High-Throughput 16S rRNA Sequencing to Assess Potentially Active Bacteria and Foodborne Pathogens: A Case Example in Ready-to-Eat Food
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Processing
2.2. Microbial Cultures
2.3. Nucleic Acid Extraction and RNA Purification
2.4. PCR and RT-PCR of 16S rRNA/rDNA and Illumina Amplicon Sequencing
2.5. Real-Time PCR of Ail and FoxA Genes of Yersinia Enterocolitica
3. Results
3.1. Microbial Cultures
3.2. High-Throughput Sequencing of 16S rRNA
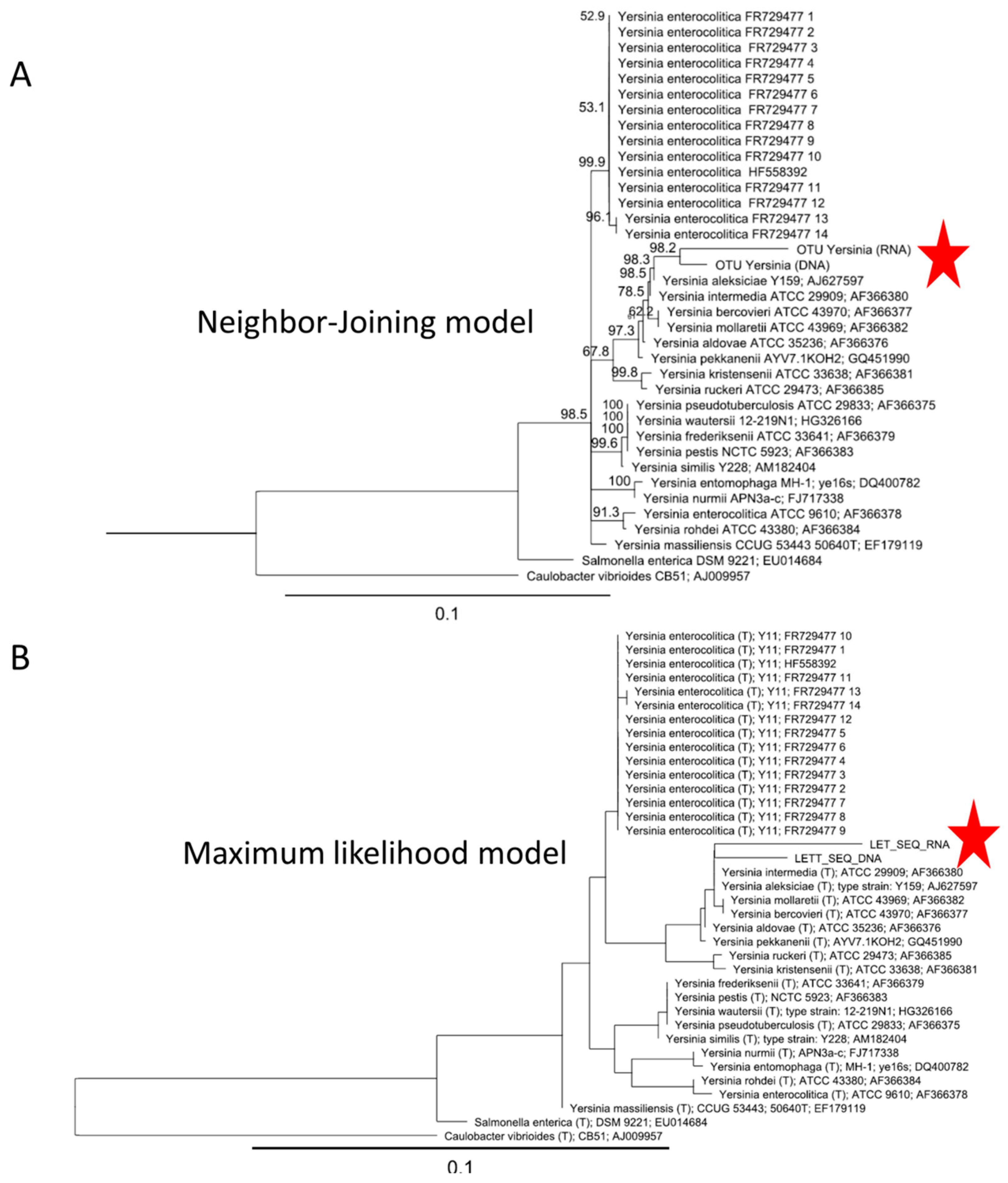
3.3. Seeking Foodborne Pathogens in the Molecular Data
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castro-Ibáñez, I.; Gil, M.I.; Allende, A. Ready-to-eat vegetables: Current problems and potential solutions to reduce microbial risk in the production chain. LWT-Food Sci. Technol. 2017, 85, 284–292. [Google Scholar] [CrossRef]
- Mercanoglu Taban, B.; Halkman, A.K. Do leafy green vegetables and their ready-to-eat [RTE] salads carry a risk of foodborne pathogens? Anaerobe 2011, 17, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Callejón, R.M.; Rodríguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported Foodborne Outbreaks Due to Fresh Produce in the United States and European Union: Trends and Causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Sagoo, S.K.; Little, C.L.; Ward, L.; Gillespie, I.A.; Mitchell, R.T. Microbiological Study of Ready-to-Eat Salad Vegetables from Retail Establishments Uncovers a National Outbreak of Salmonellosis. J. Food Prot. 2003, 66, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Althaus, D.; Kiefer, S.; Lehner, A.; Hatz, C.; Schmutz, C.; Jost, M.; Gerber, N.; Baumgartner, A.; Hächler, H.; et al. Foodborne transmission of Listeria monocytogenes via ready-to-eat salad: A nationwide outbreak in Switzerland, 2013–2014. Food Control 2015, 57, 14–17. [Google Scholar] [CrossRef]
- Hardstaff, J.L.; Clough, H.E.; Lutje, V.; McIntyre, K.M.; Harris, J.P.; Garner, P.; O’Brien, S.J. Foodborne and Food-Handler Norovirus Outbreaks: A Systematic Review. Foodborne Pathog. Dis. 2018, 15, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Little, C.L.; Gillespie, I.A. Prepared salads and public health. J. Appl. Microbiol. 2008, 105, 1729–1743. [Google Scholar] [CrossRef] [PubMed]
- Caradonna, T.; Marangi, M.; Del Chierico, F.; Ferrari, N.; Reddel, S.; Bracaglia, G.; Normanno, G.; Putignani, L.; Giangaspero, A. Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 2017, 67, 67–75. [Google Scholar] [CrossRef]
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar] [CrossRef]
- Valderrama, W.B.; Dudley, E.G.; Doores, S.; Cutter, C.N. Commercially Available Rapid Methods for Detection of Selected Food-borne Pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56, 1519–1531. [Google Scholar] [CrossRef]
- Hoorfar, J. Rapid detection, characterization, and enumeration of foodborne pathogens. APMIS 2011, 119, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Noyes, N.R.; Doster, E.; Martin, J.N.; Linke, L.M.; Magnuson, R.J.; Yang, H.; Geornaras, I.; Woerner, D.R.; Jones, K.L.; et al. Use of Metagenomic Shotgun Sequencing Technology To Detect Foodborne Pathogens within the Microbiome of the Beef Production Chain. Appl. Environ. Microbiol. 2016, 82, 2433–2443. [Google Scholar] [CrossRef]
- Ercolini, D. High-throughput sequencing and metagenomics: Moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Ercolini, D. Zooming into food-associated microbial consortia: A ‘cultural’ evolution. Curr. Opin. Food Sci. 2015, 2, 43–50. [Google Scholar] [CrossRef]
- De Filippis, F.; Parente, E.; Zotta, T.; Ercolini, D. A comparison of bioinformatic approaches for 16S rRNA gene profiling of food bacterial microbiota. Int. J. Food Microbiol. 2018, 265, 9–17. [Google Scholar] [CrossRef]
- Flores, G.E.; Bates, S.T.; Caporaso, J.G.; Lauber, C.L.; Leff, J.W.; Knight, R.; Fierer, N. Diversity, distribution and sources of bacteria in residential kitchens. Environ. Microbiol. 2013, 15, 588–596. [Google Scholar] [CrossRef]
- Humblot, C.; Guyot, J.-P. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl. Environ. Microbiol. 2009, 75, 4354–4361. [Google Scholar] [CrossRef]
- Li, R.; Tun, H.M.; Jahan, M.; Zhang, Z.; Kumar, A.; Dilantha Fernando, W.G.; Farenhorst, A.; Khafipour, E. Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep. 2017, 7, 5752. [Google Scholar] [CrossRef]
- Zordan, M.D.; Grafton, M.M.G.; Acharya, G.; Reece, L.M.; Cooper, C.L.; Aronson, A.I.; Park, K.; Leary, J.F. Detection of pathogenic E. coli O157:H7 by a hybrid microfluidic SPR and molecular imaging cytometry device. Cytom. Part A 2009, 75, 155–162. [Google Scholar] [CrossRef]
- Flekna, G.; Štefanič, P.; Wagner, M.; Smulders, F.J.M.; Možina, S.S.; Hein, I. Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR. Res. Microbiol. 2007, 158, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, S.L.; Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Duan, R.; Liang, J.-R.; Huang, Y.; Xiao, Y.-C.; Qiu, H.-Y.; Wang, X.; Jing, H.-Q. Real-time TaqMan PCR for Yersinia enterocolitica detection based on the ail and foxA genes. J. Clin. Microbiol. 2014, 52, 4443–4444. [Google Scholar] [CrossRef]
- Buchholz, U.; Bernard, H.; Werber, D.; Böhmer, M.M.; Remschmidt, C.; Wilking, H.; Deleré, Y.; an der Heiden, M.; Adlhoch, C.; Dreesman, J.; et al. German Outbreak of Escherichia coli O104:H4 Associated with Sprouts. N. Engl. J. Med. 2011, 365, 1763–1770. [Google Scholar] [CrossRef]
- van Dyk, B.N.; de Bruin, W.; du Plessis, E.M.; Korsten, L. Microbiological Food Safety Status of Commercially Produced Tomatoes from Production to Marketing. J. Food Prot. 2016, 79, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef] [PubMed]
- Leff, J.W.; Fierer, N. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Flateland, S.L.; Hanssen, J.F.; Bengtsson, G.; Nissen, H. Development and evaluation of a 16S ribosomal DNA array-based approach for describing complex microbial communities in ready-to-eat vegetable salads packed in a modified atmosphere. Appl. Environ. Microbiol. 2002, 68, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Sitaraman, R. Pseudomonas spp. as models for plant-microbe interactions. Front. Plant Sci. 2015, 6, 787. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Grimont, F. The Genus Serratia. Annu. Rev. Microbiol. 2006, 32, 221–248. [Google Scholar] [CrossRef]
- Halpern, M.; Waissler, A.; Dror, A.; Lev-Yadun, S. Biological Warfare of the Spiny Plant: Introducing Pathogenic Microorganisms into Herbivore’s Tissues. Adv. Appl. Microbiol. 2011, 74, 97–116. [Google Scholar] [CrossRef]
- Althaus, D.; Hofer, E.; Corti, S.; Julmi, A.; Stephan, R. Bacteriological Survey of Ready-to-Eat Lettuce, Fresh-Cut Fruit, and Sprouts Collected from the Swiss Market. J. Food Prot. 2012, 75, 1338–1341. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef]
- Morin, N.J.; Gong, Z.; Li, X.-F. Reverse Transcription-Multiplex PCR Assay for Simultaneous Detection of Escherichia coli O157:H7, Vibrio cholerae O1, and Salmonella Typhi. Clin. Chem. 2004, 50, 2037–2044. [Google Scholar] [CrossRef]
- Brandt, J.; Albertsen, M. Investigation of Detection Limits and the Influence of DNA Extraction and Primer Choice on the Observed Microbial Communities in Drinking Water Samples Using 16S rRNA Gene Amplicon Sequencing. Front. Microbiol. 2018, 9, 2140. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.K.; Tsen, H.Y. Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of Salmonella in foods. J. Appl. Bacteriol. 1996, 80, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Cao, W.W.; Johnson, M.G. 16S rRNA-based probes and polymerase chain reaction method to detect Listeria monocytogenes cells added to foods. Appl. Environ. Microbiol. 1992, 58, 2827–2831. [Google Scholar] [PubMed]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M.; Shaharoona, B. Hiding in Fresh Fruits and Vegetables: Opportunistic Pathogens May Cross Geographical Barriers. Int. J. Microbiol. 2016, 2016, 4292417. [Google Scholar] [CrossRef] [PubMed]
- Abadias, M.; Usall, J.; Anguera, M.; Solsona, C.; Viñas, I. Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int. J. Food Microbiol. 2008, 123, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Godziszewska, J.; Guzek, D.; Pogorzelska, E.; Brodowska, M.; Górska-Horczyczak, E.; Sakowska, A.; Wojtasik-Kalinowska, I.; Gantner, M.; Wierzbicka, A. A simple method of the detection of pork spoilage caused by Rahnella aquatilis. LWT-Food Sci. Technol. 2017, 84, 248–255. [Google Scholar] [CrossRef]
- Jensen, N.; Varelis, P.; Whitfield, F.B. Formation of guaiacol in chocolate milk by the psychrotrophic bacterium Rahnella aquatilis. Lett. Appl. Microbiol. 2001, 33, 339–343. [Google Scholar] [CrossRef]
- Lindberg, A.M.; Ljungh, Å.; Ahrné, S.; Löfdahl, S.; Molin, G. Enterobacteriaceae found in high numbers in fish, minced meat and pasteurised milk or cream and the presence of toxin encoding genes. Int. J. Food Microbiol. 1998, 39, 11–17. [Google Scholar] [CrossRef]
- Ragaert, P.; Devlieghere, F.; Debevere, J. Role of microbiological and physiological spoilage mechanisms during storage of minimally processed vegetables. Postharvest Biol. Technol. 2007, 44, 185–194. [Google Scholar] [CrossRef]
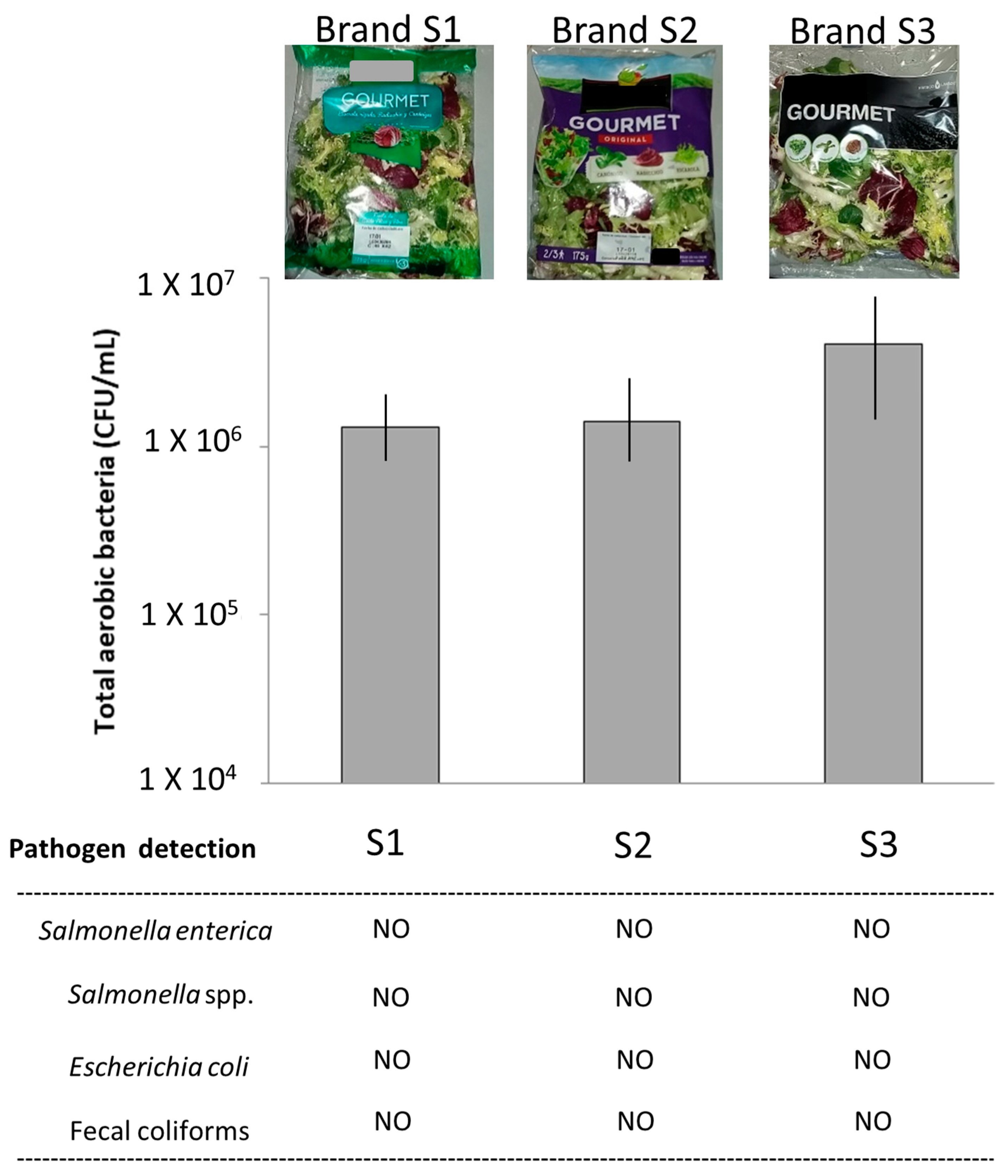
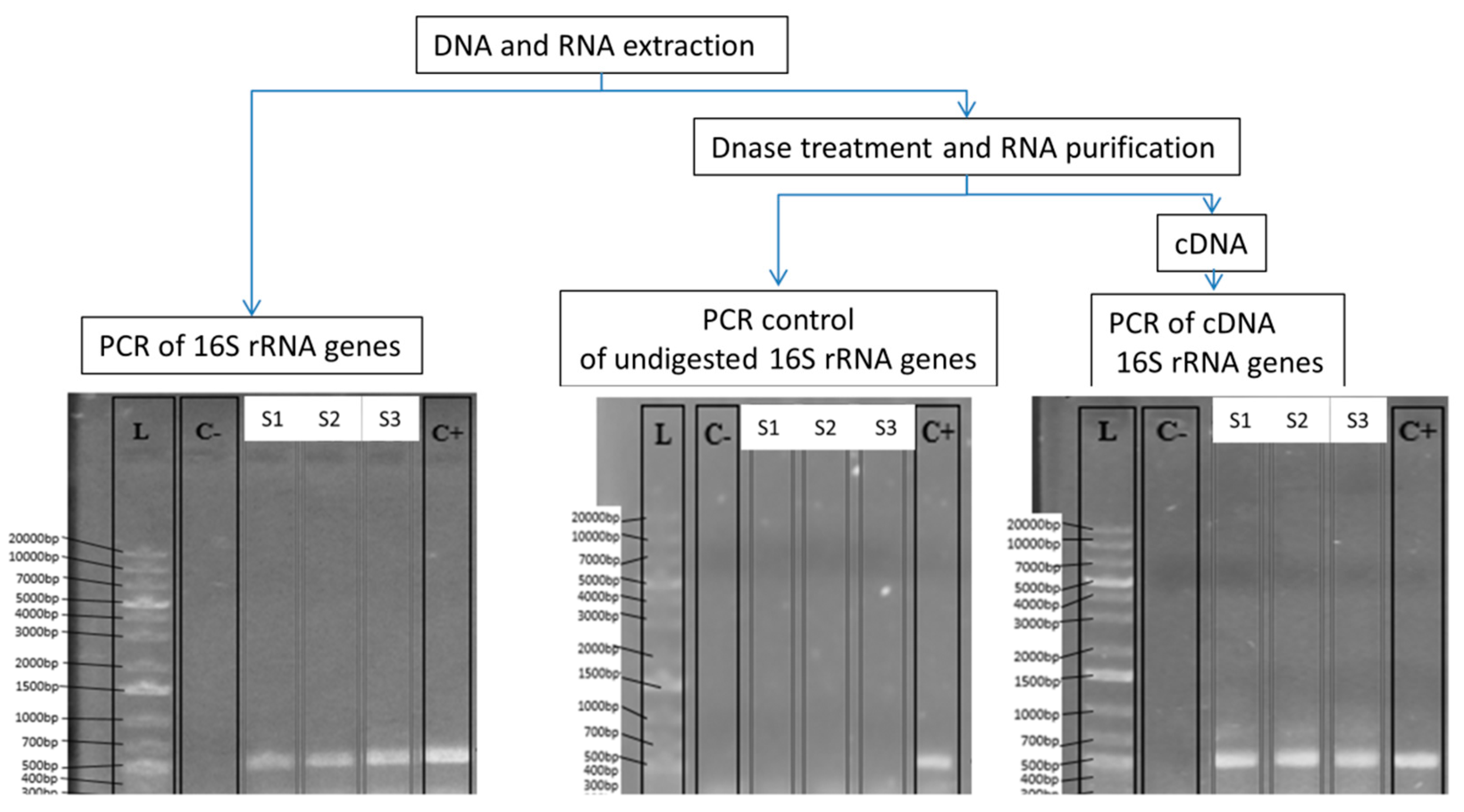
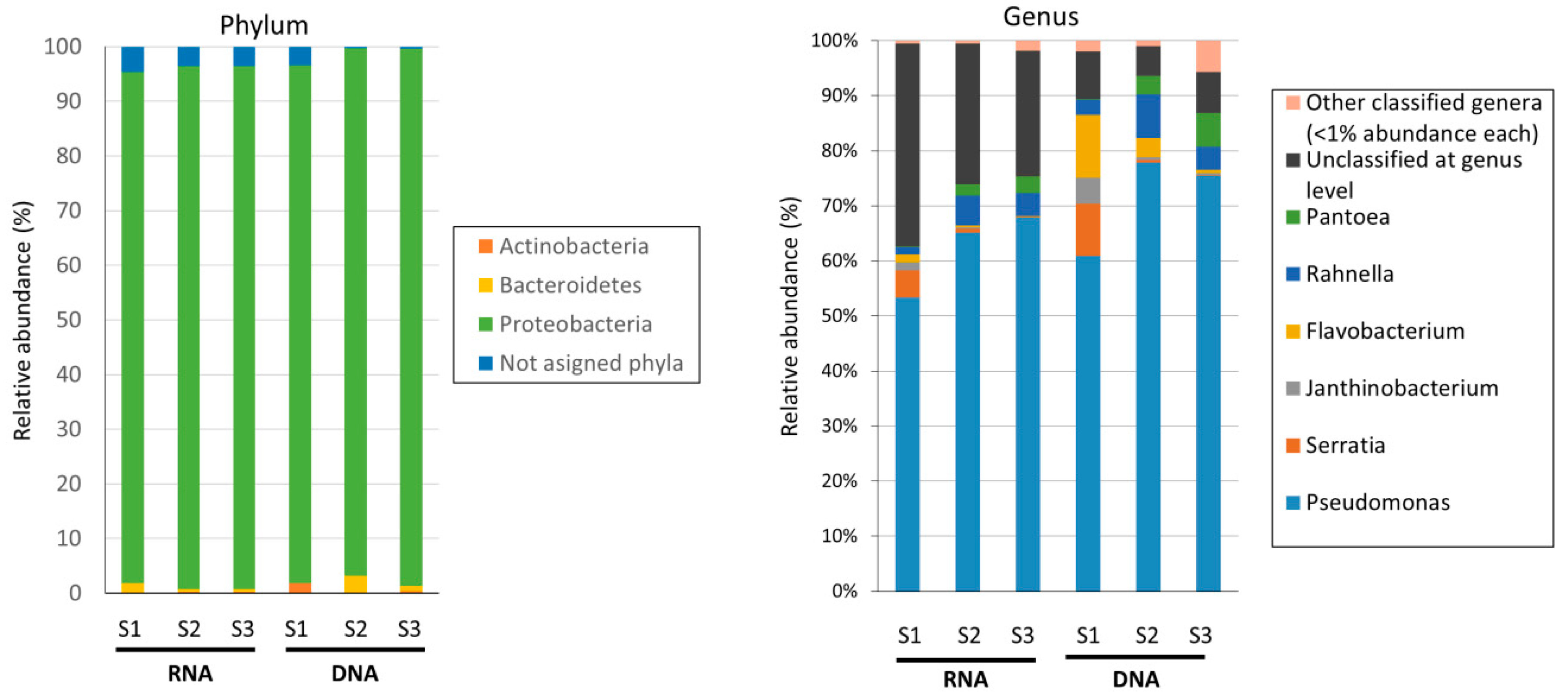
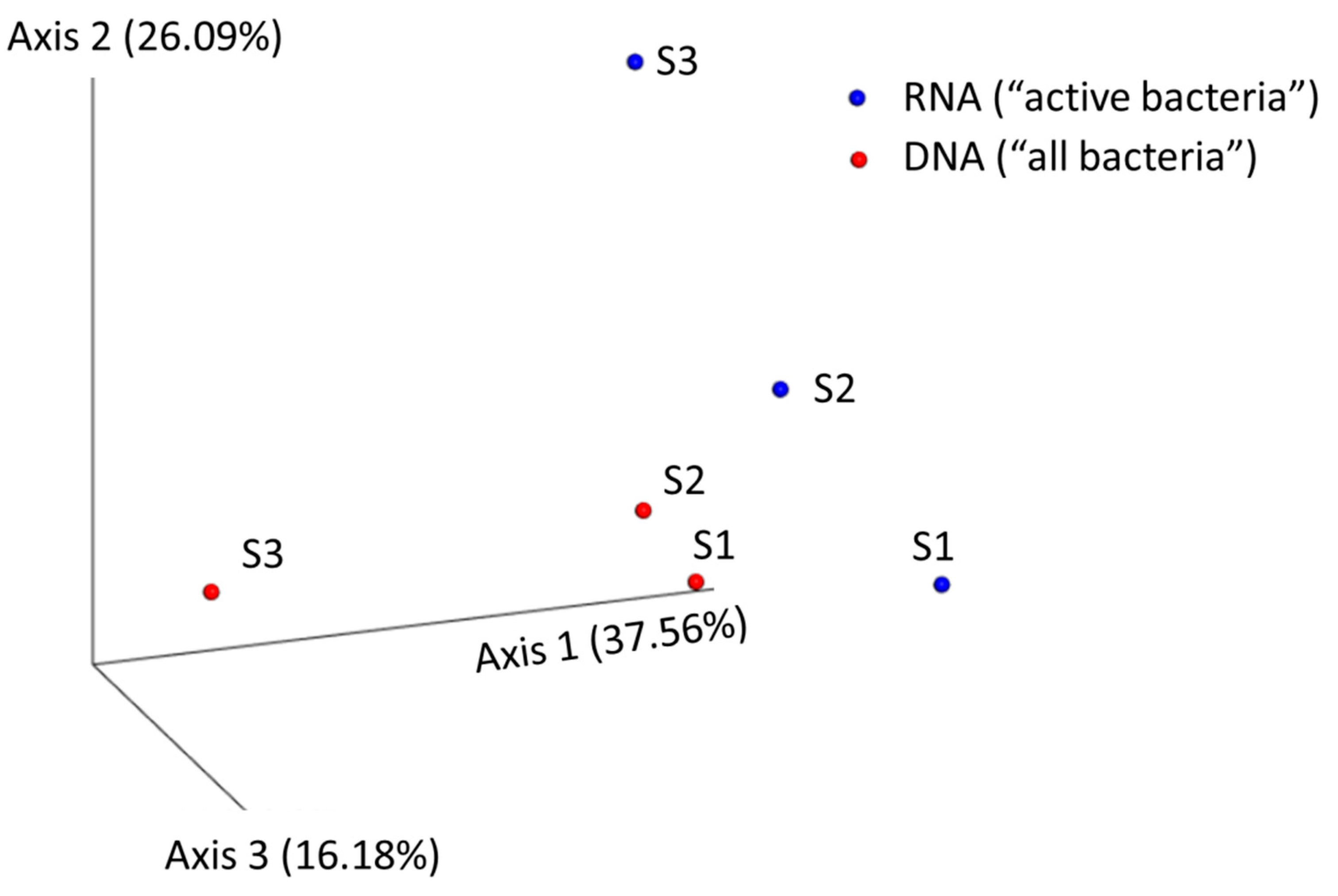
| Sample | No. of Reads | Average Length of Joined Reads (bp) | SD of Length | Detected Genera by Qiime |
|---|---|---|---|---|
| S1 (DNA) | 97,745 | 456.14 | 15.81 | 30 |
| S2 (DNA) | 78,252 | 454.41 | 16.17 | 32 |
| S3 (DNA) | 69,805 | 451.70 | 16.39 | 37 |
| S1 (RNA) | 86,122 | 238.47 | 166.95 | 28 |
| S2 (RNA) | 73,897 | 303.27 | 173.09 | 57 |
| S3 (RNA) | 58,758 | 338.65 | 167.04 | 46 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mira Miralles, M.; Maestre-Carballa, L.; Lluesma-Gomez, M.; Martinez-Garcia, M. High-Throughput 16S rRNA Sequencing to Assess Potentially Active Bacteria and Foodborne Pathogens: A Case Example in Ready-to-Eat Food. Foods 2019, 8, 480. https://doi.org/10.3390/foods8100480
Mira Miralles M, Maestre-Carballa L, Lluesma-Gomez M, Martinez-Garcia M. High-Throughput 16S rRNA Sequencing to Assess Potentially Active Bacteria and Foodborne Pathogens: A Case Example in Ready-to-Eat Food. Foods. 2019; 8(10):480. https://doi.org/10.3390/foods8100480
Chicago/Turabian StyleMira Miralles, Marina, Lucia Maestre-Carballa, Monica Lluesma-Gomez, and Manuel Martinez-Garcia. 2019. "High-Throughput 16S rRNA Sequencing to Assess Potentially Active Bacteria and Foodborne Pathogens: A Case Example in Ready-to-Eat Food" Foods 8, no. 10: 480. https://doi.org/10.3390/foods8100480
APA StyleMira Miralles, M., Maestre-Carballa, L., Lluesma-Gomez, M., & Martinez-Garcia, M. (2019). High-Throughput 16S rRNA Sequencing to Assess Potentially Active Bacteria and Foodborne Pathogens: A Case Example in Ready-to-Eat Food. Foods, 8(10), 480. https://doi.org/10.3390/foods8100480





