Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Culture Conditions and Inoculum Preparation
2.2. Grape Pomace Extracts Medium Preparation
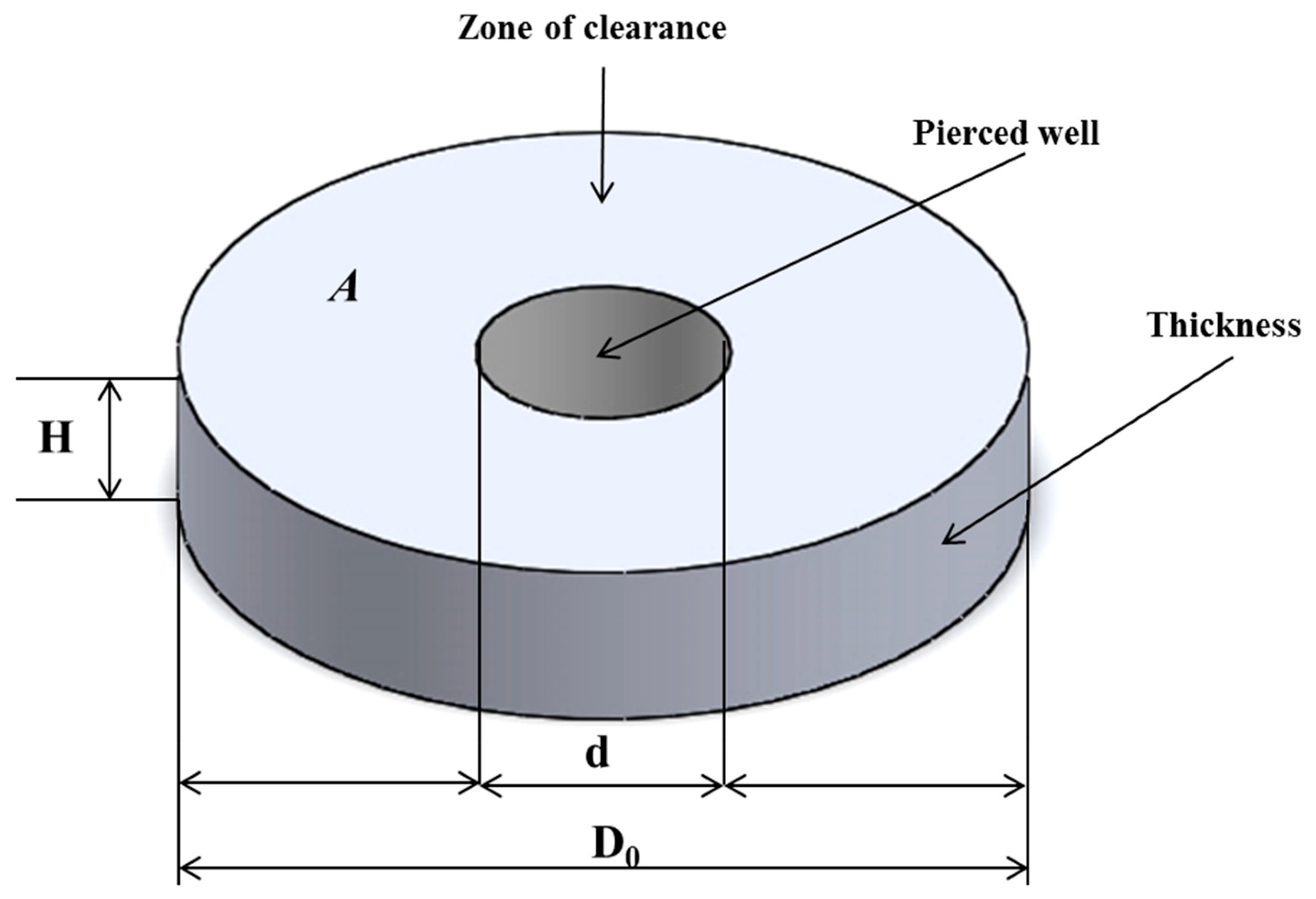
2.3. Concept of Volumetric Zone of Inhibition and Calculation
2.4. Preliminary Assessments for Biopreservation Compounds Production, Chemical Analysis and Growth Inhibition Assay
2.5. Kinetic Studies for Production of Potential Biopreservation Compounds from GPE
2.6. Response Surface Methodology (RSM) for the Optimization of Biopreservation Compounds Production using GPE Broth as Fermentation Medium
2.7. Identification and Quantification of VOCs Produced by C. pyralidae Y1117, P. kluyveri Y1125 and P. kluyveri Y1164
3. Results and Discussion
3.1. Growth Inhibition Assay on Beverage Spoilage Yeasts
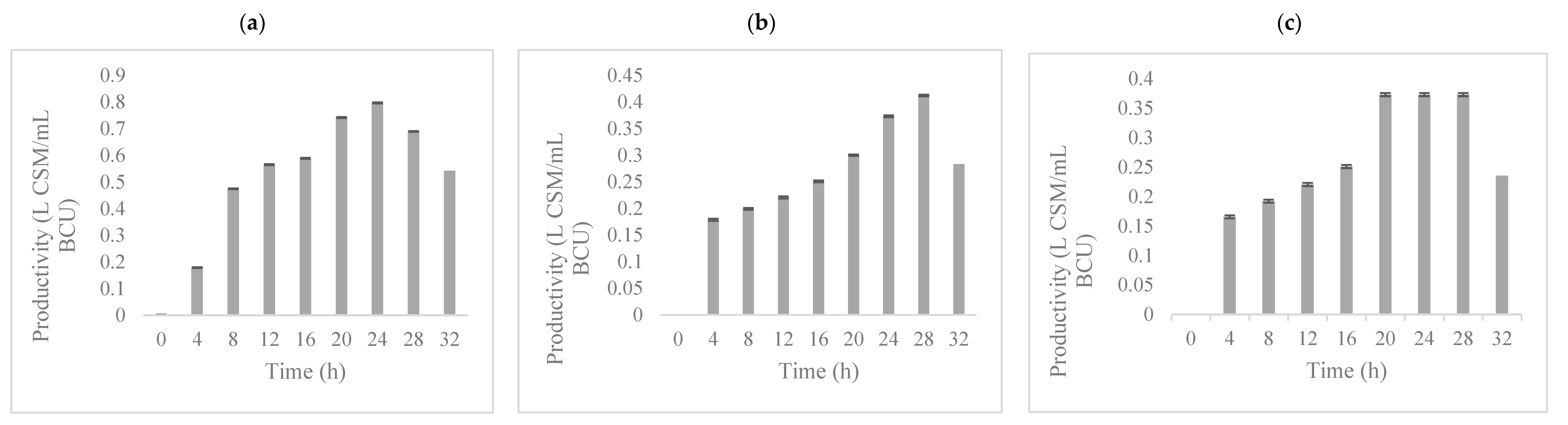
3.2. Fermentation Kinetics of Potential Biopreservation Compounds Produced in GPE Broth
3.3. Response Surface, Model Validation and Optimum Conditions for the Production of Biopreservation Compounds
3.4. Identification and Quantification of VOCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Salman, M.A.M. Biological Control of Rhizopus Soft Rot on Apple, Pear and Peach by Trichoderma harzianum. Ph.D. Thesis, An-Najah National University, Nablus, Palestine, 2005. [Google Scholar]
- Parveen, S.; Wani, A.H.; Bhat, M.Y.; Koka, J.A.; Wani, F.A. Management of postharvest fungal rot of peach (Prunus persica) caused by Rhizopus stolonifer in Kashmir Valley, India. Plant Pathol. Quar. 2016, 6, 19–29. [Google Scholar] [CrossRef]
- Abdullah, Q.; Mahmoud, A.; Al-Harethi, A. Isolation and identification of fungal post-harvest rot of some fruits in Yemen. PSM Microbiol. 2016, 1, 36–44. [Google Scholar]
- Comitini, F.; Di Pietro, N.; Zacchi, L.; Mannazzu, I.; Ciani, M. Kluyveromyces phaffii killer toxin active against wine spoilage yeasts: Purification and characterization. Microbiology 2004, 150, 2535–2541. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, M.; Pretorius, I.S. Microbial spoilage and preservation of wine: Using weapons from nature’s own arsenal—A review. S. Afr. J. Enol. Vitic. 2000, 21, 74–96. [Google Scholar]
- Sáez, J.S.; Lopes, C.A.; Kirs, V.C.; Sangorrín, M.P. Enhanced volatile phenols in wine fermented with Saccharomyces cerevisiae and spoiled with Pichia guilliermondii and Dekkera bruxellensis. Lett. Appl. Microbiol. 2010, 51, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Droby, S. Improving quality and safety of fresh fruits and vegetables after harvest by the use of biocontrol agents and natural materials. In Proceedings of the 1st International Symposium on Natural Preservatives in Food Systems, Princeton, NJ, USA, 30–31 March 2005; pp. 45–52. [Google Scholar]
- Ciani, M.; Fatichenti, F. Killer Toxin of Kluyveromyces phaffii DBVPG 6076 as a Biopreservative Agent to Control Apiculate Wine Yeasts. Appl. Environ. Microbiol. 2001, 67, 3058–3063. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; De, J.I.; Pepe, L.; Mannazzu, I.; Ciani, M. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 2004, 238, 235–240. [Google Scholar] [CrossRef]
- Mehlomakulu, N.N.; Setati, M.E.; Divol, B. Characterization of novel killer toxins secreted by wine-related non-Saccharomyces yeasts and their action on Brettanomyces spp. Int. J. Food Microbiol. 2014, 188, 83–91. [Google Scholar] [CrossRef]
- Grzegorczyk, M.; Żarowska, B.; Restuccia, C.; Cirvilleri, G. Postharvest biocontrol ability of killer yeasts against Monilinia fructigena and Monilinia fructicola on stone fruit. Food Microbiol. 2017, 61, 93–101. [Google Scholar] [CrossRef]
- Ngongang, M.M.; Ntwampe, S.K.O.; du Plessis, H.W.; Jolly, N.P.; Mekuto, L. Biopreservatives from yeasts with antimicrobial activity against common food, agricultural produce and beverage spoilage organisms. In Book Antimicrobial Research: Novel Bioknowledge and Educational Programs, Series No 6; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2017; pp. 219–228. [Google Scholar]
- Chanchaichaovivat, A.; Ruenwongsa, P.; Panijpan, B. Screening and identification of yeast strains from fruits and vegetables: Potential for biological control of postharvest chilli anthracnose (Colletotrichum capsici). Biol. Control 2007, 42, 326–335. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Wisniewski, M.; Droby, S.; Liu, Y. Review: Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 2013, 167, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Muccilli, S.; Restuccia, C. Bioprotective Role of Yeasts. Microorganisms 2015, 3, 588–611. [Google Scholar] [CrossRef] [PubMed]
- Lategan, B.W.; Pentz, C.D.; du Preez, R. Importance of wine attributes: A South African Generation Y perspective. Br. Food J. 2017, 119, 1536–1546. [Google Scholar] [CrossRef]
- Angadam, J.O.; Ntwampe, S.K.O.; Dlangamandla, N. Phanerochaete chrysosporium supported biovalorisation of grape pomace for hyper reducible sugar extraction. In Proceedings of the 10th International Conference on Advances in Science, Engineering, Technology & Healthcare (ASETH-18), Cape Town, South Africa, 19–20 November 2018; pp. 190–194. [Google Scholar] [CrossRef]
- He, X.; Sun, F.; Zhao, H.; Liu, M.; Gu, L. Optimization of fermentation of potato starch by RSM during itanonic acid production. In Proceedings of the World Automation Congress (WAC), Puerto Vallarta, Mexico, 24–28 June 2012; pp. 1–4. [Google Scholar]
- Galonde, N.; Brostaux, Y.; Richard, G.; Nott, K.; Jerôme, C.; Fauconnier, M.-L. Use of response surface methodology for the optimization of the lipase-catalyzed synthesis of mannosyl myristate in pure ionic liquid. Process Biochem. 2013, 48, 1914–1920. [Google Scholar] [CrossRef]
- Uzoh, C.F.; Onukwuli, O.D.; Nwabanne, J.T. Characterization, kinetics and statistical screening analysis of gmelina seed oil extraction process. Mater. Renew. Sustain. Energy 2014, 3, 38. [Google Scholar] [CrossRef]
- Nwabueze, T.U. Review article: Basic steps in adapting response surface methodology as mathematical modelling for bioprocess optimisation in the food systems. Int. J. Food Sci. Technol. 2010, 45, 1768–1776. [Google Scholar] [CrossRef]
- Demirel, M.; Kayan, B. Application of response surface methodology and central composite design for the optimization of textile dye degradation by wet air oxidation. Int. J. Ind. Chem. 2012, 3, 24. [Google Scholar] [CrossRef]
- Mewa-Ngongang, M.; du Plessis, H.W.; Hutchinson, U.F.; Mekuto, L.; Ntwampe, S.K. Kinetic modelling and optimisation of antimicrobial compound production by Candida pyralidae KU736785 for control of Candida Guilliermondii. Food Sci. Technol. Int. 2017, 23, 358–370. [Google Scholar] [CrossRef]
- Malthus, T.R. The Works of Thomas Robert Malthus; Wrigley, E.A., Soudenlondon, D., Eds.; William Pickering: London, UK, 1986. [Google Scholar]
- Cheigh, C.-I.; Choi, H.-J.; Park, H.; Kim, S.-B.; Kook, M.-C.; Kim, T.-S.; Hwang, J.-K.; Pyun, Y.-R. Influence of growth conditions on the production of a nisin-like bacteriocin by Lactococcus lactis subsp. lactis A164 isolated from kimchi. J. Biotechnol. 2002, 95, 225–235. [Google Scholar] [CrossRef]
- Messens, W.; De Vuyst, L. Inhibitory substances produced by Lactobacilli isolated from sourdoughs—A review. Int. J. Food Microbiol. 2002, 72, 31–43. [Google Scholar] [CrossRef]
- Narendranath, N.V.; Power, R. Relationship between pH and medium dissolved solids in terms of growth and metabolism of lactobacilli and Saccharomyces cerevisiae during ethanol production. Appl. Environ. Microbiol. 2005, 71, 2239–2243. [Google Scholar] [CrossRef] [PubMed]
- Masoud, W.; Poll, L.; Jakobsen, M. Influence of volatile compounds produced by yeasts predominant during processing of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. Yeast 2005, 22, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, G.Q.; Zhang, J.; Yang, L.; Che, H.J.; Jiang, D.H.; Huang, H.C. Control of postharvest Botrytis fruit rot of strawberry by volatile organic compounds of Candida intermedia. Phytopathology 2011, 101, 859–869. [Google Scholar] [CrossRef] [PubMed]



| Biopreservation Compounds Producer Strains | Beverage Spoilage Strains |
|---|---|
| Candida pyralidae Y1117 | Dekkera anomala |
| Pichia kluyveri Y1125 | Dekkera bruxellensis |
| Pichia kluyveri Y1164 | Zygosaccharomyces bailii |
| Fermentation Parameters | Model | Antimicrobial Compound Producing Yeasts | ||
|---|---|---|---|---|
| C. pyralidae Y1117 | P. kluyveri Y1125 | P. kluyveri Y1164 | ||
| Substrate (total sugar) utilization rate (g L−1 h−1) | 0.333 | 1.912 | 1.947 | |
| Biomass formation rate (×107 cells mL−1 h−1) | 4.542 | 5.208 | 3.917 | |
| Biomass yield (×108 cells g−1) | 1.365 | 0.272 | 0.201 | |
| Specific growth rate (h−1) | 0.196 | 0.202 | 0.190 | |
| Biopreservation compound formation rate (×103 L VZI mL−1 BCU h−1) | 33.209 | 15.547 | 15.547 | |
| Biopreservation compound formation based on cell concentration (×10−12 L VZI cells−1) | 73.121 | 29.850 | 39.694 | |
| Biopreservation compound formation based on substrate (total sugar) utilization (×10−3 L VZI g−1) | 99.840 | 8.130 | 7.985 | |
| Total sugar utilization rate (g L−1 h−1) proportional to cellular growth and formation of biopreservation compounds | 0.333 | 1.912 | 1.947 | |
| VOCs and Concentrations (mg/L) | |||
|---|---|---|---|
| Compound | C. pyralidae Y1117 | P. kluyveri Y1125 | P. kluyveri Y1164 |
| Isoamyl acetate | not detected | 16.51 | 17.73 |
| Isoamyl alcohol | 1.73 | 1.74 | 1.89 |
| Butyric acid | 1.24 | 1.25 | 1.25 |
| 2-Phenyl ethylacetate | 1.47 | 1.97 | 1.99 |
| Hexanoic acid | 0.93 | 0.93 | 0.93 |
| 2-Phenyl ethanol | 1.61 | 1.66 | 1.68 |
| Octanoic acid | 1.32 | 1.32 | 1.32 |
| Decanoic acid | 1.44 | 1.44 | 1.44 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mewa-Ngongang, M.; du Plessis, H.W.; Ntwampe, S.K.O.; Chidi, B.S.; Hutchinson, U.F.; Mekuto, L.; Jolly, N.P. Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds. Foods 2019, 8, 51. https://doi.org/10.3390/foods8020051
Mewa-Ngongang M, du Plessis HW, Ntwampe SKO, Chidi BS, Hutchinson UF, Mekuto L, Jolly NP. Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds. Foods. 2019; 8(2):51. https://doi.org/10.3390/foods8020051
Chicago/Turabian StyleMewa-Ngongang, Maxwell, Heinrich W. du Plessis, Seteno K. O. Ntwampe, Boredi S. Chidi, Ucrecia F. Hutchinson, Lukhanyo Mekuto, and Neil P. Jolly. 2019. "Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds" Foods 8, no. 2: 51. https://doi.org/10.3390/foods8020051
APA StyleMewa-Ngongang, M., du Plessis, H. W., Ntwampe, S. K. O., Chidi, B. S., Hutchinson, U. F., Mekuto, L., & Jolly, N. P. (2019). Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds. Foods, 8(2), 51. https://doi.org/10.3390/foods8020051









