Celiac Antigenicity of Ancient Wheat Species
Abstract
1. Introduction
2. Materials and Methods
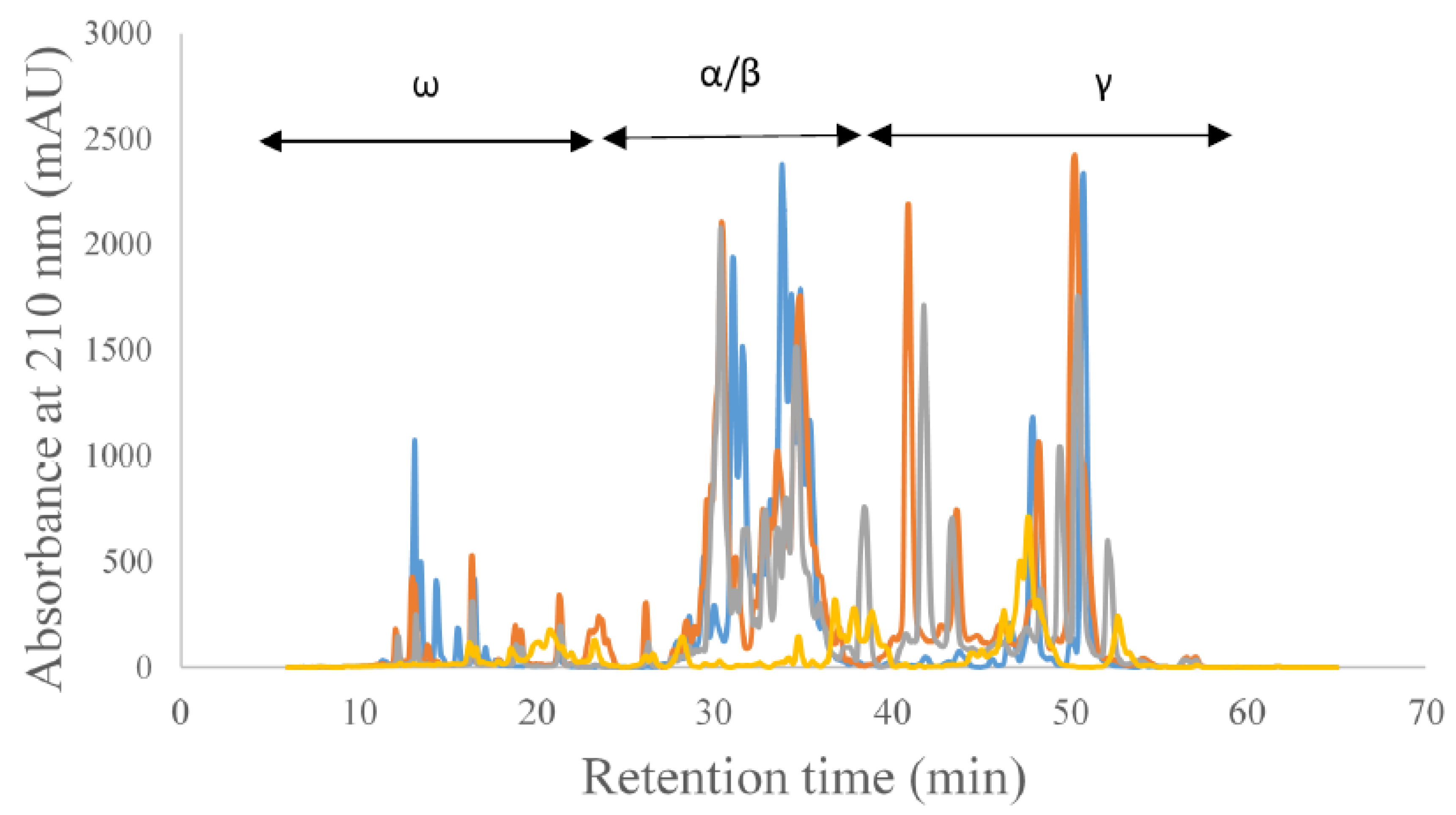
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shewry, P.R.; Gebruers, K.; Andersson, A.A.M.; Åman, P.; Piironen, V.; Lampi, A.-M.; Boros, D.; Rakszegi, M.; Bedő, Z.; Ward, J.L. Relationship between the contents of bioactive components in grain and the release dates of wheat lines in the HEALTHGRAIN diversity screen. J. Agric. Food Chem. 2011, 59, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Pellny, T.K.; Lovegrove, A. Is modern wheat bad for health? Nat. Plants 2016, 2, 16097. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Corol, D.I.; Jones, H.D.; Beale, M.H.; Ward, J.L. Defining genetic and chemical diversity in wheat grain by 1H-NMR spectroscopy of polar metabolites. Mol. Nutr. Food Res. 2017, 61, 1600807. [Google Scholar] [CrossRef] [PubMed]
- Malalgoda, M.; Simsek, S. Celiac disease and cereal proteins. Food Hydrocoll. 2017, 68, 108–113. [Google Scholar] [CrossRef]
- Zanini, B.; Villanacci, V.; De Leo, L.; Lanzini, A. Triticum monococcum in patients with celiac disease: A phase II open study on safety of prolonged daily administration. Eur. J. Nutr. 2015, 54, 1027–1029. [Google Scholar] [CrossRef] [PubMed]
- Pizzuti, D.; Buda, A.; D’Odorico, A.; D’Inca, R.; Chiarelli, S.; Curioni, A.; Martines, D. Lack of intestinal mucosal toxicity of Triticum monococcum in celiac disease patients. Scand. J. Gastroenterol. 2006, 41, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Colomba, M.S.; Gregorini, A. Are ancient durum wheats less toxic to celiac patients? A study of alpha-gliadin from Graziella ra and Kamut. Sci. World J. 2012, 8. [Google Scholar] [CrossRef]
- Dubois, B.; Bertin, P.; Mingeot, D. Molecular diversity of alpha-gliadin expressed genes in genetically contrasted spelt (Triticum aestivum ssp. spelta) accessions and comparison with bread wheat (T. aestivum ssp. aestivum) and related diploid Triticum and Aegilops species. Mol. Breed. New Strateg. Plant Improv. 2016, 36, 152. [Google Scholar] [CrossRef]
- Dubois, B.; Bertin, P.; Hautier, L.; Muhovski, Y.; Escarnot, E.; Mingeot, D. Genetic and environmental factors affecting the expression of alpha-gliadin canonical epitopes involved in celiac disease in a wide collection of spelt (Triticum aestivum ssp. spelta) cultivars and landraces. BMC Plant Biol. 2018, 18, 262. [Google Scholar] [CrossRef] [PubMed]
- Escarnot, E.; Gofflot, S.; Sinnaeve, G.; Dubois, B.; Bertin, P.; Mingeot, D. Reactivity of gluten proteins from spelt and bread wheat accessions towards A1 and G12 antibodies in the framework of celiac disease. Food Chem. 2018, 268, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H.; Koehler, P. The biochemical basis of celiac disease. Cereal Chem. 2008, 85, 1–13. [Google Scholar] [CrossRef]
- Malalgoda, M.; Manthey, F.; Simsek, S. Reducing the celiac disease antigenicity of wheat. Cereal Chem. 2018, 95, 49–58. [Google Scholar] [CrossRef]
- Tanner, G.J.; Blundell, M.J.; Colgrave, M.L.; Howitt, C.A. Creation of the first ultra-low gluten barley (Hordeum vulgare L.) for coeliac and gluten-intolerant populations. Plant Biotechnol. J. 2016, 14, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Malalgoda, M.; Meinhardt, S.W.; Simsek, S. Detection and quantitation of immunogenic epitopes related to celiac disease in historical and modern hard red spring wheat cultivars. Food Chem. 2018, 264, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Lin-Moshier, Y.; Sebastian, P.J.; Higgins, L.; Sampson, N.D.; Hewitt, J.E.; Marchant, J.S. Re-evaluation of the Role of Calcium Homeostasis Endoplasmic Reticulum Protein (CHERP) in Cellular Calcium Signaling. J. Biol. Chem. 2013, 288, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Qiao, S.W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Tatham, A.S. Improving wheat to remove coeliac epitopes but retain functionality. J. Cereal Sci. 2016, 67, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; Stewart, J.A.; Dromey, J.A.; Beissbarth, T.; van Heel, D.A.; Tatham, A.; Henderson, K.; Mannering, S.I.; Gianfrani, C.; Jewell, D.P.; et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2010, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Gessendorfer, B.; Koehler, P.; Wieser, H. Preparation and characterization of enzymatically hydrolyzed prolamins from wheat, rye, and barley as references for the immunochemical quantitation of partially hydrolyzed gluten. Anal. Bioanal. Chem. 2009, 395, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
| Immunogenic Peptide | Amaranth | Einkorn | Emmer | Kamut | Quinoa | Rye | Sorghum | Teff | |
|---|---|---|---|---|---|---|---|---|---|
| Type of Gliadin | Sequence * | ||||||||
| Alpha gliadin | FRPQQPYPQ | ✓ | ✓ | ✓ | |||||
| PFPQPQLPY | ✓ | ✓ | |||||||
| PYPQPQLPY | |||||||||
| PQPQLPYPQ | |||||||||
| QGSFQPSQQ | ✓ | ✓ | |||||||
| Gamma gliadin | PQQSFPQQQ | ||||||||
| IQPQQPAQL | ✓ | ||||||||
| QQPQQPYPQ | |||||||||
| SQPQQQFPQ | |||||||||
| PQPQQQFPQ | ✓ | ||||||||
| QQPQQPFPQ | |||||||||
| PQPQQPFCQ | |||||||||
| QQPFPQQPQ | ✓ | ||||||||
| Omega gliadin | PFPQPQQPF | ||||||||
| PQPQQPFPW | |||||||||
| Omega secalin | PFPQPQQPF | ✓ | |||||||
| PQPQQPFPQ | |||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malalgoda, M.; Ohm, J.-B.; Simsek, S. Celiac Antigenicity of Ancient Wheat Species. Foods 2019, 8, 675. https://doi.org/10.3390/foods8120675
Malalgoda M, Ohm J-B, Simsek S. Celiac Antigenicity of Ancient Wheat Species. Foods. 2019; 8(12):675. https://doi.org/10.3390/foods8120675
Chicago/Turabian StyleMalalgoda, Maneka, Jae-Bom Ohm, and Senay Simsek. 2019. "Celiac Antigenicity of Ancient Wheat Species" Foods 8, no. 12: 675. https://doi.org/10.3390/foods8120675
APA StyleMalalgoda, M., Ohm, J.-B., & Simsek, S. (2019). Celiac Antigenicity of Ancient Wheat Species. Foods, 8(12), 675. https://doi.org/10.3390/foods8120675





