Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Bacteriophage Culture and Propagation
2.3. Mammalian Cell and Culture Conditions
2.4. Isolation of Phage-Resistant L. plantarum (LP+PR) Mutant
2.5. Growth and Survival of LP and LP+PR Strains
2.6. Co-Culture of LP or LP+PR Strain with Zoonotic Bacterial Pathogens
2.7. Cell-Free Cultural Supernatants (CFCSs) of LP and LP+PR Strains on Growth of Zoonotic Pathogens
2.8. Host Cells-LP or LP+PR Interactions
2.9. Inhibition of Adherence and Invasion of Zoonotic Enteric Pathogens with CFCSs Collected from LP and LP+PR Strains
2.10. RNA Extraction and cDNA Synthesis
2.11. Quantitative RT-PCR Assay
2.12. Statistical Analysis
3. Results
3.1. Confirmation of Bacteriophage Resistant LP+PR Strain
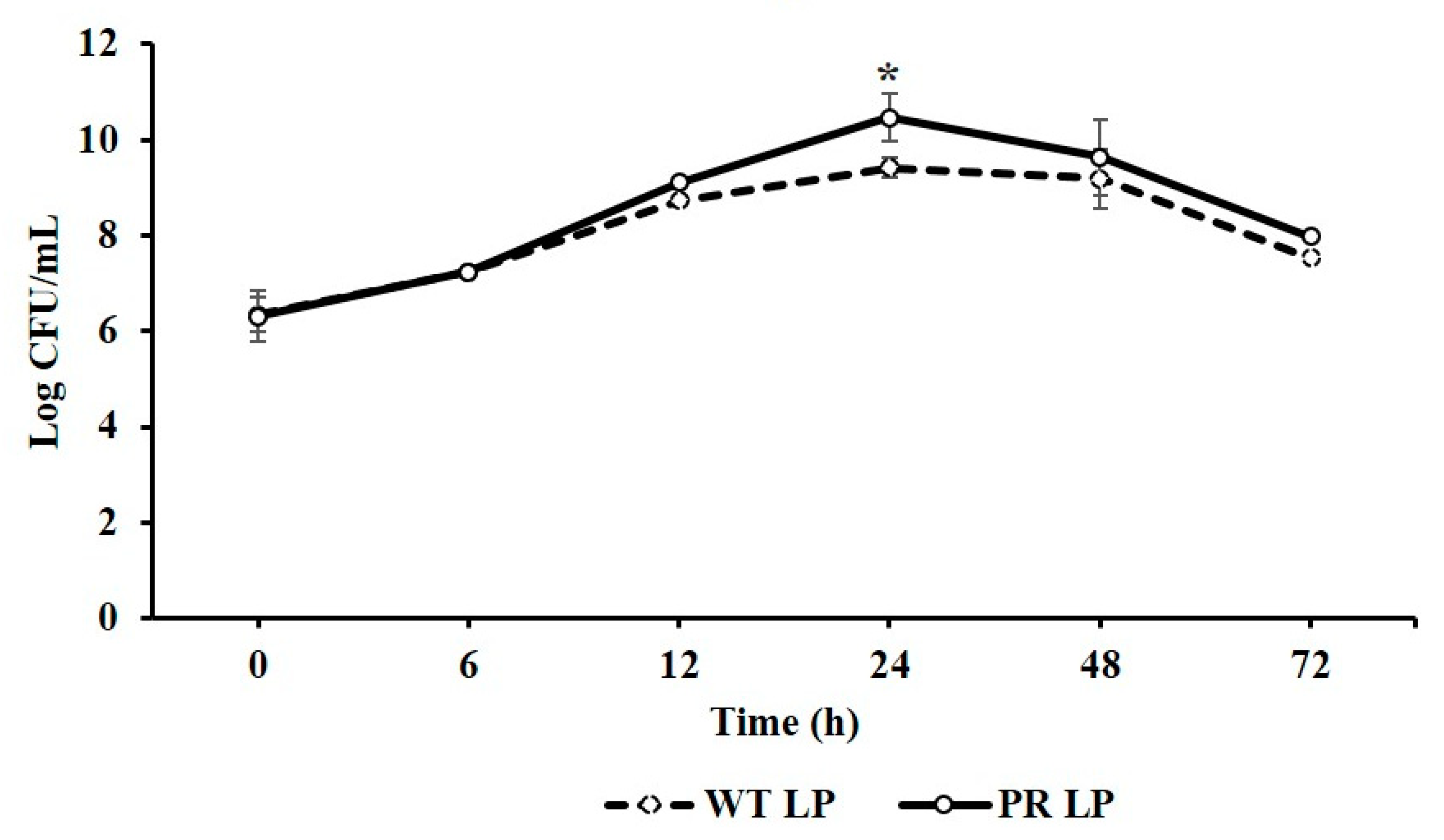
3.2. Comparative Growth and Survival Ability of LP and LP+PR Strains
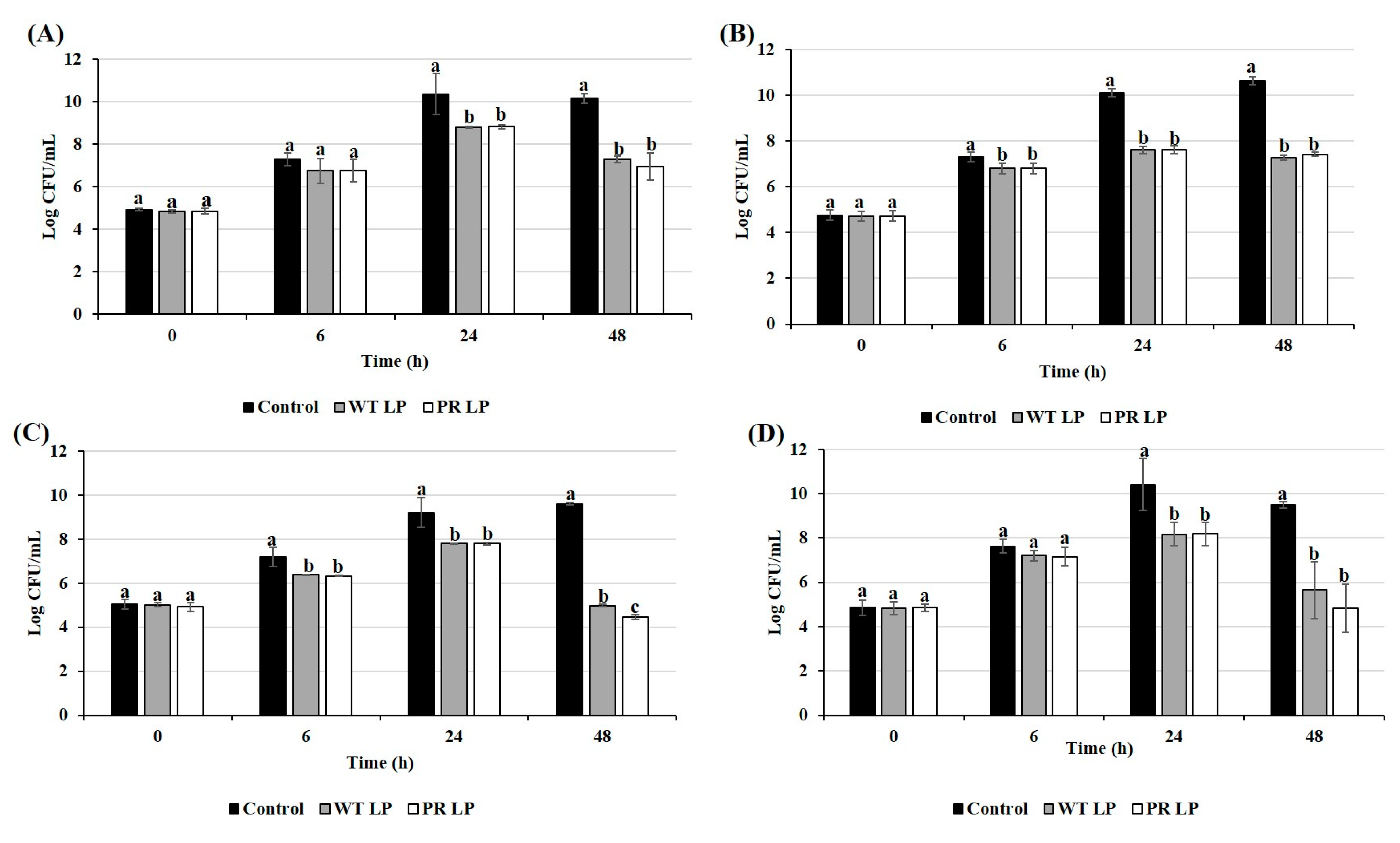
3.3. Competitive Inhibition of Zoonotic Pathogens in Co-Culture Condition
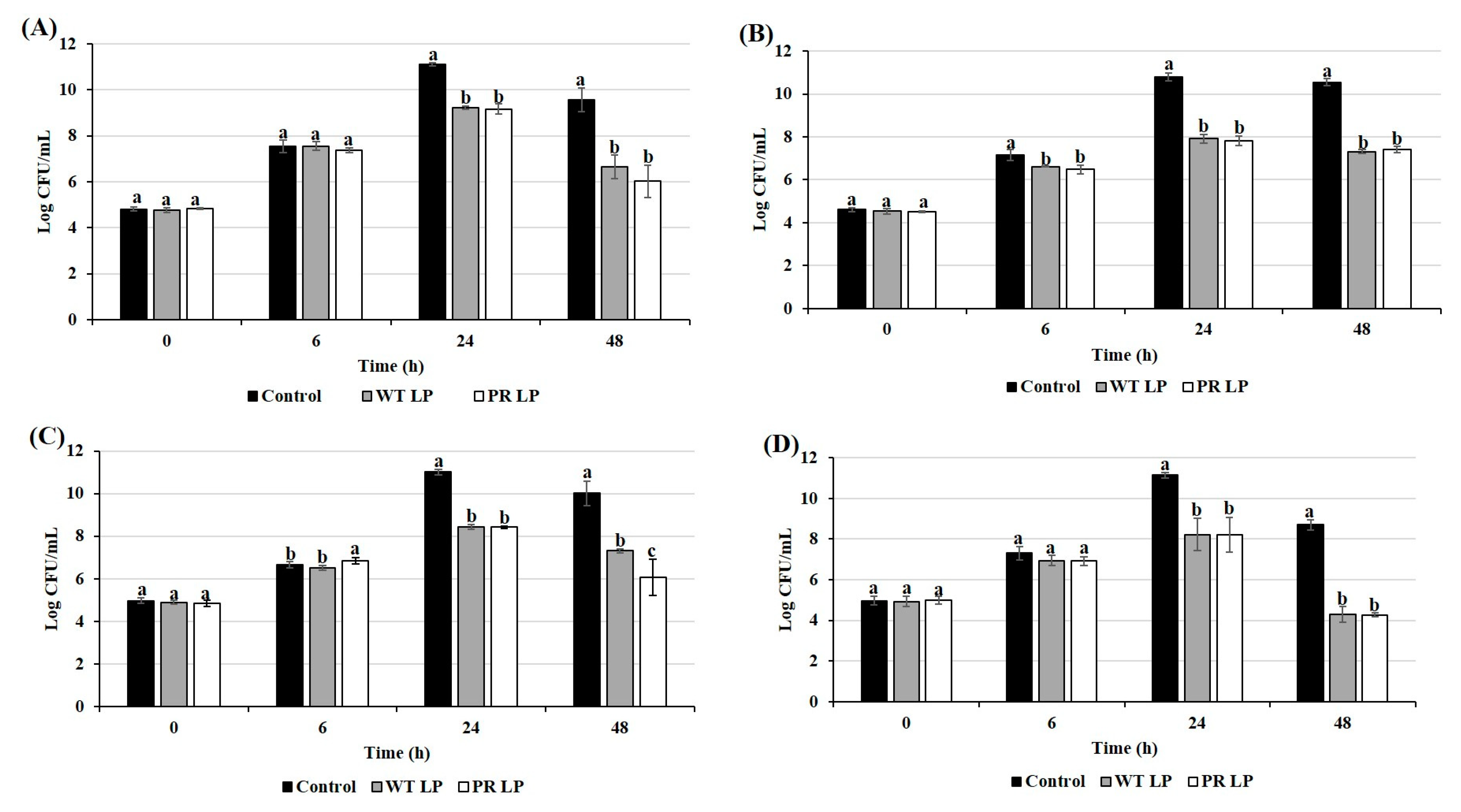
3.4. Effects of CFCSs of LP and LP+PR on Growth of Zoonotic Pathogens
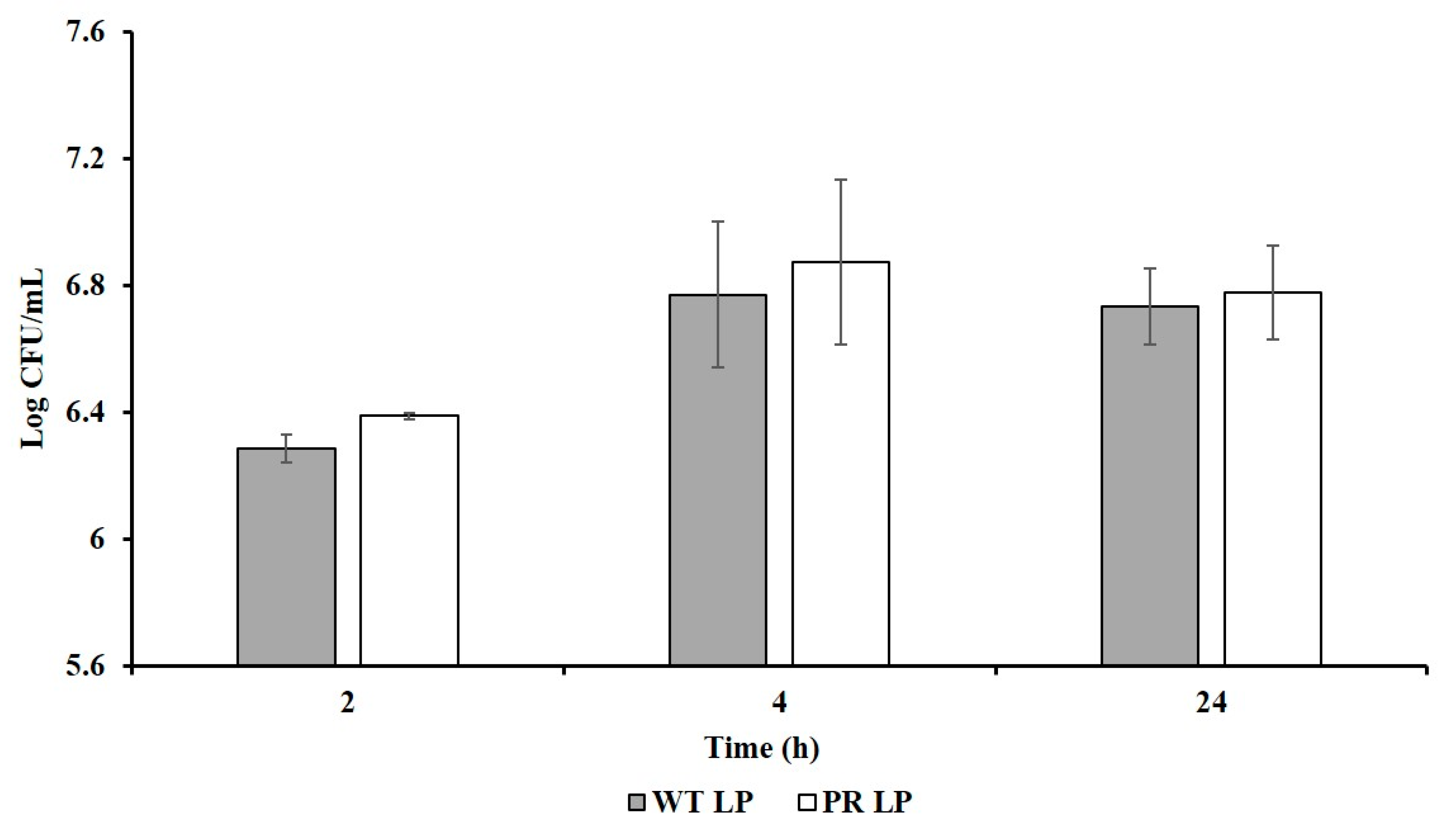
3.5. Adhesion Abilities of LP and LP+PR to Mammalian Cells
3.6. Role of LP and LP+PR in Alteration of Pathogen-Host Cell Interactions
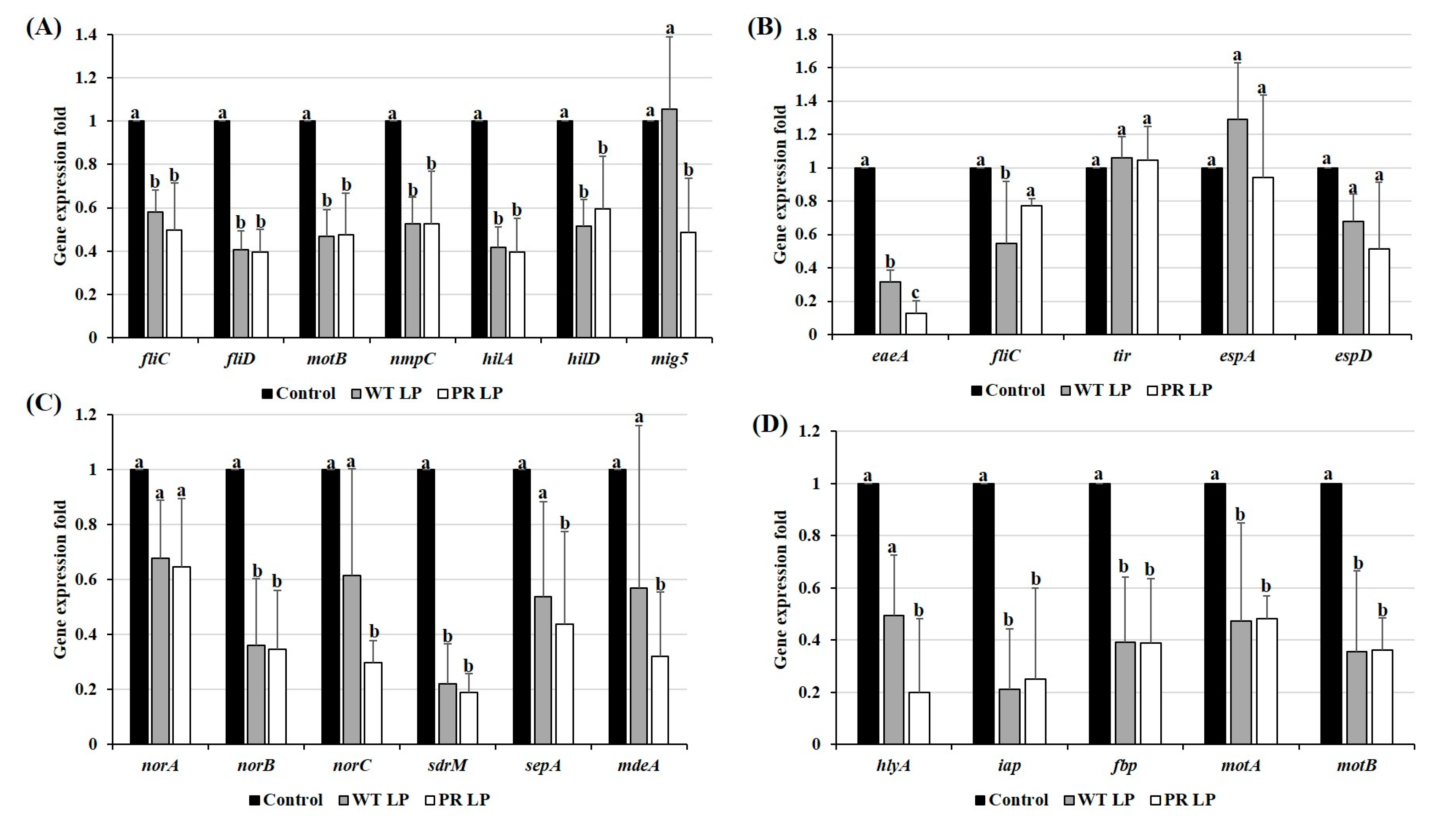
3.7. Alteration of Virulence Gene Expression of Zoonotic Pathogens in Presence of CFCSs Collected from LP and LP+PR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Asmahan, A. Beneficial role of lactic acid bacteria in food preservation and human health: A review. Res. J. Microbiol. 2010, 5, 1213–1221. [Google Scholar] [CrossRef][Green Version]
- Fijan, S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed]
- Upadrasta, A.; Stanton, C.; Hill, C.; Fitzgerald, G.F.; Ross, R.P. Improving the stress tolerance of probiotic cultures: Recent trends and future directions. In Stress Responses of Lactic Acid Bacteria; Tsakalidou, E., Papadimitriou, K., Eds.; Springer: New York, NY, USA, 2011; pp. 395–438. [Google Scholar]
- Šuškoviæ, J.; Kos, B.; Beganoviæ, J.; Pavunc, A.L.; Habjaniè, K.; Matošiæ, S. Antimicrobial activity-The most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 2010, 48, 296–307. [Google Scholar]
- Peng, M.; Biswas, D. Short chain and polyunsaturated fatty acids in host gut health and foodborne bacterial pathogen inhibition. Crit. Rev. Food Sci. Nutr. 2016, 57, 3987–4002. [Google Scholar] [CrossRef] [PubMed]
- Duary, R.K.; Bhausaheb, M.A.; Batish, V.K.; Grover, S. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactobacillus plantarum Lp91 in colitis mouse model. Mol. Biol. Rep. 2012, 39, 4765–4775. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Grover, S.; Batish, V.K. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br. J. Nutr. 2011, 105, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, A.; Shrier, I.; Heilpern, D.; Je, J.; Park, S.; Chong, G.; Lalonde, C.; Cote, L.F.; Lee, B. Differential impact of lactose/lactase phenotype on colonic microflora. Can. J. Gastroenterol. 2010, 24, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Rerksuppaphol, S.; Rerksuppaphol, L. A randomized double-blind controlled trial of Lactobacillus acidophilus plus Bifidobacterium bifidum versus placebo in patients with hypercholesterolemia. J. Clin. Diagn. Res. 2015, 9, KC01–KC04. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.M. The Role of Probiotics in the Poultry Industry. Int. J. Mol. Sci. 2009, 10, 3531–3546. [Google Scholar] [CrossRef]
- Zago, M.; Fornasari, M.E.; Carminati, D.; Bums, P.; Suàrez, V.; Vinderola, G.; Reinheimer, J.; Giraffa, G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011, 28, 1033–1040. [Google Scholar] [CrossRef]
- Georgieva, R.; Iliev, I.; Haertlé, T.; Chobert, J.M.; Ivanova, I.; Danova, S. Technological properties of candidate probiotic Lactobacillus plantarum strains. Int. Dairy J. 2009, 19, 696–702. [Google Scholar] [CrossRef]
- Karimi, S.; Azizi, F.; Nayeb-Aghaee, M.; Mahmoodnia, L. The antimicrobial activity of probiotic bacteria Escherichia coli isolated from different natural sources against hemorrhagic E. coli O157:H7. Electron. Physician 2018, 10, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, L.; Lv, Y.; Chen, Q.; Feng, J.; Zhao, X. Lactobacillus plantarum Restores Intestinal Permeability Disrupted by Salmonella Infection in Newly-hatched Chicks. Sci. Rep. 2018, 8, 2229. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, H.; Smoragiewicz, W. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int. J. Antimicrob. Agents 2013, 42, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Geeta, P.; Yadav, A.S. Antioxidant and antimicrobial profile of chicken sausages prepared after fermentation of minced chicken meat with Lactobacillus plantarum and with additional dextrose and starch. LWT-Food Sci. Technol. 2017, 77, 249–258. [Google Scholar] [CrossRef]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Giochetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar] [CrossRef]
- Emond, E.; Moineau, S. Bacteriophages and food fermentations. In Bacteriophage: Genetics and Molecular Biology; McGrath, S., van Sinderen, D., Eds.; Horizon Scientific Press/Caister Academic Press: Norfolk, UK, 2007; pp. 93–124. [Google Scholar]
- Samson, J.E.; Moineau, S. Bacteriophages in food fermentations: New frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 2013, 4, 347–368. [Google Scholar] [CrossRef]
- Ahn, J.; Kim, S.; Jung, L.; Biswas, D. Effect of bacteriophage on the transcriptional and translational expression of inflammatory mediators in chicken macrophage. J. Poult. Sci. 2014, 51, 96–103. [Google Scholar] [CrossRef][Green Version]
- Peng, M.; Reichmann, G.; Biswas, D. Lactobacillus casei and its byproducts alter the virulence factors of foodborne bacterial pathogens. Funct. Foods 2015, 15, 418–428. [Google Scholar] [CrossRef]
- Guglielmotti, D.M.; Reinheimer, J.A.; Binetti, A.G.; Giraffa, G.; Carminati, D.; Quiberoni, A. Characterization of spontaneous phage-resistant derivatives of Lactobacillus delbrueckii commercial strains. Int. J. Food Microbiol. 2006, 111, 126–133. [Google Scholar] [CrossRef]
- Peng, M.; Aryal, U.; Cooper, B.; Biswas, D. Metabolites produced during the growth of probiotics in cocoa supplementation and the limited role of cocoa in host-enteric bacterial pathogen interactions. Food Control 2015, 53, 124–133. [Google Scholar] [CrossRef]
- Tabashsum, Z.; Peng, M.; Salaheen, S.; Comis, C.; Biswas, D. Competitive elimination and virulence property alteration of Campylobacter jejuni by genetically engineered Lactobacillus casei. Food Control 2018, 85, 283–291. [Google Scholar] [CrossRef]
- Guglielmotti, D.M.; Marco, M.B.; Golowczyb, M.; Treinherimer, J.A.; Quiberoni, A.L. Probiotic potential of Lactobacillus delbrueckii strains and their phage resistant mutants. Int. Dairy J. 2007, 17, 916–925. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tufail, M.; Hussain, S.; Malik, F.; Mirza, T.; Parveen, G.; Shafaat, S.; Wajid, A.; Mahmood, R.; Channa, R.A.; Sadiq, A. Isolation and evaluation of antibacterial activity of bacteriocin produced by Lactobacillus bulgaricus from yogurt. Afr. J. Microbiol. Res. 2011, 5, 3842–3847. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef]
- Oelschlaeger, T.A. Mechanisms of probiotic actions—A Review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Haas, B.; Gottschalk, M.; Grenier, D. Antimicrobial potential of bacteriocins in poultry and swine production. Vet. Res. 2017, 48, 22. [Google Scholar] [CrossRef]
- Neal-McKinney, J.M.; Lu, X.; Duong, T.; Larson, C.L.; Call, D.R.; Shah, D.H.; Konkel, M.E. Production of organic acids by probiotic Lactobacilli can be used to reduce pathogen load in poultry. PLoS ONE 2012, 7, e43928. [Google Scholar] [CrossRef]
- Das, J.K.; Mishra, D.; Ray, P.; Tripathy, P.; Beuria, T.K.; Singh, N.; Suar, M. In vitro evaluation of anti-infective activity of a Lactobacillus plantarum strain against Salmonella enterica serovar Enteritidis. Gut Pathog. 2013, 5, 11. [Google Scholar] [CrossRef]
- Ikeda, J.S.; Schmitt, C.K.; Darnell, S.C.; Watson, P.R.; Bispham, J.; Wallis, T.S.; Weinstein, D.L.; Metcalf, E.S.; Adams, P.; O’Connor, C.D.; et al. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 2001, 69, 3021–3030. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Westerlund-Wikstrom, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [PubMed]
- Donnenberg, M.S.; Tzipori, S.; McKee, M.L.; O’Brien, A.D.; Alroy, J.; Kaper, J.B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 1993, 92, 1418–1424. [Google Scholar] [CrossRef]
- Fournier, B.; Aras, R.; Hooper, D.C. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 2000, 182, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Strahilevitz, J.; Hooper, D.C. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 1104–1107. [Google Scholar] [CrossRef]
- Mazurkiewicz, P.; Sakamoto, K.; Poelarends, G.J.; Konings, W.N. Multidrug transporters in lactic acid bacteria. Mini Rev. Med. Chem. 2005, 5, 173–181. [Google Scholar] [CrossRef]
- Salaheen, S.; Peng, M.; Joo, J.; Teramoto, H.; Biswas, D. Eradication and sensitization of methicillin resistant Staphylococcus aureus to methicillin with bioactive extracts of berry pomace. Front. Microbiol. 2017, 8, 253. [Google Scholar] [CrossRef]
- Fournier, B.; Hooper, D.C. A new two-component regulatory system involved in adhesion, autolysis and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 2000, 182, 3955–3964. [Google Scholar] [CrossRef]
- Marini, E.; Magi, G.; Ferretti, G.; Bacchetti, T.; Giuliani, A.; Pugnaloni, A.; Rippo, M.R.; Facinelli, B. Attenuation of Listeria monocytogenes virulence by Cannabis sativa L. essential oil. Front. Cell. Infect. Microbiol. 2018, 8, 293. [Google Scholar] [CrossRef]
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 2007, 189, 4418–4424. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagarajan, V.; Peng, M.; Tabashsum, Z.; Salaheen, S.; Padilla, J.; Biswas, D. Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens. Foods 2019, 8, 194. https://doi.org/10.3390/foods8060194
Nagarajan V, Peng M, Tabashsum Z, Salaheen S, Padilla J, Biswas D. Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens. Foods. 2019; 8(6):194. https://doi.org/10.3390/foods8060194
Chicago/Turabian StyleNagarajan, Vinod, Mengfei Peng, Zajeba Tabashsum, Serajus Salaheen, Joselyn Padilla, and Debabrata Biswas. 2019. "Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens" Foods 8, no. 6: 194. https://doi.org/10.3390/foods8060194
APA StyleNagarajan, V., Peng, M., Tabashsum, Z., Salaheen, S., Padilla, J., & Biswas, D. (2019). Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens. Foods, 8(6), 194. https://doi.org/10.3390/foods8060194



