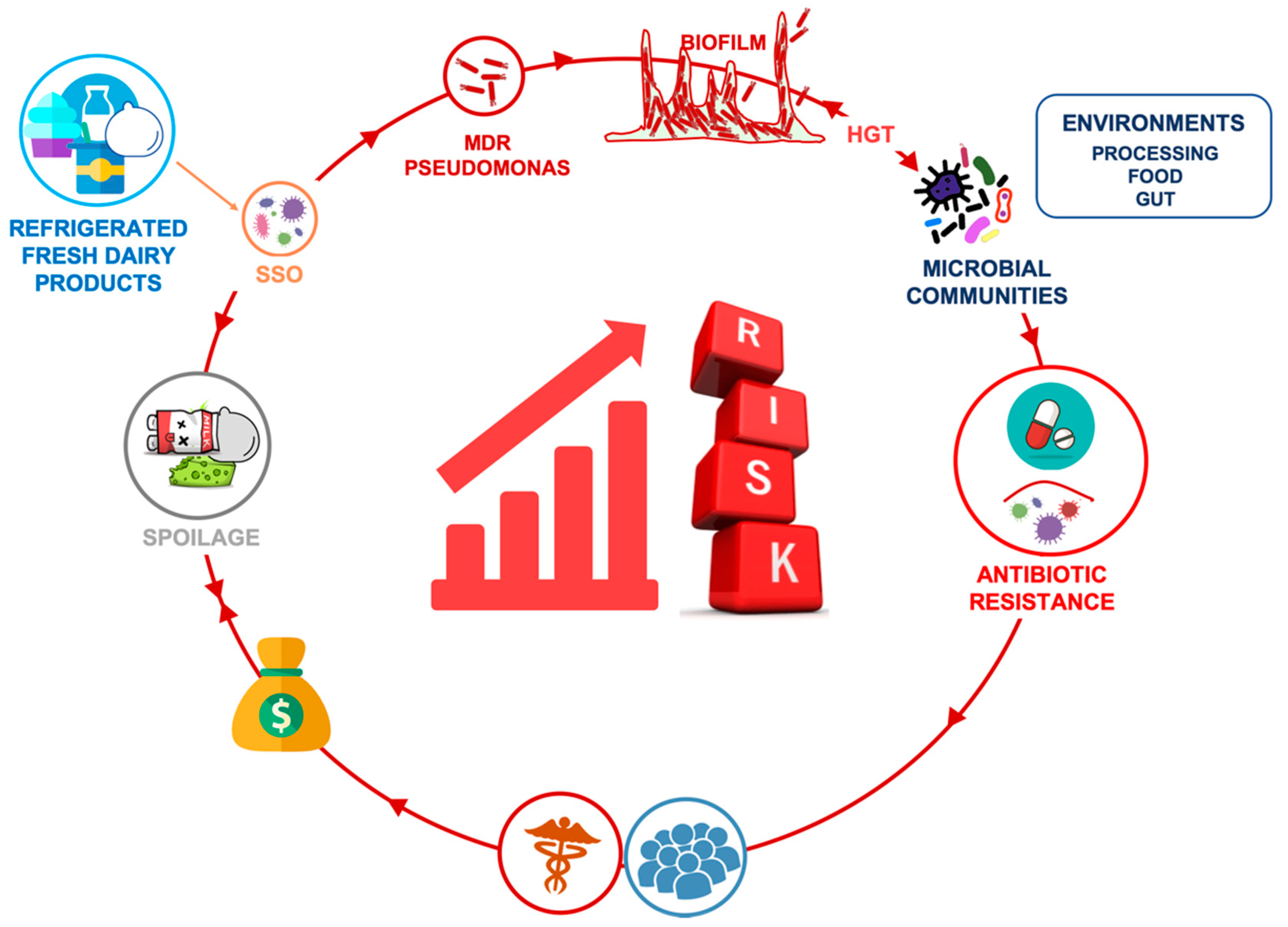
Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies
Abstract
1. Introduction
2. Pseudomonas spp. Genus and Species Occurrence in Dairy Products
3. Antibiotic Resistance in Pseudomonas spp. Spoiler: Mechanisms and Influencing Factors in Dairy Sector
4. Sequencing-Based Tools and Database for AR Prediction
5. Strategies to Control the Spread of Antibiotic Resistant Pseudomonas spp. in Dairy Sector
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vaishali, G.M.; Geetha, R.V. The superbug threat. Res. J. Pharm. Technol. 2015, 8, 343. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2017, 42, fux053. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Niederman, M.S.; Chastre, J.; Ewig, S.; Fernandez-Vandellos, P.; Hanberger, H.; Paiva, J.A. Summary of the international clinical guidelines for the management of hospital-acquired and ventilator-acquired pneumonia. ERJ Open Res. 2018, 4, 00028–02018. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. The cost of antimicrobial resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014; WHO: Geneva, Switzerland, 2014; ISBN 978-92-4-156474-8. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 26 August 2019).
- ECDC, European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe—Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. ECDC: Stockholm. 2018. Available online: https://ecdc.europa.eu/sites/portal/files/documents/EARS-Net-report-2017-update-jan-2019.pdf (accessed on 26 August 2019).
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States; Atlanta: CDC. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (accessed on 26 August 2019).
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Genet. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Hall, J.P.J.; Brockhurst, M.A.; Harrison, E. Sampling the mobile gene pool: Innovation via horizontal gene transfer in bacteria. Philos. Trans. R. Soc. B Boil. Sci. 2017, 372, 20160424. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 305. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, Present, and Future. Microbiol. Mol. Boil. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Anju, C.; Biswas, L.; Kumar, V.A.; Mohan, C.G.; Biswas, R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int. J. Med. Microbiol. 2016, 306, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Hattori, K.; Inoue, A.; Ishii, T.; Yumoto, T.; Tsukahara, K.; Nakao, A.; Ishihara, S.; Nakayama, S. Bacteremia or pseudobacteremia? Review of Pseudomonas fluorescens infections. World J. Emerg. Med. 2017, 8, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.L.; Singh, O.V. Foodborne Pathogens and Their Apparent Linkage with Antibiotic Resistance. In Foodborne Pathogens and Antibiotic Resistance; Wiley: Hoboken, NJ, USA, 2017; pp. 247–274. [Google Scholar]
- Mulet, M.; Gomila, M.; Scotta, C.; Sánchez, D.; Lalucat, J.; García-Valdés, E. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst. Appl. Microbiol. 2012, 35, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [PubMed]
- Baruzzi, F.; Lagonigro, R.; Quintieri, L.; Morea, M.; Caputo, L. Occurrence of non-lactic acid bacteria populations involved in protein hydrolysis of cold-stored high moisture Mozzarella cheese. Food Microbiol. 2012, 30, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Zühlke, D.; Fanelli, F.; Caputo, L.; Liuzzi, V.C.; Logrieco, A.F.; Hirschfeld, C.; Becher, D.; Riedel, K.; Laura, Q. Proteomic analysis of the food spoiler Pseudomonas fluorescens ITEM 17298 reveals the antibiofilm activity of the pepsin-digested bovine lactoferrin. Food Microbiol. 2019, 82, 177–193. [Google Scholar] [CrossRef]
- Raposo, A.; Pérez, E.; de Faria, C.T.; Ferrús, M.A.; Carrascosa, C. Food Spoilage by Pseudomonas spp.—An overview. In Foodborne Pathogens and Antibiotic Resistance; Singh, O.V., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 41–58. [Google Scholar]
- Stellato, G.; Utter, D.R.; Voorhis, A.; De Angelis, M.; Eren, A.M.; Ercolini, D. A Few Pseudomonas Oligotypes Dominate in the Meat and Dairy Processing Environment. Front. Microbiol. 2017, 8, 148. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2005, 50, 1–26. [Google Scholar]
- Carminati, D.; Bonvini, B.; Rossetti, L.; Zago, M.; Tidona, F.; Giraffa, G. Investigation on the presence of blue pigment-producing Pseudomonas strains along a production line of fresh mozzarella cheese. Food Control 2019, 100, 321–328. [Google Scholar] [CrossRef]
- Lerma, L.L.; Benomar, N.; Muñoz, M.D.C.C.; Gálvez, A.; Abriouel, H. Antibiotic Multiresistance Analysis of Mesophilic and Psychrotrophic Pseudomonas spp. Isolated from Goat and Lamb Slaughterhouse Surfaces throughout the Meat Production Process. Appl. Environ. Microbiol. 2014, 80, 6792–6806. [Google Scholar] [CrossRef] [PubMed]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Heal. 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Møretrø, T.; Langsrud, S. Residential Bacteria on Surfaces in the Food Industry and Their Implications for Food Safety and Quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef]
- Teh, K.H.; Flint, S.; Palmer, J.; Andrewes, P.; Bremer, P.; Lindsay, D. Biofilm—An unrecognised source of spoilage enzymes in dairy products? Int. Dairy J. 2014, 34, 32–40. [Google Scholar] [CrossRef]
- Palleroni, N.J. Pseudomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [PubMed]
- Prasad, G.; Minakshi, P.; Virdi, J.S. Normal microbial flora of human body and host parasite relationship. In Immunology and Medical Microbiology; National Science Digital Library: Boulder, CO, USA, 2007; pp. 1–23. [Google Scholar]
- Scales, B.S.; Dickson, R.P.; Lipuma, J.J.; Huffnagle, G.B. Microbiology, Genomics, and Clinical Significance of the Pseudomonas fluorescens Species Complex, an Unappreciated Colonizer of Humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Schroth, M.N.; Hildebrand, D.C.; Panopoulos, N. Phytopathogenic Pseudomonads and Related Plant-Associated Pseudomonads; Springer Science and Business Media LLC: Berlin, Germany, 2006; pp. 714–740. [Google Scholar]
- Ercolini, D.; Russo, F.; Torrieri, E.; Masi, P.; Villani, F. Changes in the Spoilage-Related Microbiota of Beef during Refrigerated Storage under Different Packaging Conditions. Appl. Environ. Microbiol. 2006, 72, 4663–4671. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Eyi, A.; Ozdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef] [PubMed]
- Decimo, M.; Cabeza, M.C.; Ordóñez, J.A.; De Noni, I.; Brasca, M. Volatile organic compounds associated with milk spoilage by psychrotrophic bacteria. Int. J. Dairy Technol. 2018, 71, 593–600. [Google Scholar] [CrossRef]
- Del Olmo, A.; Calzada, J.; Nuñez, M. The blue discoloration of fresh cheeses: A worldwide defect associated to specific contamination by Pseudomonas fluorescens. Food Control. 2018, 86, 359–366. [Google Scholar] [CrossRef]
- Carrascosa, C.; Millán, R.; Jaber, J.R.; Lupiola, P.; Del Rosario-Quintana, C.; Mauricio, C.; Sanjuán, E. Blue pigment in fresh cheese produced by Pseudomonas fluorescens. Food Control 2015, 54, 95–102. [Google Scholar] [CrossRef]
- Caputo, L.; Quintieri, L.; Bianchi, D.M.; Decastelli, L.; Monaci, L.; Visconti, A.; Baruzzi, F. Pepsin-digested bovine lactoferrin prevents Mozzarella cheese blue discoloration caused by Pseudomonas fluorescens. Food Microbiol. 2015, 46, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, C.; Soncini, G.; Milesi, S.; Cocolin, L.; Iacumin, L. Colorazioni anomale e rigonfiamento di formaggi fusi e mozzarelle. Ind. Aliment. 2006, 45, 276–281. [Google Scholar]
- Riva, M.; Franzetti, L.; Galli, A. Microbiological Quality and Shelf Life Modeling of Ready-to-Eat Cicorino. J. Food Prot. 2001, 64, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Federico, B.; Pinto, L.; Quintieri, L.; Carito, A.; Calabrese, N.; Caputo, L. Efficacy of lactoferricin B in controlling ready-to-eat vegetable spoilage caused by Pseudomonas spp. Int. J. Food Microbiol. 2015, 215, 179–186. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, V.; Coorevits, A.; Van Hoorde, K.; Messens, W.; Van Landschoot, A.; De Vos, P.; Heyndrickx, M. Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 2011, 77, 460–470. [Google Scholar] [CrossRef]
- Orellana-Saez, M.; Pacheco, N.; Costa, J.I.; Mendez, K.N.; Miossec, M.J.; Meneses, C.; Castro-Nallar, E.; Marcoleta, A.E.; Poblete-Castro, I. In-Depth Genomic and Phenotypic Characterization of the Antarctic Psychrotolerant Strain Pseudomonas sp. MPC6 Reveals Unique Metabolic Features, Plasticity, and Biotechnological Potential. Front. Microbiol. 2019, 10, 10. [Google Scholar] [CrossRef]
- Liu, X.; Xu, J.; Zhu, J.; Du, P.; Sun, A. Combined Transcriptome and Proteome Analysis of RpoS Regulon Reveals Its Role in Spoilage Potential of Pseudomonas fluorescens. Front. Microbiol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Guidone, A.; Zotta, T.; Matera, A.; Ricciardi, A.; De Filippis, F.; Ercolini, D.; Parente, E. The microbiota of high-moisture mozzarella cheese produced with different acidification methods. Int. J. Food Microbiol. 2016, 216, 9–17. [Google Scholar] [CrossRef]
- Brasca, M.; Decimo, M.; Morandi, S.; Machado, S.G.; Bagliniére, F.; Vanetti, M.C.D. Psychrotrophic bacteria. In Microbiology in Dairy Processing: Challenges and Opportunities; Poltronieri, P., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2018; Chapter 37; pp. 37–61. [Google Scholar]
- Langsrud, S.; Sundheim, G.; Borgmann-Strahsen, R.; Borgmann-Strahsen, R. Intrinsic and acquired resistance to quaternary ammonium compounds in food-related Pseudomonas spp. J. Appl. Microbiol. 2003, 95, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Palleroni, N.J.; Ballard, R.W.; Ralston, E.; Doudoroff, M. Deoxyribonucleic Acid Homologies Among Some Pseudomonas Species. J. Bacteriol. 1972, 110, 1–11. [Google Scholar] [PubMed]
- Meyer, J.M.; Geoffroy, V.K.; Baida, N.; Gardan, L.; Izard, D.; Lemanceau, P.; Achouak, W.; Palleroni, N.J. Siderophores typing: A powerful tool for the identification of fluorescent and non-fluorescent Pseudomonads. Appl. Environ. Microbiol. 2002, 68, 2745–2753. [Google Scholar] [CrossRef] [PubMed]
- Kidd, T.J.; Grimwood, K.; Ramsay, K.A.; Rainey, P.B.; Bell, S.C. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 2011, 49, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, L.; Scarpellini, M. Characterisation of Pseudomonas spp. isolated from foods. Ann. Microbiol. 2007, 57, 39–47. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Genus, I. Pseudomonas Migula 1984, 237AL. In Bergey’s Manual of Systematic Bacteriology; Krieg, N.R., Holt. J., G., Eds.; Williams & Wilkins: Baltimore, MA, USA, 1984; Volume 1, pp. 141–199. [Google Scholar]
- Peix, Á.; Ramírez-Bahena, M.-H.; Velázquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef]
- Anzai, Y.; Kim, H.; Park, J.Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589. [Google Scholar] [CrossRef] [PubMed]
- Palleroni, N.J.; Genus, I. Pseudomonas. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: East Lansing, MI, USA, 2005; Volume 2, pp. 323–379. [Google Scholar]
- Yamamoto, S.; Harayama, S.; Arnold, D.L.; Jackson, R.W.; Kasai, H.; Vivian, A. Phylogeny of the genus Pseudomonas: Intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 2000, 146, 2385–2394. [Google Scholar] [CrossRef]
- Hilario, E.; Buckley, T.R.; Young, J.M. Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Leeuwenhoek 2004, 86, 51–64. [Google Scholar] [CrossRef]
- Liao, C.-H.; McCallus, D.E. Biochemical and Genetic Characterization of an Extracellular Protease from Pseudomonas fluorescens CY091. Appl. Environ. Microbiol. 1998, 64, 914–921. [Google Scholar]
- Caldera, L.; Franzetti, L.; Van Coillie, E.; De Vos, P.; Stragier, P.; De Block, J.; Heyndrickx, M. Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol. 2016, 54, 142–153. [Google Scholar] [CrossRef]
- Decimo, M.; Morandi, S.; Silvetti, T.; Brasca, M. Characterization of Gram-Negative Psychrotrophic Bacteria isolated from Italian Bulk Tank Milk. J. Food Sci. 2014, 79, M2081–M2090. [Google Scholar] [CrossRef] [PubMed]
- Andreani, N.A.; Carraro, L.; Martino, M.E.; Fondi, M.; Fasolato, L.; Miotto, G.; Magro, M.; Vianello, F.; Cardazzo, B. A genomic and transcriptomic approach to investigate the blue pigment phenotype in Pseudomonas fluorescens. Int. J. Food Microbiol. 2015, 213, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Nogarol, C.; Acutis, P.L.; Bianchi, D.M.; Maurella, C.; Peletto, S.; Gallina, S.; Adriano, D.; Zuccon, F.; Borrello, S.; Caramelli, M.; et al. Molecular Characterization of Pseudomonas fluorescens Isolates Involved in the Italian “Blue Mozzarella” Event. J. Food Prot. 2013, 76, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Dogan, B.; Boor, K.J. Genetic Diversity and Spoilage Potentials among Pseudomonas spp. Isolated from Fluid Milk Products and Dairy Processing Plants. Appl. Environ. Microbiol. 2003, 69, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martin, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, 0150183. [Google Scholar] [CrossRef]
- Raab, V.; Bruckner, S.; Beierle, E.; Kampmann, Y.; Petersen, B.; Kreyenschmidt, J. Generic model for the prediction of remaining shelf life in support of cold chain management in pork and poultry supply chains. J. Chain Netw. Sci. 2008, 8, 59–73. [Google Scholar] [CrossRef]
- Palleroni, N.J. The Pseudomonas story. Environ. Microbiol. 2010, 12, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Tran, Q.T.; Nawaz, M.S.; Deck, J.; Nguyen, K.T.; Cerniglia, C.E. Plasmid-mediated quinolone resistance in Pseudomonas putida isolates from imported shrimp. Appl. Environ. Microbiol. 2011, 77, 1885–1887. [Google Scholar] [CrossRef]
- Andreani, N.A.; Fasolato, L. Pseudomonas and related genera. In The Microbiological Quality of Food; Bevilacqua, A., Corbo, M.R., Sinigaglia, M., Eds.; Woodhead Publishing: Sawston, UK, 2017; pp. 25–59. [Google Scholar]
- Cocolin, L.; Mataragas, M.; Bourdichon, F.; Doulgeraki, A.; Pilet, M.-F.; Jagadeesan, B.; Rantsiou, K.; Phister, T. Next generation microbiological risk assessment meta-omics: The next need for integration. Int. J. Food Microbiol. 2018, 287, 10–17. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Baruch, M.Q.; Escobar, A. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Duru, I.C.; Laine, P.; Andreevskaya, M.; Paulin, L.; Kananen, S.; Tynkkynen, S.; Auvinen, P.; Smolander, O.-P. Metagenomic and metatranscriptomic analysis of the microbial community in Swiss-type Maasdam cheese during ripening. Int. J. Food Microbiol. 2018, 281, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, D.J.; Daly, D.; O’Sullivan, O.; Burdikova, Z.; Vana, R.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; McSweeney, P.L.H.; et al. Thermus and the Pink Discoloration Defect in Cheese. MSystems 2016, 1, e00023-16. [Google Scholar] [CrossRef] [PubMed]
- Bezanson, G.; MacInnis, R.; Pötter, G.; Hughes, T. Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int. J. Food Microbiol. 2008, 127, 37–42. [Google Scholar] [CrossRef] [PubMed]
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. The Mechanisms of Resistance to β-Lactam Antibiotics. In Handbook of Antimicrobial Resistance; Springer Science and Business Media LLC: Berlin, Germany, 2017; Volume 67, pp. 177–201. [Google Scholar]
- Walsh, C.C.; McIntosh, M.P.; Peleg, A.Y.; Kirkpatrick, C.M.; Bergen, P.J. In vitro pharmacodynamics of fosfomycin against clinical isolates of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015, 70, 3042–3050. [Google Scholar] [CrossRef]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Treviño, M.; Moldes, L.; Hernández, M.; Martínez-Lamas, L.; García-Riestra, C.; Regueiro, B.J.; Regueiro, B. Nosocomial infection by VIM-2 metallo- -lactamase-producing Pseudomonas putida. J. Med. Microbiol. 2010, 59, 853–855. [Google Scholar] [CrossRef]
- Koh, T.H.; Wang, G.C.Y.; Sng, L.-H. IMP-1 and a Novel Metallo-β-Lactamase, VIM-6, in Fluorescent Pseudomonads Isolated in Singapore. Antimicrob. Agents Chemother. 2004, 48, 2334–2336. [Google Scholar] [CrossRef]
- Wong, M.H.-Y.; Chan, E.W.C.; Chen, S. Isolation of carbapenem-resistant Pseudomonas spp. from food. J. Glob. Antimicrob. Resist. 2015, 3, 109–114. [Google Scholar] [CrossRef]
- Munsch-Alatossava, P.; Alatossava, T. Antibiotic resistance of raw-milk-associated psychrotrophic bacteria. Microbiol. Res. 2007, 162, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Decimo, M.; Silvetti, T.; Brasca, M. Antibiotic Resistance Patterns of Gram-Negative Psychrotrophic Bacteria from Bulk Tank Milk. J. Food Sci. 2016, 81, M944–M951. [Google Scholar] [CrossRef] [PubMed]
- Coton, M.; Delbès-Paus, C.; Irlinger, F.; Desmasures, N.; Le Flèche, A.; Stahl, V.; Montel, M.-C.; Coton, E. Diversity and assessment of potential risk factors of Gram-negative isolates associated with French cheeses. Food Microbiol. 2012, 29, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Joyeeta, S.B. Analysis of Enteric Pathogenicity of Multidrug-Resistant Pseudomonas spp. Isolated from Commercial UHT Milk. Ph.D. Thesis, Bachelor of Science in Biotechnology, Brac University, Dhaka, Bangladesh, September 2018. Available online: http://hdl.handle.net/10361/11064 (accessed on 26 August 2019).
- Brown, M.H.; Paulsen, I.T.; Skurray, R.A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 1999, 31, 394–395. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Livermore, D.M.; Nikaido, H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: Resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 1994, 38, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Adebusuyi, A.A.; Foght, J.M. An alternative physiological role for the EmhABC efflux pump in Pseudomonas fluorescens cLP6a. BMC Microbiol. 2011, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Terán, W.; Felipe, A.; Segura, A.; Rojas, A.; Ramos, J.-L.; Gallegos, M.-T. Antibiotic-Dependent Induction of Pseudomonas putida DOT-T1E TtgABC Efflux Pump Is Mediated by the Drug Binding Repressor TtgR. Antimicrob. Agents Chemother. 2003, 47, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Baysse, C.; O’Gara, F. Role of Membrane Structure during Stress Signalling and Adaptation in Pseudomonas. In Pseudomonas; Springer Science and Business Media LLC: Berlin, Germany, 2007; pp. 193–224. [Google Scholar]
- Cebrián, G.; Sagarzazu, N.; Pagán, R.; Condón, S.; Mañas, P. Resistance of Escherichia coli grown at different temperatures to various environmental stresses. J. Appl. Microbiol. 2008, 105, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Wheat, P.F.; Winstanley, T.G.; Spencer, R.C. Effect of temperature on antimicrobial susceptibilities of Pseudomonas maltophilia. J. Clin. Pathol. 1985, 38, 1055–1058. [Google Scholar] [CrossRef]
- Fanelli, F.; Liuzzi, V.C.; Quintieri, L.; Mulè, G.; Baruzzi, F.; Logrieco, A.F.; Caputo, L. Draft Genome Sequence of Pseudomonas fluorescens Strain ITEM 17298, Associated with Cheese Spoilage. Genome Announc. 2017, 5, 01141-17. [Google Scholar] [CrossRef]
- Licciardello, G.; Caruso, A.; Bella, P.; Gheleri, R.; Strano, C.P.; Anzalone, A.; Trantas, E.A.; Sarris, P.F.; Almeida, N.F.; Catara, V. The LuxR Regulators PcoR and RfiA co-regulate antimicrobial peptide and alginate production in Pseudomonas corrugata. Front. Microbiol. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Ocampo-Sosa, A.A.; Guzmán-Gómez, L.P.; Fernández-Martínez, M.; Román, E.; Rodríguez, C.; Marco, F.; Vila, J.; Martínez-Martínez, L. Isolation of VIM-2-Producing Pseudomonas monteilii Clinical Strains Disseminated in a Tertiary Hospital in Northern Spain. Antimicrob. Agents Chemother. 2015, 59, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, Z.; Zhang, G.; Mo, X.; Ding, X.; Xia, L.; Hu, S. A rifampicin-resistant (rpoB) mutation in Pseudomonas protegens Pf-5 strain leads to improved antifungal activity and elevated production of secondary metabolites. Res. Microbiol. 2016, 167, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.; Arca, P.; Suárez, J.E. Product of fosC, a gene from Pseudomonas syringae, mediates fosfomycin resistance by using ATP as cosubstrate. Antimicrob. Agents Chemother. 1995, 39, 1569–1573. [Google Scholar] [CrossRef]
- Sarris, P.F.; Trantas, E.A.; Baltrus, D.A.; Bull, C.T.; Wechter, W.P.; Yan, S.; Ververidis, F.; Almeida, N.F.; Jones, C.D.; Dangl, J.L.; et al. Comparative Genomics of Multiple Strains of Pseudomonas cannabina pv. alisalensis, a Potential Model Pathogen of Both Monocots and Dicots. PLoS ONE 2013, 8, e59366. [Google Scholar] [CrossRef]
- Montaña, S.; Lazzaro, T.; Uong, S.; Place, K.; Iriarte, A.; Ocampo, C.V.; Vay, C.; Ramírez, M.S. Genomics helps to decipher the resistance mechanisms present in a Pseudomonas chlororaphis strain recovered in an HIV patient. New Microbes New Infect. 2018, 25, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.; Schmid, M.; Gekenidis, M.-T.; Pelludat, C.; Frey, J.E.; Ahrens, C.H.; Drissner, D. Complete genome sequence of Pseudomonas citronellolis P3B5, a candidate for microbial phyllo-remediation of hydrocarbon-contaminated sites. Stand. Genom. Sci. 2016, 11, 75. [Google Scholar] [CrossRef]
- Meier, M.J.; Subasinghe, R.M.; Beaudette, L.A. Draft Genome Sequence of the Industrially Significant Bacterium Pseudomonas fluorescens ATCC 13525. Microbiol. Resour. Announc. 2018, 7, e01368-18. [Google Scholar] [CrossRef]
- Kenzaka, T.; Tani, K. Draft Genome Sequence of Carbapenem-Resistant Pseudomonas fluorescens Strain BWKM6, Isolated from Feces of Mareca penelope. Genome Announc. 2018, 6, e00186-18. [Google Scholar] [CrossRef]
- Girlich, D.; Poirel, L.; Nordmann, P. Novel ambler class A carbapenem-hydrolyzing beta-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2010, 54, 328–332. [Google Scholar] [CrossRef]
- Maravić, A.; Skočibušić, M.; Šamanić, I.; Puizina, J. Antibiotic susceptibility profiles and first report of TEM extended-spectrum β-lactamase in Pseudomonas fluorescens from coastal waters of the Kaštela Bay, Croatia. World J. Microbiol. Biotechnol. 2012, 28, 2039. [Google Scholar] [CrossRef] [PubMed]
- Hearn, E.M.; Dennis, J.J.; Gray, M.R.; Foght, J.M. Identification and Characterization of the emhABC Efflux System for Polycyclic Aromatic Hydrocarbons in Pseudomonas fluorescens cLP6a. J. Bacteriol. 2003, 185, 6233–6240. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wu, X.G.; Duan, H.M.; Zhang, L.Q. The resistance-nodulation-division efflux pump EmhABC influences the production of 2, 4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Microbiology 2010, 156, 39–48. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Strain-Level Diversity Analysis of Pseudomonas fragi after In Situ Pangenome Reconstruction Shows Distinctive Spoilage-Associated Metabolic Traits Clearly Selected by Different Storage Conditions. Appl. Environ. Microbiol. 2018, 85, e02212-18. [Google Scholar] [CrossRef]
- Yamamoto, M.; Matsumura, Y.; Gomi, R.; Matsuda, T.; Nakano, S.; Nagao, M.; Tanaka, M.; Ichiyama, S. Molecular Analysis of a blaIMP-1-Harboring Class 3 Integron in Multidrug-Resistant Pseudomonas fulva. Antimicrob. Agents Chemother. 2018, 62, e00701-18. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.; Park, G.-S.; Shin, J.-H. High quality draft genome sequence of the type strain of Pseudomonas lutea OK2T, a phosphate-solubilizing rhizospheric bacterium. Stand. Genom. Sci. 2016, 11, 51. [Google Scholar] [CrossRef]
- Doublet, B.; Robin, F.; Casin, I.; Fabre, L.; Le Fleche, A.; Bonnet, R.; Weill, F.X. Molecular and biochemical characterization of the natural chromosome-encoded class A beta-lactamase from Pseudomonas luteola. Antimicrob. Agents Chemother. 2010, 54, 45–51. [Google Scholar] [CrossRef]
- Giani, T.; Marchese, A.; Coppo, E.; Kroumova, V.; Rossolini, G.M. VIM-1-Producing Pseudomonas mosselii Isolates in Italy, Predating Known VIM-Producing Index Strains. Antimicrob. Agents Chemother. 2012, 56, 2216–2217. [Google Scholar] [CrossRef]
- Thaller, M.C.; Borgianni, L.; Di Lallo, G.; Chong, Y.; Lee, K.; Dajcs, J.; Stroman, D.; Rossolini, G.M. Metallo-beta-lactamase production by Pseudomonas otitidis: A species-related trait. Antimicrob. Agents Chemother. 2011, 55, 118–123. [Google Scholar] [CrossRef]
- Ibáñez, M.I.; Cabello, P.; Luque-Almagro, V.M.; Sáez, L.P.; Olaya, A.; De Medina, V.S.; De Castro, M.D.L.; Moreno-Vivián, C.; Roldán, M.D. Quantitative proteomic analysis of Pseudomonas pseudoalcaligenes CECT5344 in response to industrial cyanide-containing wastewaters using Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS). PLoS ONE 2017, 12, e0172908. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Vollmers, J.; Mäusezahl, I.; Kaster, A.-K.; Müller, J.A. Detection of the carbapenemase gene blaVIM-5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Sci. Rep. 2018, 8, 15116. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, L.; Peters, B.M.; Liu, J.; Li, L.; Li, B.; Xu, Z.; Shirtliff, M.E. Complete Sequence of a Novel Multidrug-Resistant Pseudomonas putida Strain Carrying Two Copies of qnrVC6. Microb. Drug Resist. 2019, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Yoon, E.-J.; Song, W.; Bin Seo, Y.; Shin, S.; Park, M.-J.; Jeong, S.H.; Lee, K. Molecular Characterization of Pseudomonas putida Group Isolates Carrying blaVIM-2 Disseminated in a University Hospital in Korea. Microb. Drug Resist. 2018, 24, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Peter, S.; Oberhettinger, P.; Schuele, L.; Dinkelacker, A.; Vogel, W.; Dörfel, D.; Bezdan, D.; Ossowski, S.; Marschal, M.; Liese, J.; et al. Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa. BMC Genom. 2017, 18, 859. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.L.; Duque, E.; Godoy, P.; Segura, A. Efflux Pumps Involved in Toluene Tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 1998, 180, 3323–3329. [Google Scholar] [PubMed]
- Kieboom, J.; De Bont, J.A.M. Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 2001, 147, 43–51. [Google Scholar] [CrossRef][Green Version]
- Yao, X.; Tao, F.; Zhang, K.; Tang, H.; Xu, P. Multiple roles for two efflux pumps in the polycyclic aromatic hydrocarbon-degrading Pseudomonas putida strain B6-2 (DSM 28064). Appl. Environ. Microbiol. 2017, 83, e01882-17. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yu, H.; Li, Q.; Wang, X.; Gai, Z.; Yin, G.; Su, F.; Tao, F.; Ma, C.; Xu, P. Genome Sequence of Pseudomonas putida Strain B6-2, a Superdegrader of Polycyclic Aromatic Hydrocarbons and Dioxin-Like Compounds. J. Bacteriol. 2011, 193, 6789–6790. [Google Scholar] [CrossRef]
- Sun, F.; Zhou, D.; Wang, Q.; Feng, J.; Feng, W.; Luo, W.; Liu, Y.; Qiu, X.; Yin, Z.; Xia, P. Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J. Antimicrob. Chemother. 2016, 71, 909–912. [Google Scholar] [CrossRef]
- Chan, X.Y.; Chua, K.O.; How, K.Y.; Yin, W.-F.; Chan, K.-G. Global Genome Comparative Analysis Reveals Insights of Resistome and Life-Style Adaptation of Pseudomonas putida Strain T2-2 in Oral Cavity. Sci. World J. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Molina, L.; Udaondo, Z.; Duque, E.; Fernández, M.; Molina-Santiago, C.; Roca, A.; Porcel, M.; De La Torre, J.; Segura, A.; Plesiat, P.; et al. Antibiotic Resistance Determinants in a Pseudomonas putida Strain Isolated from a Hospital. PLoS ONE 2014, 9, e81604. [Google Scholar] [CrossRef] [PubMed]
- Marchiaro, P.M.; Brambilla, L.; Morán-Barrio, J.; Revale, S.; Pasteran, F.; Vila, A.J.; Viale, A.M.; Limansky, A.S. The Complete Nucleotide Sequence of the Carbapenem Resistance-Conferring Conjugative Plasmid pLD209 from a Pseudomonas putida Clinical Strain Reveals a Chimeric Design Formed by Modules Derived from Both Environmental and Clinical Bacteria. Antimicrob. Agents Chemother. 2014, 58, 1816–1821. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.P.S.; Kosmopoulou, M.; Poirel, L.; Nordmann, P.; Spencer, J. Crystal Structure of DIM-1, an Acquired Subclass B1 Metallo-β-Lactamase from Pseudomonas stutzeri. PLoS ONE 2015, 10, e0140059. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Rodríguez-Martínez, J.-M.; Al Naiemi, N.; Debets-Ossenkopp, Y.J.; Nordmann, P. Characterization of DIM-1, an Integron-Encoded Metallo-β-Lactamase from a Pseudomonas stutzeri Clinical Isolate in the Netherlands. Antimicrob. Agents Chemother. 2010, 54, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Grell, E.; Malviya, V.N.; Xie, H.; Wang, J.; Michel, H. Identification of the High-affinity Substrate-binding Site of the Multidrug and Toxic Compound Extrusion (MATE) Family Transporter from Pseudomonas stutzeri. J. Boil. Chem. 2016, 291, 15503–15514. [Google Scholar] [CrossRef] [PubMed]
- Bashar, S.; Sanyal, S.K.; Sultana, M.; Hossain, M.A. Emergence of IntI1 associated blaVIM-2 gene cassette-mediated carbapenem resistance in opportunistic pathogen Pseudomonas stutzeri. Emerg. Microbes Infect. 2017, 6, e29. [Google Scholar] [CrossRef] [PubMed]
- Stoitsova, S.O.; Braun, Y.; Ullrich, M.S.; Weingart, H. Characterization of the RND-Type Multidrug Efflux Pump MexAB-OprM of the Plant Pathogen Pseudomonas syringae. Appl. Environ. Microbiol. 2008, 74, 3387–3393. [Google Scholar] [CrossRef] [PubMed]
- Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G.; Scortichini, M. Pseudomonas syringae pv. actinidiae Draft Genomes Comparison Reveal Strain-Specific Features Involved in Adaptation and Virulence to Actinidia Species. PLoS ONE 2011, 6, e27297. [Google Scholar] [CrossRef]
- Feil, H.; Feil, W.S.; Chain, P.; Larimer, F.; DiBartolo, G.; Copeland, A.; Lykidis, A.; Trong, S.; Nolan, M.; Goltsman, E.; et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2005, 102, 11064–11069. [Google Scholar] [CrossRef]
- Sundin, G.W.; Bender, C.L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 1993, 59, 1018–1024. [Google Scholar]
- Crovadore, J.; Cochard, B.; Calmin, G.; Chablais, R.; Schulz, T.; Lefort, F. Whole-Genome Sequence of Pseudomonas xanthomarina Strain UASWS0955, a Potential Biological Agent for Agricultural and Environmental Uses. Genome Announc. 2016, 4, e01136-16. [Google Scholar] [CrossRef] [PubMed]
- Gifford, D.R.; Moss, E.; MacLean, R.C. Environmental variation alters the fitness effects of rifampicin resistance mutations in Pseudomonas aeruginosa. Evolution 2016, 70, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Loftie-Eaton, W.; Bashford, K.; Quinn, H.; Dong, K.; Millstein, J.; Hunter, S.; Thomason, M.K.; Merrikh, H.; Ponciano, J.M.; Top, E.M. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 2017, 1, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Cherif-Antar, A.; Moussa–Boudjemâa, B.; Didouh, N.; Medjahdi, K.; Mayo, B.; Flórez, A.B. Diversity and biofilm-forming capability of bacteria recovered from stainless steel pipes of a milk-processing dairy plant. Dairy Sci. Technol. 2016, 96, 27–38. [Google Scholar] [CrossRef]
- De La Fuente-Núñez, C.; Reffuveille, F.; Fernandez, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Serio, A.; Chaves-López, C.; Anniballi, F.; Auricchio, B.; Goffredo, E.; Paparella, A. Biofilm formation, pigment production and motility in Pseudomonas spp. isolated from the dairy industry. Food Control. 2018, 86, 241–248. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.R.K.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in oxidative stress at low temperature in an Antarctic bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Murray, T.S.; Kazmierczak, B.I.; He, C. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 2010, 75, 76–91. [Google Scholar] [CrossRef]
- Gillespie, S.H. Antibiotic resistance in the absence of selective pressure. Int. J. Antimicrob. Agents 2001, 17, 171–176. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Akhova, A.V.; Shumkov, M.S.; Nesterova, L.Y. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res. Microbiol. 2012, 163, 83–91. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, J.; Yuan, Y.; Yue, T. Diversity and characterization of spoilage-associated psychrotrophs in food in cold chain. Int. J. Food Microbiol. 2019, 290, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Brock, M.T.; Fedderly, G.C.; Borlee, G.I.; Russell, M.M.; Filipowska, L.K.; Hyatt, D.R.; Ferris, R.A.; Borlee, B.R. Pseudomonas aeruginosa variants obtained from veterinary clinical samples reveal a role for cyclic di-GMP in biofilm formation and colony morphology. Microbiology 2017, 163, 1613–1625. [Google Scholar] [CrossRef]
- Xavier, B.B.; Das, A.J.; Cochrane, G.; De Ganck, S.; Kumar-Singh, S.; Aarestrup, F.M.; Goossens, H.; Malhotra-Kumar, S. Consolidating and Exploring Antibiotic Resistance Gene Data Resources. J. Clin. Microbiol. 2016, 54, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Boisvert, S.; Brettin, T.; Kenyon, R.W.; Mao, C.; Olson, R.; Overbeek, R.; Santerre, J.; Shukla, M.; Wattam, A.R.; et al. Antimicrobial Resistance Prediction in PATRIC and RAST. Sci. Rep. 2016, 6, 27930. [Google Scholar] [CrossRef] [PubMed]
- Drouin, A.; Giguere, S.; Sagatovich, V.; Deraspe, M.; Laviolette, F.; Marchand, M.; Corbeil, J. Learning interpretable models of phenotypes from whole genome sequences with the set covering machine. arXiv 2014, arXiv:1412.1074. [Google Scholar]
- Santerre, J.; Boisvert, S.; Davis, J.; Xia, F.; Stevens, R. Gene Identification and Strain Classification Using Random Forests. In Proceedings of the Great Lakes Bioinformatics Conference, West Lafayette, IN, USA, 18 May 2015. [Google Scholar]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef]
- Danishuddin, M.; Baig, M.H.; Kaushal, L.; Khan, A.U. BLAD: A comprehensive database of widely circulated beta-lactamases. Bioinformatics 2013, 29, 2515–2516. [Google Scholar] [CrossRef][Green Version]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-Lactamase DataBase (BLDB)—Structure and Function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Sing, O.V. (Ed.) Foodborne Pathogens and Antibiotic Resistance; Wiley Blackwell: Hoboken, NJ, USA, 2017; p. 512. [Google Scholar]
- Schmidt, K.; Mwaigwisya, S.; Crossman, L.C.; Doumith, M.; Munroe, D.; Pires, C.; Khan, A.M.; Woodford, N.; Saunders, N.J.; Wain, J.; et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 2017, 72, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Oniciuc, E.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.; Alvarez-Ordóñez, A. The present and future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.F.; Riley, L.W. Identification of novel antimicrobial resistance genes from microbiota on retail spinach. BMC Microbiol. 2013, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Florez, C.; Raab, J.E.; Cooke, A.C.; Schertzer, J.W. Membrane Distribution of the Pseudomonas Quinolone Signal Modulates Outer Membrane Vesicle Production in Pseudomonas aeruginosa. mBio 2017, 8, e01034-17. [Google Scholar] [CrossRef] [PubMed]
- Devirgiliis, C.; Zinno, P.; Stirpe, M.; Barile, S.; Perozzi, G. Functional Screening of Antibiotic Resistance Genes from a Representative Metagenomic Library of Food Fermenting Microbiota. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; Van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Zankari, E. Comparison of the Web Tools ARG-ANNOT and ResFinder for Detection of Resistance Genes in Bacteria. Antimicrob. Agents Chemother. 2014, 58, 4986. [Google Scholar] [CrossRef]
- FAO/WHO Food and Agriculture Organization of the United Nations/World Health Organization. Microbiological Hazards in Fresh Leafy Vegetables and Herbs: Meeting Report; Microbiological Risk Assessment Series No. 14; FAO/WHO: Rome, Italy, 2008; ISSN 1726-5274. [Google Scholar]
- Akbas, M.Y. Bacterial biofilms and their new control strategies in food industry. In The Battle against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs, Badajoz: Formatex; Méndez-Vilas, A., Ed.; FORMATEX: Badajoz, Spain, 2015; Volume 1, pp. 383–394. [Google Scholar]
- Caputo, L.; Quintieri, L.; Cavalluzzi, M.M.; Lentini, G.; Habtemariam, S. Antimicrobial and Antibiofilm Activities of Citrus Water-Extracts Obtained by Microwave-Assisted and Conventional Methods. Biomedicines 2018, 6, 70. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Sahari, M.A.; Asgari, S. Effects of plants bioactive compounds on foods microbial spoilage and lipid oxidation. Food Sci. Technol. 2013, 1, 52–61. [Google Scholar]
- Pedonese, F.; Fratini, F.; Pistelli, L.; Porta, F.M.; Di Ciccio, P.; Fischetti, R.; Turchi, B.; Nuvoloni, R. Antimicrobial activity of four essential oils against pigmenting Pseudomonas fluorescens and biofilm producing Staphylococcus aureus of dairy origin. Ital. J. Food Saf. 2017, 6, 6939. [Google Scholar] [PubMed]
- Roila, R.; Branciari, R.; Ranucci, D.; Ortenzi, R.; Urbani, S.; Servili, M.; Valiani, A. Antimicrobial Activity of Olive Mill Wastewater Extract Against Pseudomonas fluorescens Isolated from Mozzarella Cheese. Ital. J. Food Saf. 2016, 5, 5760. [Google Scholar] [CrossRef] [PubMed]
- Roila, R.; Valiani, A.; Ranucci, D.; Ortenzi, R.; Servili, M.; Veneziani, G.; Branciari, R.; Rossana, R. Antimicrobial efficacy of a polyphenolic extract from olive oil by-product against “Fior di latte” cheese spoilage bacteria. Int. J. Food Microbiol. 2019, 295, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.M.; Howes, T.; Bhandari, B.R. Methods to extend the shelf-life of cottage cheese—A review. Int. J. Dairy Technol. 2016, 69, 313–327. [Google Scholar] [CrossRef]
- Lewies, A.; Du Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2018, 11, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Caputo, L.; Monaci, L.; DeSerio, D.; Morea, M.; Baruzzi, F. Antimicrobial efficacy of pepsin-digested bovine lactoferrin on spoilage bacteria contaminating traditional Mozzarella cheese. Food Microbiol. 2012, 31, 64–71. [Google Scholar] [CrossRef]
- Quintieri, L.; Caputo, L.; Morea, M.; Baruzzi, F. Control of Mozzarella spoilage bacteria by using bovine lactoferrin pepsin-digested hydrolysate. In Worldwide Research Efforts in the Fighting against Microbial Pathogens: From Basic Research to Technological Developments; Mendèz-Vilas, A., Ed.; Brown Walker Press: Boca Raton, FL, USA, 2013; pp. 118–122. [Google Scholar]
- CFSAN/Office of Food Additive Safety. Agency Response Letter: GRAS Notice No.GRN 000077; US Food and Drug Administration Web Site. Available online: http://www.fda.gov/Food/ FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/GRASListings/ucm154188.htm (accessed on 14 August 2001).
- Tomita, M.; Wakabayashi, H.; Shin, K.; Yamauchi, K.; Yaeshima, T.; Iwatsuki, K. Twenty-five years of research on bovine lactoferrin applications. Biochimie 2009, 91, 52–57. [Google Scholar] [CrossRef]
- Quintieri, L.; Carito, A.; Pinto, L.; Calabrese, N.; Baruzzi, F.; Caputo, L. Application of lactoferricin B to control microbial spoilage in cold stored fresh foods. In Multidisciplinary Approach for Studying and Combating Microbial Pathogens. Microbiology Series; Mendèz-Vilas, A., Ed.; Brown Walker Press: Boca Raton, FL, USA, 2015; Volume 3, pp. 58–62. [Google Scholar]
- Quintieri, L.; Pistillo, B.R.; Caputo, L.; Favia, P.; Baruzzi, F. Bovine lactoferrin and lactoferricin on plasma-deposited coating against spoilage Pseudomonas spp. Innov. Food Sci. Emerg. Technol. 2013, 20, 215–222. [Google Scholar] [CrossRef]
- McPhee, J.B.; Lewenza, S.; Hancock, R.E.W. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 205–217. [Google Scholar] [CrossRef]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B Boil. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef] [PubMed]
- Nesse, L.L.; Simm, R. Biofilm: A Hotspot for Emerging Bacterial Genotypes. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2018; Volume 103, pp. 223–246. [Google Scholar]
- Beaudoin, T.; Stone, T.A.; Glibowicka, M.; Adams, C.; Yau, Y.; Ahmadi, S.; Bear, C.E.; Grasemann, H.; Waters, V.; Deber, C.M. Activity of a novel antimicrobial peptide against Pseudomonas aeruginosa biofilms. Sci. Rep. 2018, 8, 14728. [Google Scholar] [CrossRef] [PubMed]
- Moussouni, M.; Nogaret, P.; Garai, P.; Ize, B.; Vivès, E.; Blanc-Potard, A.-B. Activity of a Synthetic Peptide Targeting MgtC on Pseudomonas aeruginosa Intramacrophage Survival and Biofilm Formation. Front. Microbiol. 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Nishino, K.; Roberts, M.C.; Tolmasky, M.; Aminov, R.I.; Zhang, L. Mechanisms of antibiotic resistance. Front. Microbiol. 2015, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G.; Senechal, A.C.; Mukherjee, A.; Ané, J.-M.; Blackwell, H.E. Plant Responses to Bacterial N-Acyl l-Homoserine Lactones are Dependent on Enzymatic Degradation to l-Homoserine. ACS Chem. Boil. 2014, 9, 1834–1845. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wei, Y.; Xia, B.; Jin, Y.; Liu, C.; Pan, X.; Shi, J.; Zhu, F.; Li, J.; Qian, L.; et al. Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 19141. [Google Scholar] [CrossRef]
- Ding, T.; Li, T.; Li, J. Identification of natural product compounds as quorum sensing inhibitors in Pseudomonas fluorescens P07 through virtual screening. Bioorg. Med. Chem. 2018, 26, 4088–4099. [Google Scholar] [CrossRef]
- Ding, T.; Li, T.; Li, J. Virtual screening for quorum sensing inhibitors of Pseudomonas fluorescens P07 from a food-derived compound database. J. Appl. Microbiol. 2019, 127, 763–777. [Google Scholar] [CrossRef]
- Li, T.; Wang, D.; Liu, N.; Ma, Y.; Ding, T.; Mei, Y.; Li, J. Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde. Int. J. Food Microbiol. 2018, 269, 98–106. [Google Scholar] [CrossRef]
- Myszka, K.; Schmidt, M.T.; Majcher, M.; Juzwa, W.; Olkowicz, M.; Czaczyk, K. Inhibition of quorum sensing-related biofilm of Pseudomonas fluorescens KM121 by Thymus vulgare essential oil and its major bioactive compounds. Int. Biodeterior. Biodegrad. 2016, 114, 252–259. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, J.; Xie, Y.; Guo, Y.; Cheng, Y.; Qian, H.; Yao, W. Essential oil components inhibit biofilm formation in Erwinia carotovora and Pseudomonas fluorescens via anti-quorum sensing activity. LWT 2018, 92, 133–139. [Google Scholar] [CrossRef]
- Dima, C.; Dima, S. Essential oils in foods: Extraction, stabilization, and toxicity. Curr. Opin. Food Sci. 2015, 5, 29–35. [Google Scholar] [CrossRef]
- Bai, A.J.; Rai Vittal, R. Quorum Sensing Regulation and Inhibition of Exoenzyme Production and Biofilm Formation in the Food Spoilage Bacteria Pseudomonas psychrophila PSPF19. Food Biotechnol. 2014, 28, 293–308. [Google Scholar] [CrossRef]
- Fothergill, J.L.; Winstanley, C.; James, C.E. Novel therapeutic strategies to counter Pseudomonas aeruginosa infections. Expert Rev. Anti-Infect. Ther. 2012, 10, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in Food Applications: From Foe to Friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef]
- Lacivita, V.; Conte, A.; Lyng, J.G.; Arroyo, C.; Zambrini, V.A.; Del Nobile, M.A. High intensity light pulses to reduce microbial load in fresh cheese. J. Dairy Res. 2018, 85, 232–237. [Google Scholar] [CrossRef]
- Lacivita, V.; Conte, A.; Musavian, H.S.; Krebs, N.H.; Zambrini, V.A.; Del Nobile, M.A. Steam-ultrasound combined treatment: A promising technology to significantly control mozzarella cheese quality. LWT 2018, 93, 450–455. [Google Scholar] [CrossRef]
- Lacivita, V.; Mentana, A.; Centonze, D.; Chiaravalle, E.; Zambrini, V.A.; Conte, A.; Del Nobile, M.A. Study of X-Ray irradiation applied to fresh dairy cheese. LWT 2019, 103, 186–191. [Google Scholar] [CrossRef]
- Codex Alimentarius. Codex Alimentarius. Code of hygienic practice for milk and milk products (CAC/RCP 57-2004). In Codex Alimentarius Milk and Milk Products. CODEX STAN 243–2003, 2nd ed.; World Health Organization Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; pp. 183–199. [Google Scholar]
- Codex Alimentarius. Recommended International Code of Practice General Principles of Food Hygiene; CAC/RCP 1-1969, Rev. 4; World Health Organization Food and Agriculture Organization of the United Nations: Rome, Italy, 2003; pp. 1–31. [Google Scholar]
- Faccia, M.; Gambacorta, G.; Natrella, G.; Caponio, F. Shelf life extension of Italian mozzarella by use of calcium lactate buffered brine. Food Control. 2019, 100, 287–291. [Google Scholar] [CrossRef]
- Varga, L.; Szigeti, J. Use of ozone in the dairy industry: A review. Int. J. Dairy Technol. 2016, 69, 157–168. [Google Scholar] [CrossRef]
- Jiménez-Pichardo, R.; Regalado, C.; Castaño-Tostado, E.; Meas-Vong, Y.; Santos-Cruz, J.; García-Almendárez, B.E. Evaluation of electrolyzed water as cleaning and disinfection agent on stainless steel as a model surface in the dairy industry. Food Control 2016, 60, 320–328. [Google Scholar] [CrossRef]
- Wang, H.; Cai, L.; Li, Y.; Xu, X.; Zhou, G. Biofilm formation by meat-borne Pseudomonas fluorescens on stainless steel and its resistance to disinfectants. Food Control. 2018, 91, 397–403. [Google Scholar] [CrossRef]
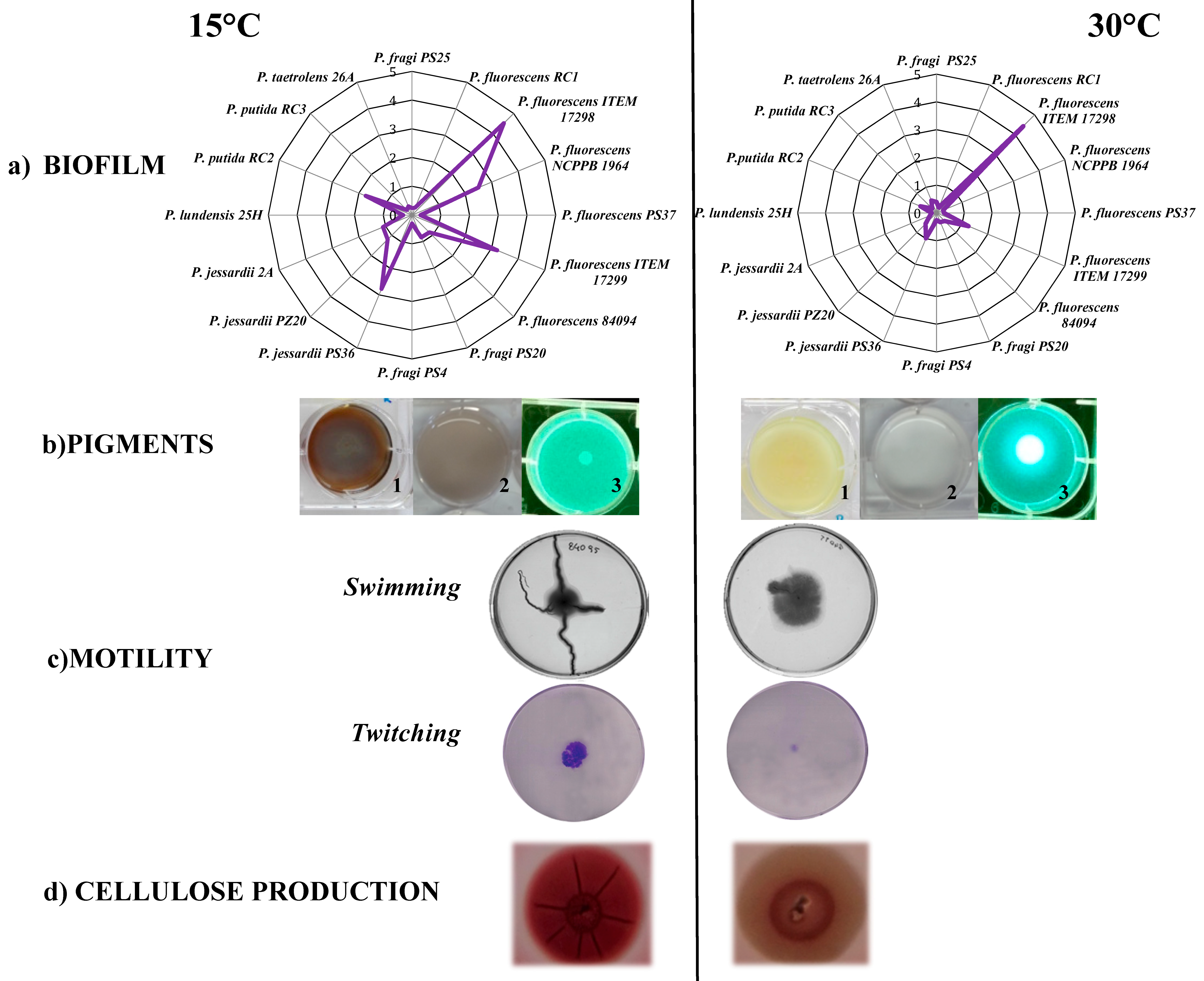
| Species | Source | Antibiotics | References | |
|---|---|---|---|---|
| Class | Molecule (μg) | |||
| P. pseudoalcaligenes, P. fluorescens biovar V, P. alcaligenes, P. pseudoalcaligenes subspecies citrulli | Turkish homemade white cheese | β-lactams and β-lactams/β-lactamas inhibitors | Penicillin G (10 μg); Piperacillin (16 μg); Piperacillin/tazobactam (64/4 μg); | [37] |
| Sulfanilamide/2,4-diaminopyrimidine | Sulfamethoxazole/trimethoprim (25 μg) | |||
| P. fluorescens, P. tolaasii | Raw milk from Finland farms | β-lactams and β-lactams/β-lactamas inhibitors | Ticarcillin (64 μg); Ticarcillin-Clavulonic acid (64/2 μg); | [84] |
| Monocyclic bacterially derived beta-lactam | Aztreonam (32 μg); | |||
| Phosphonic acid derivative | Fosfomycin (32 μg); | |||
| Third-generation cephalosporins | Ceftazidim (32 μg); | |||
| Aminoglycosides | Tobramycin (8 μg); Amikacin (16 μg); Netilmicin (4 μg), Gentamicin (8 μg); | |||
| Fluoroquinolones | Ofloxacin (1 μg); ciprofloxacin (4 μg); | |||
| Lipopeptides | Colistin (2 μg); | |||
| Sulfanilamide/2,4-diaminopyrimidine | Sulfamethoxazole/trimethoprim (2/38 μg) | |||
| P. putida, P. fulva, P. fragi, P. mosselii, P. rhodesiae, P. libanensis, P. teatrolens, P. chlororaphis, P. fluorescens | Italian bulk tank milk | Penicillin | Piperacillin (100 ug), Ticarcillin/ clavulanic acid (85 ug); | [85] |
| Monocyclic bacterially derived beta-lactam | Aztreonam (30 μg); | |||
| Third and Fourth-generation cephalosporin | Ceftazidim (30 μg); Cefepime (30 μg); | |||
| Aminoglycosides | Tobramycin (10 μg); Amikacin (30 μg); Netilmicin (10 μg); | |||
| Fluoroquinolones | Ciprofloxacin (5 μg); Levofloxacin (5 μg); | |||
| Carbapenems | Meropenem (10 μg); Imipenem (10 μg); | |||
| Lipopeptides | Colistin sulphate (10 μg); | |||
| P. fluorescens, P. taetrolens, P. putida, P. fragi, P. alcaligenes, P. lundensis | French milks or semi-hard and soft, smear-ripened cheeses | Penicillin | Ticarcillin (75 μg); Amoxicillin (25 μg); Ampicillin (10 μg); Mecillinam (10 μg); Amoxicillin/Clavulanic acid (20/10 μg); | [86] |
| Monocyclic bacterially derived beta-lactam | Aztreonam (30 μg); | |||
| First and Third-generation cephalosporin | Cefalotin (30 μg); Cefotaxime (30 μg); | |||
| Lipopeptides | Colistin sulphate (50 μg); | |||
| Polyketide antibiotics | Tetracycline (30 μg); | |||
| Amphenicol-class | Chloramphenicol (30 μg); | |||
| Pseudomonas spp. | Commercial UHT milk | Monocyclic bacterially derived beta-lactam | Aztreonam | [87] |
| Carbapenems | Meropenem | |||
| Aminoglycosides | Amikacin; Gentamicin | |||
| Third and Fourth generation cephalosporins | Ceftazidime; Cefepime | |||
| Fluoroquinolones | Levofloxacin | |||
| P. fluorescens, P. gessardii, P. fragi | Italian high moisture mozzarella cheese | Aminoglycosides | Tobramycin (10 μg); Kanamycin (30 μg); Gentamicin (10 μg); Streptomycin (10 µg); | This work (see Table 2) |
| Fluoroquinolones | Ofloxacin (5 μg); Ciprofloxacin (5 μg); | |||
| Quinolones | Nalidixic acid (30 µg); | |||
| Nitrofurans | Nitrofurantoin (300 µg) | |||
| P. fluorescens | P. gessardii | P. fragi | ||||
|---|---|---|---|---|---|---|
| ITEM 17299 | ITEM 17298 | NCCP 1964 | ITEM 17295 | PS36 | PS4 | |
| Ampicillin (10 μg) ** | N.D.* | N.D. | N.D. | N.D. | N.D. | N.D. |
| Methicillin (10 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Oxacillin (1 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Penicillin G (10 μg) | N.D. | N.D. | 126 (117–131) | N.D. | N.D. | N.D. |
| Ceftizoxime (30 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Gentamicin (10 μg) | 286 a (266–297) | N.D. | 632 c (588–657) | 318 a (296–331) | 502 b (467–522) | 545 b (507–567) |
| Tobramicin (10 μg) | 355 a (330–369) | 352 a (328–367) | 678 d (631–705) | 424 b (394–441) | 544 c (506–566) | 457 b (425–475) |
| Kanamicin (30 μg) | 80 a (75–84) | 161 b (179–167) | 424 d (394–441) | 502 e (467–522) | 544 e (507–565) | 326 c (303–339) |
| Ciprofloxacin (5 μg) | 443 a (412–461) | 405 a (377–422) | 776 d (721–807) | 678 c (631–705) | 726 c (675–755) | 458 a (426–476) |
| Ofloxacin (5 μg) | 314 a (292–327) | 611 d (568–636) | 776 e (723–810) | 632 d(588–657) | 544 c (506–566) | 435 b(405–452) |
| Streptomycin (10 μg) | 85 b (79–88) | 76 a (70–79) | 462 e (430–481) | 85 b (79–88) | 424 e (394–441) | 377 d (351–392) |
| Nalidixic acid (30 μg) | 256 d (238–266) | 186 c (173–193) | 387 e (260–402) | 173 c (161–180) | 148 b (138–154) | 235 (218–244) |
| Tetracycline (30 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Vancomycin (30 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Clindamycin (2 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Lincomycin (2 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Erythromycin (15 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Fusidic acid (10 μg) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Nitrofurantoin (300 μg) | N.D. | N.D. | 35 (33–37) | N.D. | N.D. | N.D. |
| Species | Source | Antibiotic Resistance Genes | Reference |
|---|---|---|---|
| P. corrugata | tomato (Italy) | arpC | [96] |
| P. monteilii | clinical isolate | β-lactamase, aac(6′)-Ib, sul1 | [97] |
| P. protegens | Pf-5 cotton rhizosphere | rpoB mutation | [98] |
| P. syringae | - | fosC | [99] |
| P. cannabina | plant | β-lactamase, multidrug efflux system transmembrane protein | [100] |
| P. chlororaphis | clinical isolate | β-lactamase, ampC, mbl, phnP, cmeABC, mexCD-oprJ, mexE-oprN, fosE | [101] |
| P. citronellolis | soil collected under pine trees in northern Virginia, USA | β-lactamase, tetA, oprM1-5, vanX, fosA | [102] |
| P. fluorescens | Industrial strain pre-filtered tanks England | uppP, mexAB | [103] |
| feces of of Mareca penelope | cmeABC, mexC-mexD-oprJ, mexE-mexF-oprN, macA, macB | [104] | |
| Siene river | β-lactamase | [105] | |
| clinical isolate | [82] | ||
| coastal water | [106] | ||
| P. fluorescens cLP6a (petroleum-contaminated soil) | emhABC | [107] | |
| wheat take-all decline soil in China | [108] | ||
| P. fluorescens cLP6a (petroleum-contaminated soil) | [90] | ||
| mozzarella | mexA, ampC, oprD, mdtL, emrB | [95] | |
| meat microbiome | tolC, mdtB | [109] | |
| clinical isolate | aacA31, fosE, β-lactamase | [110] | |
| P. lutea | rhizosphera | ampG, ampE, ampD, mrcA, mrcB, β-lactamase, pbpC, mdrA, acrB, mexB, adeJ, smeE, mtrD, cmeB, marC, mdtC, mdtB, bcr, fsr | [111] |
| P. luteola | clinical isolate | β-lactamase | [112] |
| P. mosselii | lower respiratory tract patient | β-lactamase, aacA4, aphA15, and aadA1 | [113] |
| P. otitidis | food (chicken and pork) | β-lactamase | [83] |
| clinical isolates | [114] | ||
| P. pseudoalcaligens | Guadalquivir River | RND efflux pump | [115] |
| P. putida | polluted Nigerian wetlands | β-lactamase | [116] |
| clinical isolate | [81] | ||
| clinical isolate | [82] | ||
| clinical isolate | β-lactamase, qnrVC6, gcu173, strA, strB, aacA4 | [117] | |
| clinical isolate | β-lactamase, fosE, aacA4, aadA1, dfrB1b | [118] | |
| clinical isolate | β-lactamase, aadA1, aph(3′)-XV, aacA4, aph3-Ib, strA, strB, sul1 | [119] | |
| toluene enrichment | ttgABC | [91,120] | |
| S12 from soil isolated styrene enrichment | arpABC | [121] | |
| B6 soil | ttgABC, srpABC, ttgGHI | [122] | |
| B6 soil | 30 efflux pump coding genes | [123] | |
| clinical isolate | aadA2, qacED1, sul1 | [124] | |
| clinical isolate | β-lactamase, pnrVC6, aacA3, ISPpu24, catB11c, Gcu56, aadA1a, dfrB2c, aacA4′, catB3 | [124] | |
| clinical isolate | RND pumps, cmeABC, MATE family efflux pump | [125] | |
| clinical isolate | ttgGHI, β-lactamase, ttgABC, sul1, strA, merA, tetA, aphA1-IAB, aadA1, ttgGHI | [126] | |
| clinical isolate | β-lactamase, aacA4 | [127] | |
| shrimp | qnrA, qnrB | [70] | |
| P. stutzeri | clinical isolate | β-lactamase | [128] |
| clinical isolate | [129] | ||
| P. stutzeri strain ZoBell (ATCC14405) marine sample taken in the Pacific Ocean | MATE efflux pump | [130] | |
| clinical isolate | β-lactamase, aacA7, dfrB5 gene, aacC-A5 | [131] | |
| P. syringae | P. syringae pv. syringae B728a snap bean leaflet in Wisconsin, and P. syringae pv. tomato DC3000 | RND-type multidrug efflux pump, mexAB-oprM | [132] |
| P. syringae pv. actinidiae | Actinidia pathogen | resistance nodulation division (RND), multi antimicrobial resistance (MAR), multidrug endosomal transporter (MET), major facilitator superfamily (MFS) | [133] |
| P. syringae pv. syringae | snap bean leaflet in Wisconsin | strA, strB | [134] |
| plant | [135] | ||
| P. xanthomarina | Strain UASWS0955 sewage sludge | fosmidomycin, polymyxin, penicillin, fluoroquinolones resistance genes (not specified) | [136] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintieri, L.; Fanelli, F.; Caputo, L. Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies. Foods 2019, 8, 372. https://doi.org/10.3390/foods8090372
Quintieri L, Fanelli F, Caputo L. Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies. Foods. 2019; 8(9):372. https://doi.org/10.3390/foods8090372
Chicago/Turabian StyleQuintieri, Laura, Francesca Fanelli, and Leonardo Caputo. 2019. "Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies" Foods 8, no. 9: 372. https://doi.org/10.3390/foods8090372
APA StyleQuintieri, L., Fanelli, F., & Caputo, L. (2019). Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies. Foods, 8(9), 372. https://doi.org/10.3390/foods8090372







