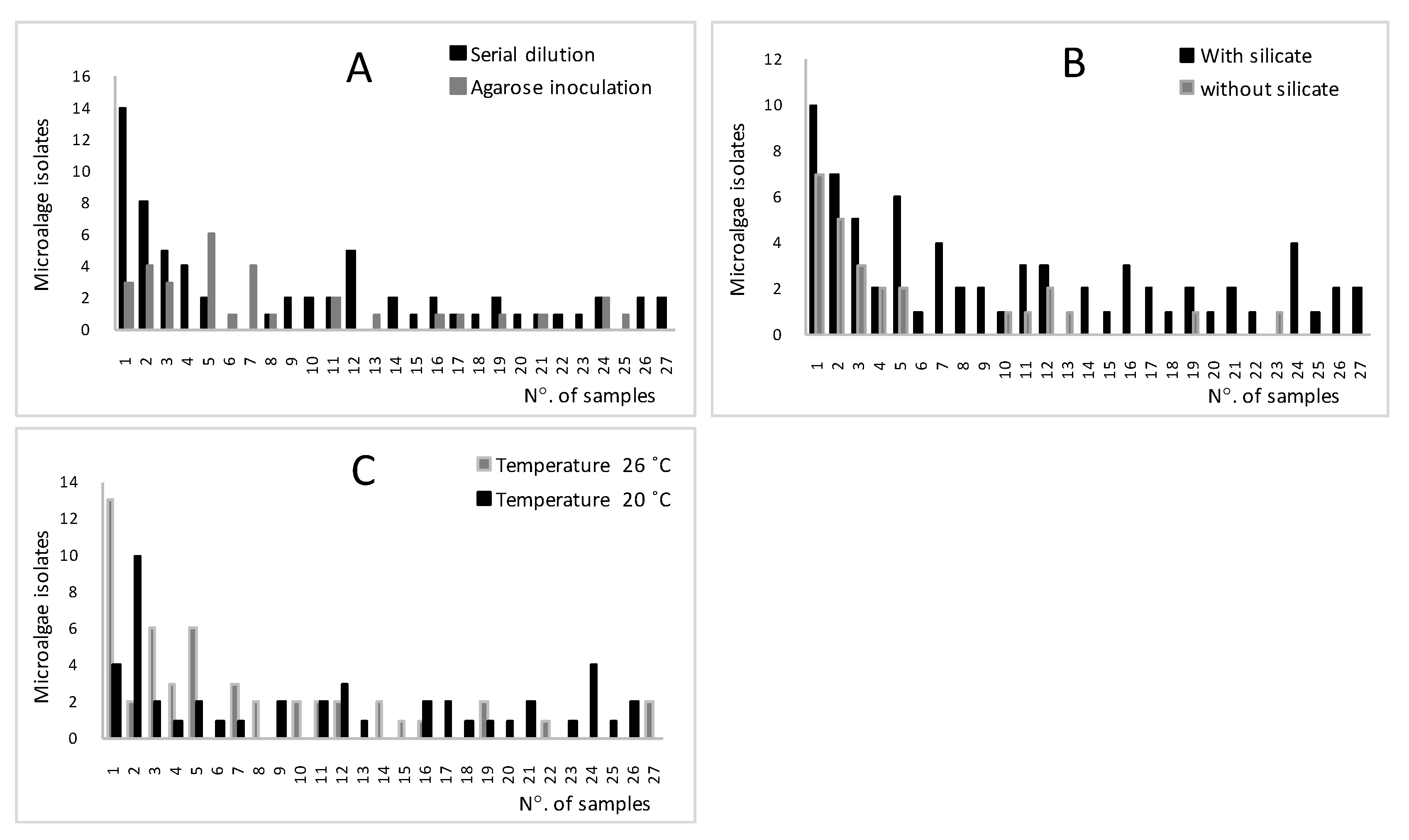
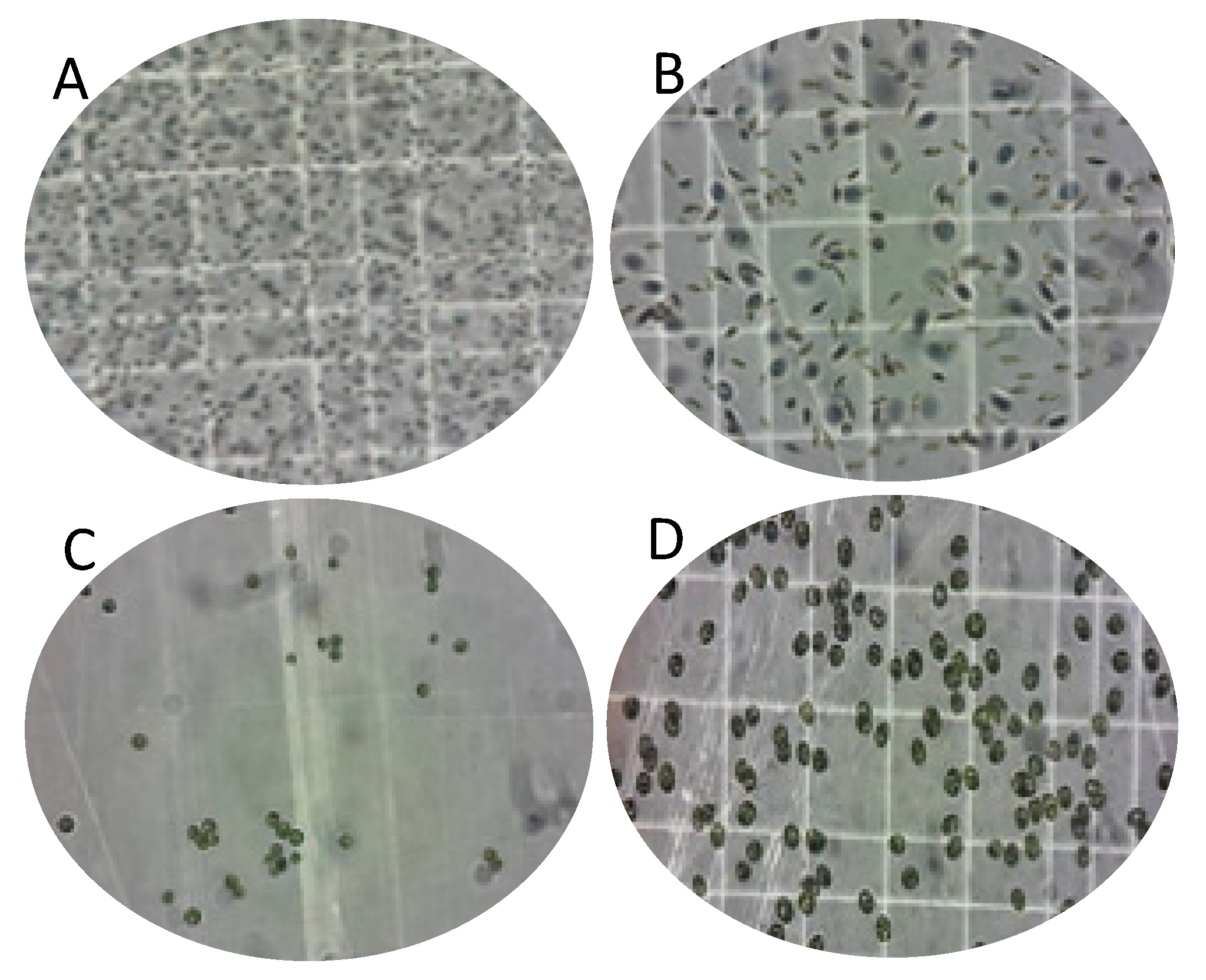
3.1. Isolation of Native Microalgae
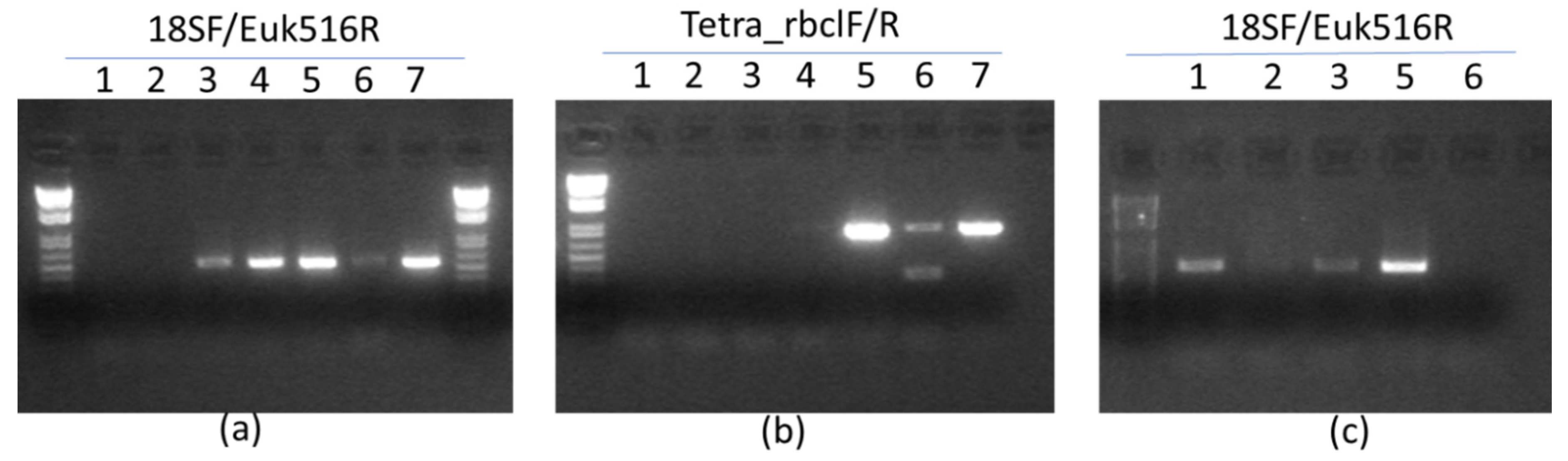
3.2. Molecular Identification of Native Microalgae
3.3. Biomass and Lipid Productivity
3.4. HPLC Analysis of Lipid Contents
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richmond, A. Handbook of Microalgal Culture: Biotechnology and Applied Phycology; Blackwell Science: Oxford, UK, 2004; p. 289. ISBN 978-1-4051-2832-2. [Google Scholar]
- Falkowski, P.G. The Evolution of Modern Eukaryotic Phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Sathasivam, R.; Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Microalgae metabolites: A rich source for food and medicine. Saudi J. Biol. Sci. 2017, 26, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Craggs, R.J.; Adey, W.H.; Jenson, K.R.; John, M.S.S.; Green, F.B.; Oswald, W.J. Phosphorus removal from wastewater using an algal turf scrubber. Water Sci. Technol. 1996, 33, 191–198. [Google Scholar] [CrossRef]
- Demirbas, A.; Demirbas, M.F. Importance of algae oil as a source of biodiesel. Energy Convers. Manag. 2011, 52, 163–170. [Google Scholar] [CrossRef]
- Enzing, C.; Ploeg, M.; Barbosa, M.; Sijtsma, L. Microalgae-based products for the food and feed sector: An outlook for Europe. JRC Sci. Policy Rep. 2014, 19–37. [Google Scholar]
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef]
- Hu, G.-P.; Yuan, J.; Sun, L.; She, Z.-G.; Wu, J.-H.; Lan, X.-J.; Zhu, X.; Lin, Y.-C.; Chen, S.-P. Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar. Drugs. 2011, 9, 514–525. [Google Scholar] [CrossRef]
- De JesusRaposo, M.F.; De Morais, R.M.S.C.; de Morais, B.; Miranda, A.M. Bioactivity and applications of sulphated polysaccharides from marine microalgae. Mar. Drugs. 2013, 11, 233–252. [Google Scholar]
- Peng, J.; Yuan, J.-P.; Wu, C.-F.; Wang, J.-H. Fucoxanthin, a marine carotenoid present in brown seaweeds and diatoms: Metabolism and bioactivities relevant to human health. Mar. Drugs. 2011, 9, 1806–1828. [Google Scholar] [CrossRef] [PubMed]
- Haimeur, A.; Ulmann, L.; Mimouni, V.; Guéno, F.; Pineau-Vincent, F.; Meskini, N.; Tremblin, G. The role of Odontellaaurita, a marine diatom rich in EPA, as a dietary supplement in dyslipidemia, platelet function and oxidative stress in high-fat fed rats. Lipids Health Dis. 2012, 11, 147. [Google Scholar] [CrossRef]
- Goiris, K.; Muylaert, K.; Fraeye, I.; Foubert, I.; De Brabanter, J.; De Cooman, L. Antioxidant potential of microalgae in relation to their phenolic and carotenoid content. J. Appl. Phycol. 2012, 24, 1477–1486. [Google Scholar] [CrossRef]
- Nuzzo, G.; Gallo, C.; d’Ippolito, G.; Cutignano, A.; Sardo, A.; Fontana, A. Composition and quantitation of microalgal lipids by ERETIC 1H NMR method. Mar. Drugs. 2013, 11, 3742–3753. [Google Scholar] [CrossRef]
- Hannon, M.; Gimpel, J.; Tran, M.; Rasala, B.; Mayfield, S. Biofuels from algae: Challenges and potential. Biofuels 2010, 1, 763–784. [Google Scholar] [CrossRef]
- Elliott, L.G.; Feehan, C.; Laurens, L.M.; Pienkos, P.T.; Darzins, A.; Posewitz, M.C. Establishment of a bioenergy-focused microalgal culture collection. Algal Res. 2012, 1, 102–113. [Google Scholar] [CrossRef]
- Abdelaziz, A.E.M.; Leite, G.B.; Belhaj, M.A.; Hallenbeck, P.C. Screening microalgae native to Quebec for wastewater treatment and biodiesel production. Bioresour. Technol. 2014, 157, 140–148. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Escapa, I.F.; Jäger, C.; Puchalka, J.; Lam, C.M.C.; Schomburg, D.; Prieto, M.A.; dos Santos, V.A.M. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single-and multiple-nutrient-limited growth: Highlights from a multi-level omics approach. Microb. Cell Factor. 2012, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Beetul, K.; Gopeechund, A.; Kaullysing, D.; Mattan-Moorgawa, S.; Puchooa, D.; Bhagooli, R. Challenges and Opportunities in the Present Era of Marine Algal Applications; InTechOpen: London, UK, 2016; pp. 237–276. [Google Scholar]
- Judd, S.; van den Broeke, L.J.P.; Shurair, M.; Kuti, Y.; Znad, H. Algal remediation of CO2 and nutrient discharges: A review. Water Res. 2015, 87, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.M.; Ismail, A.F.; Mustafa, A.; Matsuura, T.; Soga, T.; Nagata, K.; Asaka, T. Qualitative and quantitative analysis of intercalated and exfoliated silicate layers in asymmetric polyethersulfone/cloisite15A® mixed matrix membrane for CO2/CH4 separation. Chem. Eng. J. 2015, 268, 371–383. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef] [PubMed]
- Ragonese, C.; Tedone, L.; Beccaria, M.; Torre, G.; Cichello, F.; Cacciola, F.; Dugo, P.; Mondello, L. Characterisation of lipid fraction of marine macroalgae by means of chromatography techniques coupled to mass spectrometry. Food Chem. 2014, 145, 932–940. [Google Scholar] [CrossRef]
- He, H.; Rodgers, R.P.; Marshall, A.G.; Hsu, C.S. Algae polar lipids characterized by online liquid chromatography coupled with hybrid linear quadrupole ion trap/fourier transform ion cyclotron resonance mass spectrometry. Energy Fuels. 2011, 25, 4770–4775. [Google Scholar] [CrossRef]
- Rigano, F.; Oteri, M.; Russo, M.; Dugo, P.; Mondello, L. Proposal of a Linear Retention Index System for Improving Identification Reliability of Triacylglycerol Profiles in Lipid Samples by Liquid Chromatography Methods. Anal. Chem. 2018, 90, 3313–3320. [Google Scholar] [CrossRef] [PubMed]
- Beccaria, M.; Inferrera, V.; Rigano, F.; Gorynski, K.; Purcaro, G.; Pawliszyn, J.; Dugo, P.; Mondello, L. Highly informative multiclass profiling of lipids by ultra-high performance liquid chromatography—Low resolution (quadrupole) mass spectrometry by using electrospray ionization and atmospheric pressure chemical ionization interfaces. J. Chromatogr. A 2017, 1509, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Morowvat, M.H.; Ghasemi, Y. Cell Growth, Lipid Production and Productivity in Photosynthetic Micro-alga Chlorella vulgaris under Different Nitrogen Concentrations and Culture Media Replacement. RecentPat. Food Nutr. Agric. 2018, 9, 142–151. [Google Scholar]
- Brahamsha, B. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 1996, 62, 1747–1751. [Google Scholar] [CrossRef]
- Singh, P.; Gupta, S.K.; Guldhe, A.; Rawat, I.; Bux, F. Microalgae Isolation and Basic Culturing Techniques. In Handbook of Marine Microalgae; Elsevier: Amsterdam, The Netherlands, 2015; pp. 43–54. ISBN 978-0-12-800776-1. [Google Scholar]
- Andersen, R.A. (Ed.) Algal Culturing Techniques; Academic Press: Cambridge, UK, 2005; ISBN 978-0-12-088426-1. [Google Scholar]
- Tischer, R.G. Pure Culture of Anabaena flos-aquae A-37. Nature 1965, 205, 419–420. [Google Scholar] [CrossRef]
- Izadpanah, M.; Gheshlaghi, R.; Mahdavi, M.A.; Elkamel, A. Effect of light spectrum on isolation of microalgae from urban wastewater and growth characteristics of subsequent cultivation of the isolated species. Algal Res. 2018, 29, 154–158. [Google Scholar] [CrossRef]
- Sakata, T.; Yoshikawa, T.; Maeda, K.; del Castillo, C.S.; Dureza, L.A. Identification of microalgae isolated from green water of tilapia culture ponds in the Philippines. Mem. Fac. Fish. Kagoshima Univ. 2005, 54, 35–44. [Google Scholar]
- Díez, B.; Pedrós-Alió, C.; Marsh, T.L.; Massana, R. Application of Denaturing Gradient Gel Electrophoresis (DGGE) To Study the Diversity of Marine Picoeukaryotic Assemblages and Comparison of DGGE with Other Molecular Techniques. Appl. Environ. Microbiol. 2001, 67, 2942–2951. [Google Scholar] [CrossRef]
- Rodríguez-Ferri, E.; Badiola-Díez, J.J.; Cepeda-Sáez, A.; Domínguez-Rodríguez, L.; Otero-Carballeira, A.; Zurera-Cosano, G. Grupo de trabajo. Informe del ComitéCientífico de la Agencia Española de SeguridadAlimentaria y Nutrición (AESAN) sobre la evisceración de loslagomorfos. RevistaComitéCientífico AESAN 2009, 9, 31–38. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Li, Y.; Horsman, M.; Wang, B.; Wu, N.; Lan, C.Q. Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochlorisoleoabundans. Appl. Microbiol. Biotechnol. 2008, 81, 629–636. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 1-119-11347-4. [Google Scholar]
- Egge, J.K. Are diatoms poor competitors at low phosphate concentrations? J. Mar. Syst. 1998, 16, 191–198. [Google Scholar] [CrossRef]
- Egge, J.K.; Aksnes, D.L. Silicate as regulating nutrient in phytoplankton competition. Mar. Ecol. Prog. Ser. Oldendorf. 1992, 83, 281–289. [Google Scholar] [CrossRef]
- Ahlgren, G. Temperature Functions in Biology and Their Application to Algal Growth Constants. Oikos 1987, 49, 177. [Google Scholar] [CrossRef]
- Thompson, P.A.; Guo, M.; Harrison, P.J.; Whyte, J.N.C. Effects of variation in temperature. ii. on the fatty acid composition of eight species of marine phytoplankton. J. Phycol. 1992, 28, 488–497. [Google Scholar] [CrossRef]
- Stirk, W.A.; Bálint, P.; Tarkowská, D.; Strnad, M.; van Staden, J.; Ördög, V. Endogenous brassinosteroids in microalgae exposed to salt and low temperature stress. Eur. J. Phycol. 2018, 53, 273–279. [Google Scholar] [CrossRef]
- Kang, E.J.; Kim, K.Y. Effects of future climate conditions on photosynthesis and biochemical component of Ulva pertusa (Chlorophyta). Algae 2016, 31, 49–59. [Google Scholar] [CrossRef]
- El Arroussi, H.; Benhima, R.; El Mernissi, N.; Bouhfid, R.; Tilsaghani, C.; Bennis, I.; Wahby, I. Screening of marine microalgae strains from Moroccan coasts for biodiesel production. Renew. Energy. 2017, 113, 1515–1522. [Google Scholar] [CrossRef]
- Griffiths, M.J.; Harrison, S.T. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J. Appl. Phycol. 2009, 21, 493–507. [Google Scholar] [CrossRef]
- Rodolfi, L.; ChiniZittelli, G.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef]
- Haixing, C.; Qian, F.; Yun, H.; Ao, X.; Qiang, L.; Xun, Z. Improvement of microalgae lipid productivity and quality in an ion-exchange-membrane photobioreactor using real municipal wastewater. Int. J. Agric. Biol. Eng. 2017, 10, 97–106. [Google Scholar]
- Illman, A.M.; Scragg, A.H.; Shales, S.W. Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb. Technol. 2000, 27, 631–635. [Google Scholar] [CrossRef]
- MacDougall, K.M.; McNichol, J.; McGinn, P.J.; O’Leary, S.J.; Melanson, J.E. Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography–high-resolution mass spectrometry. Anal. Bioanal. Chem. 2011, 401, 2609–2616. [Google Scholar] [CrossRef]
- Alves, E.; Domingues, M.R.M.; Domingues, P. Polar lipids from olives and olive oil: A review on their identification, significance and potential biotechnological applications. Foods 2018, 7, 109. [Google Scholar] [CrossRef]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgaltriacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, Y.; Wu, N.; Lan, C.Q. CO2biomitigation using microalgae. Appl. Microbiol. Biotechnol. 2008, 79, 707–718. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]


| Primer | Molecular Marker | Sequence | Annealing T° | Reference |
|---|---|---|---|---|
| 18SF | 18S rDNA | AACCTGGTTGATYCTGCCAG | 56 °C, 60 °C | [32] |
| Euk516r | 18S rDNA | ACCAGACTTGCCCTCC | 56 °C, 60 °C | [33] |
| Tetra_rbcL_F | rbcl | GKACTTGGACAACTGTATGGACKGATGGT | 56 °C | IFAPA |
| Tetra_rbcL_R | rbcL | GRTCTTTTTCWACRTAAGCATCACGCATTA | 56 °C | IFAPA |
| One-Factor ANOVA | |||||
|---|---|---|---|---|---|
| Parameter | Source | df | Sum of Squares | F | P |
| Silicate | Inter-group | 1 | 35.85 | 9.07 | 0.004 * |
| Intra-group | 52 | 205.48 | |||
| Total | 53 | 241.33 | |||
| Species | Weight of dry biomass (mg/400 mL) | Concentration of biomass (mg/L) | Productivity of biomass (mg/L/day) | Total lipids (mg/400mL) | Lipids (%) | Lipid productivity (mg/L/day) |
|---|---|---|---|---|---|---|
| P. tricornutum | 146.82 ± 1.1 b | 367.05 ± 0.15 b | 24.47 ± 0.15 b | 27.85 ± 0.1 b | 47.43 ± 0.68 d | 4.64 ± 0.1 b |
| T. suecica | 75.25 ± 0.2 a | 188.13 ± 0.9 a | 12.54 ± 0.03 a | 5.50 ± 0.04 a | 18.28 ± 0.7 a | 0.92 ± 0.02 a |
| N. gaditana | 582.53 ± 0.15 c | 1456.33 ± 1.43 c | 97.09 ± 0.68 c | 33.29 ± 0.8 c | 14.28 ± 0.32 b | 5.55 ± 0.01 b |
| Nannochloris sp., | 581.52 ± 1.4 c | 1453.8 ± 1.3 c | 96.92 ± 0.72 c | 95.58 ± 0.81 d | 41.08 ± 0.26 c | 15.93 ± 0.9 c |
| Peak | Compounds | [M+H]+ | [M+H]− | N. gaditana | T. suecica | P. tricornutum | Nannochloris sp., |
|---|---|---|---|---|---|---|---|
| 1 | MG 20:0 | 369.5 | + | - | - | - | |
| 2 | MG 16:0 | 313.2 | + | - | - | - | |
| 3 | DG (36:4) | 599.5 | + | + | - | - | |
| 4 | PG (34:3) | 762.3 | - | - | + | - | |
| 5 | DG (34:2) | 617.3 | + | - | - | - | |
| 6 | SQDG (32:1) | 549.5 | 791.5 | - | - | + | - |
| 7 | DG (36:2) | 603.4 | - | + | - | - | |
| 8 | DG (32:1) | 549.5 | - | - | + | - | |
| 9 | Neoxanthin | 601.7 | + | - | - | - | |
| 10 | DG (32:0) | 551.4 | - | - | - | + | |
| 11 | PG (34:4) | 741.4 | - | + | - | + | |
| 12 | PG (34:3) | 762.3 | - | + | - | - | |
| 13 | MGDG (34:5) | 766.5 | + | - | - | - | |
| 14 | DG (32:5) | 583.6 | - | + | - | - | |
| 15 | TG (ArArO/SArEp) | 928.5 | 909.7 | + | - | - | - |
| 16 | PE (34:3) | 762.4 | + | - | - | - | |
| 17 | DG (32:4) | 585.5 | - | - | - | + | |
| 18 | DGDG (36:6) | 954.6 | 935.4 | + | - | - | - |
| 19 | SQDG (34:4) | 813.6 | - | - | + | - | |
| 20 | DGDG (34:4) | 930.6 | - | - | - | + | |
| 21 | DGDG (36:6) | 954.6 | 935.4 | - | - | + | - |
| 22 | Antheraxanthin | 585.9 | - | + | - | - | |
| 23 | PC (36:1) | 788.5 | 812.7 | + | - | + | - |
| 24 | SQDG (34:0) | 821.5 | + | - | - | - | |
| 25 | PI (36:4) | 857.5 | + | - | - | - | |
| 26 | trans-Lutein | 569.9 | - | - | - | + | |
| 27 | TG (C20:3LL) | 907.6 | + | - | - | - | |
| 28 | SQDG (34:3) | 815.5 | - | - | + | - | |
| 29 | TG (20:3LL) | 907.5 | - | - | + | - | |
| 30 | PG (34:1) | 766.3 | - | + | - | - | |
| 31 | PC (36:3) | 784.5 | + | + | - | - | |
| 32 | MGDG (36:5) | 794.5 | + | - | - | - | |
| 33 | MGDG (34:6) | 764.5 | + | - | - | - | |
| 34 | MGDG (36:6) | 792.5 | - | - | + | - | |
| 35 | PG (36:5) | 786.5 | - | - | - | + | |
| 36 | PG (34:1) | 766.3 | - | - | + | - | |
| 37 | TG (SOAr) | 908.6 | + | - | - | - | |
| 38 | PE (39:6) | 776.5 | + | - | - | - | |
| 39 | PE (38:3) | 768.5 | + | - | - | - | |
| 40 | PG (36:4) | 788.4 | - | - | - | + | |
| 41 | DGDG (34:2) | 934.5 | 915.6 | - | + | - | + |
| 42 | PC (38:3) | 814.5 | + | - | - | - | |
| 43 | PG (34:0) | 768.5 | - | - | + | - | |
| 44 | SQDG (34:1) | 819.4 | - | - | + | - | |
| 45 | PC (38:5) | 808.8 | - | + | - | - | |
| 46 | DGDG (36:1) | 964.7 | - | + | - | - | |
| 47 | hydroxychlorophyllide b | 645.1 | - | + | - | - | |
| 48 | PC (33:2) | 744.5 | + | - | - | - | |
| 49 | PE (38:5) | 764.5 | + | - | - | - | |
| 50 | b-carotene | 537.9 | + | - | - | - | |
| 51 | TG (LnLnPo) | 849.4 | - | - | - | + | |
| 52 | MGDG (34:2) | 772.5 | 753.6 | - | - | - | + |
| 53 | TG (LnGG/OGLn/SGAr) | 936.8 | - | + | - | - | |
| 54 | PC (33:1)–PC (O-16:0/18:1) | 746.5 | - | - | + | - | |
| 55 | TG (GEpD) | 981.5 | - | + | - | - | |
| 56 | PI (40:8) | 905.5 | - | + | - | - | |
| 57 | TG (GLL) | 909.5 | - | + | - | - | |
| 58 | TG (ArArAr) | 950.5 | - | + | - | - | |
| 59 | PC (32:2) | 730.5 | + | - | - | - | |
| 60 | TG (OLC18:4) | 877.5 | - | - | + | - | |
| 61 | TG (C17:0LAr/C17:0OEp–OOP/SLnP) | 893.6–859.8 | + | - | - | - | |
| 62 | TG (EpC18:4C18:4) | 893.4 | - | + | - | - | |
| 63 | TG (GLL/LLnA/OLnG) | 909.5 | - | - | + | - | |
| 64 | TG (EpEpL) | 923.5 | - | - | + | - | |
| 65 | PI (38:1) | 892.6 | - | - | + | - | |
| 66 | TG (SSS) | 891.4 | - | - | + | - | |
| 67 | Anhydroeschscholtzxanthin | 529.3 | - | + | - | + | |
| 68 | TG (C18:4C18:4Ep) | 893.6 | + | - | - | + | |
| 69 | TG (OOP/PPoG/PoOL) | 925.6 | - | + | - | - | |
| 70 | PC (44:5) | 890.3 | - | - | + | - | |
| 71 | TG (EpC18:4C18:4) | 893.6 | - | - | + | - | |
| 72 | TG (ArArLn/OEpEp/ArEpL) | 925.6 | - | + | - | - | |
| 73 | TG (SOO/SSL/PLA) | 887.5 | + | - | - | - | |
| 74 | TG (MMDh) | 823.5 | - | + | - | - | |
| 75 | TG (SOO/SSL/PLA) | 887.5 | - | - | + | - | |
| 76 | TG (OOMo/LnLn18:4) | 871.4 | + | - | - | - | |
| 77 | Pheophytin a (C55H74N405) | 871.4 | 870.6 | + | - | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haoujar, I.; Cacciola, F.; Manchado, M.; Abrini, J.; Haoujar, M.; Chebbaki, K.; Oteri, M.; Rigano, F.; Mangraviti, D.; Mondello, L.; et al. Isolation of Microalgae from Mediterranean Seawater and Production of Lipids in the Cultivated Species. Foods 2020, 9, 1601. https://doi.org/10.3390/foods9111601
Haoujar I, Cacciola F, Manchado M, Abrini J, Haoujar M, Chebbaki K, Oteri M, Rigano F, Mangraviti D, Mondello L, et al. Isolation of Microalgae from Mediterranean Seawater and Production of Lipids in the Cultivated Species. Foods. 2020; 9(11):1601. https://doi.org/10.3390/foods9111601
Chicago/Turabian StyleHaoujar, Imane, Francesco Cacciola, Manuel Manchado, Jamal Abrini, Mohammed Haoujar, Kamal Chebbaki, Marianna Oteri, Francesca Rigano, Domenica Mangraviti, Luigi Mondello, and et al. 2020. "Isolation of Microalgae from Mediterranean Seawater and Production of Lipids in the Cultivated Species" Foods 9, no. 11: 1601. https://doi.org/10.3390/foods9111601
APA StyleHaoujar, I., Cacciola, F., Manchado, M., Abrini, J., Haoujar, M., Chebbaki, K., Oteri, M., Rigano, F., Mangraviti, D., Mondello, L., Essafi, A., Chairi, H., & Skali Senhaji, N. (2020). Isolation of Microalgae from Mediterranean Seawater and Production of Lipids in the Cultivated Species. Foods, 9(11), 1601. https://doi.org/10.3390/foods9111601








