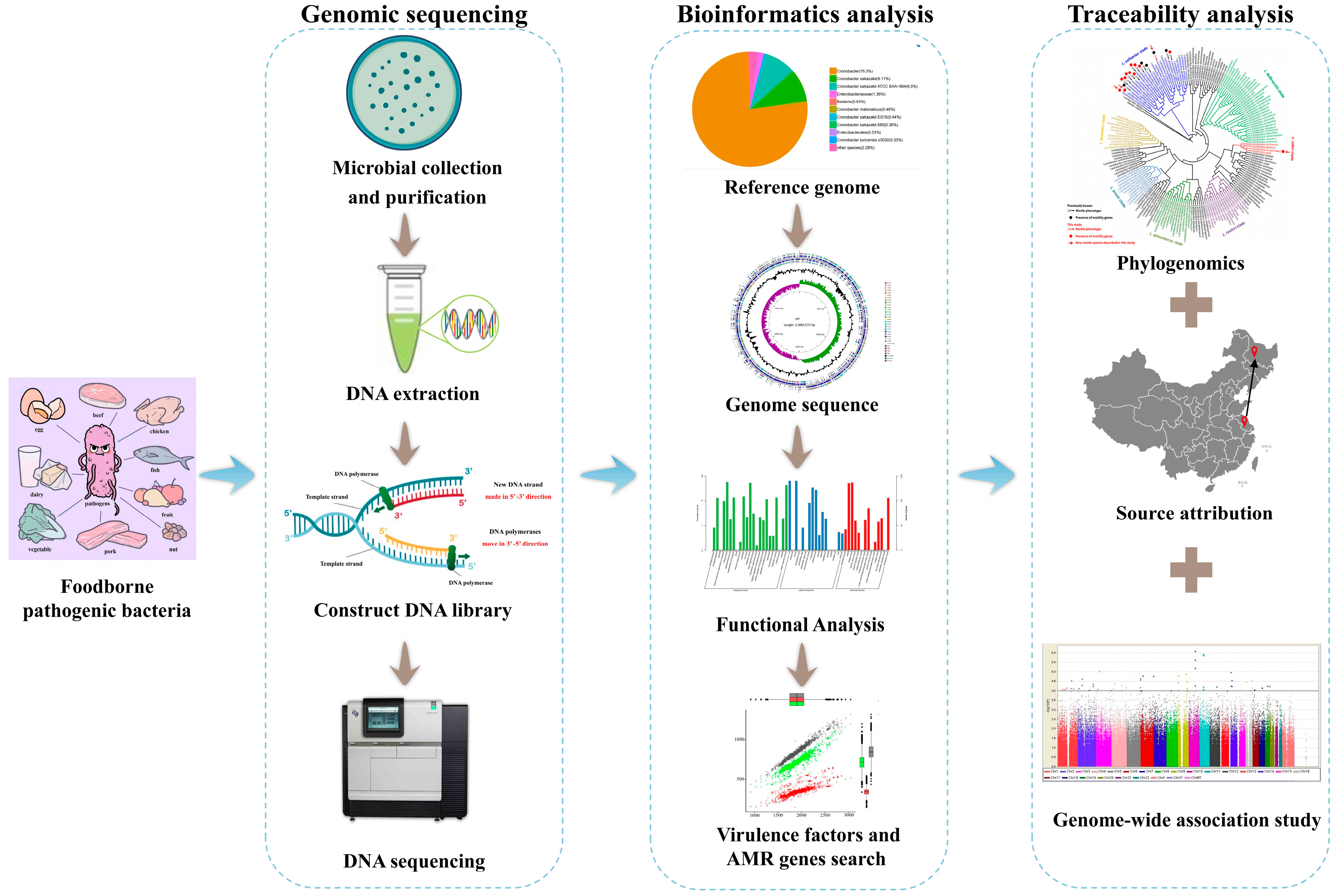
Significance of Whole-Genome Sequencing for the Traceability of Foodborne Pathogens: During the Processing of Meat and Dairy Products
Abstract
1. Introduction
2. Typing Traceability Technology
2.1. Traditional Technology
2.1.1. Biochemical Typing
2.1.2. Molecular Typing Techniques
| Methods | Advantages | Drawbacks | References | ||
|---|---|---|---|---|---|
| Traditional pathogen typing | Serotyping | Simple Fast Good repeatability | Difficult to find new antibody antigens Lower resolution No effective identification the pathogens | [31] | |
| Antibiotic sensitivity typing | Strain typing and evolutionary studies can be performed based on multi-drug resistance characteristics | The resistance genes can change Different strains may have same resistance map | [31] | ||
| Phage typing | Phage-lysing bacterial cells exhibit host specificity | Phage type incomplete Phage typing may have multiple results | [32] | ||
| Genotyping methods | Based on enzyme digestion technology | PFGE | High accuracy Good repeatability High typing rate The ‘gold standard’ for bacterial typing | Complex operation Take a long time Low sensitivity and specificity Affected by manmade mistakes | [33] |
| Based on PCR technology | RFLP | Good repeatability High stability High specificity | Cumbersome experimental operation Long testing period High cost Not convenient for large-scale operation | [25] | |
| AFLP | High sensitivity Good accuracy Good repeatability High information content | High requirements for personnel High quality of formwork Expensive | [27] | ||
| Based on sequencing technology | MLVA | High sensitivity Good repeatability Simple and efficient | Certain requirements for tandem repeat sequences High operational requirements for personnel screening sequences The sequences screened can affect the results | [34] | |
| MLST | Great typing ability Good repeatability Storable results Easy data sharing | High sequencing costs Genes may be mutated in conserved sequences | [35] | ||
| WGS | Greater discriminatory Complete information Analyze the source of foodborne pathogens Surveillance of foodborne pathogens | Large amount of data Lack of correct interpretation High cost of use and maintenance | [12] | ||
2.2. WGS Technology
2.2.1. The Development of Gene Sequencing Technology
2.2.2. The Development and Application of WGS
3. Application of WGS in the Traceability of Foodborne Pathogens
3.1. Application of WGS in the Meat Industry Chain
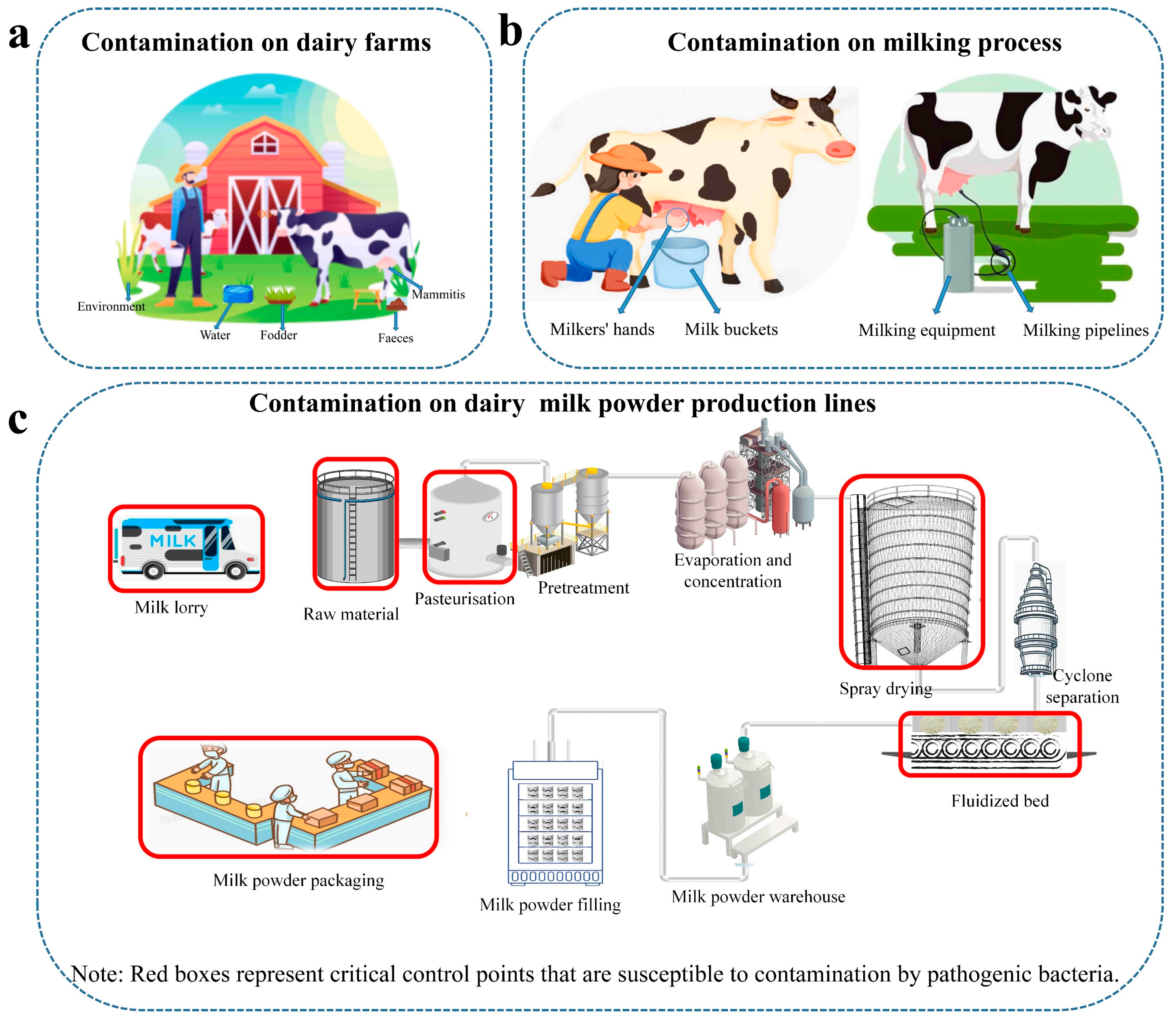
3.2. Application of WGS in the Dairy Chain
3.3. Other Applications of WGS
4. Challenges and Opportunities
- (1)
- Analysis of the vast amount of WGS data is necessary, but there are no reasonable solutions.
- (2)
- WGS methods and measurement orders need to be standardized.
- (3)
- Lack of sufficient epidemiological and food traceability evidence to properly interpret WGS findings.
- (4)
- WGS requires the selection of isolates for culture by traditional laboratory techniques.
- (5)
- The high cost of maintaining and operating WGS.
5. Conclusions and Prospect
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhaskar, S.V. Foodborne diseases—Disease burden—ScienceDirect. In Food Safety in the 21st Century; Academic Press: Cambridge, MA, USA, 2017; pp. 1–10. [Google Scholar] [CrossRef]
- Somorin, Y.M.; Odeyemi, O.A.; Ateba, C.N. Salmonella is the most common foodborne pathogen in African food exports to the European Union: Analysis of the Rapid Alert System for Food and Feed (1999–2019). Food Control 2021, 123, 107849. [Google Scholar] [CrossRef]
- Ortega, D.L.; Tschirley, D.L. Demand for food safety in emerging and developing countries: A research agenda for Asia and Sub-Saharan Africa. J. Agribus. Dev. Emerg. Econ. 2017, 7, 21–34. [Google Scholar] [CrossRef]
- Larsen, M.H.; Dalmasso, M.; Ingmer, H.; Langsrud, S.; Malakauskas, M.; Mader, A.; Møretrø, T.; Smole Možina, S.; Rychli, K.; Wagner, M.; et al. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control 2014, 44, 92–109. [Google Scholar] [CrossRef]
- Park, J.; Bae, D.; Kim, S.A. Microbial trace investigation throughout the entire chicken supply chain based on metagenomic high-throughput sequencing. Food Res. Int. 2023, 169, 112775. [Google Scholar] [CrossRef]
- Cuervo, M.P.; Castillo, A.; Santiago-Connolly, L.M. Overview of Biological Hazards and Foodborne Diseases. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar] [CrossRef]
- Mir, S.A.; Shah, M.A.; Mir, M.M.; Dar, B.N.; Greiner, R.; Roohinejad, S. Microbiological contamination of ready-to-eat vegetable salads in developing countries and potential solutions in the supply chain to control microbial pathogens. Food Control 2018, 85, 235–244. [Google Scholar] [CrossRef]
- Wei, X.; Zhao, X. Advances in typing and identification of foodborne pathogens. Curr. Opin. Food Sci. 2021, 37, 52–57. [Google Scholar] [CrossRef]
- Kang, X.; Wang, M.; Meng, C.; Li, A.; Jiao, X.; Pan, Z. Prevalence and whole-genome sequencing analysis of Salmonella reveal its spread along the duck production chain. Poult. Sci. 2022, 101, 101993. [Google Scholar] [CrossRef]
- Wellman, A.; Bazaco, M.C.; Blessington, T.; Pightling, A.; Dwarka, A.; Hintz, L.; Wise, M.E.; Gieraltowski, L.; Conrad, A.; Nguyen, T.-A.; et al. An Overview of Foodborne Sample-Initiated Retrospective Outbreak Investigations and Interagency Collaboration in the United States. J. Food Prot. 2023, 86, 100089. [Google Scholar] [CrossRef]
- Rantsiou, K.; Kathariou, S.; Winkler, A.; Skandamis, P.; Saint-Cyr, M.J.; Rouzeau-Szynalski, K.; Amézquita, A. Next generation microbiological risk assessment: Opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int. J. Food Microbiol. 2018, 287, 3–9. [Google Scholar] [CrossRef]
- Zhang, R.; Yang, T.; Zhang, Q.; Liu, D.; Elhadidy, M.; Ding, T. Whole-genome sequencing: A perspective on sensing bacterial risk for food safety. Curr. Opin. Food Sci. 2022, 47, 100888. [Google Scholar] [CrossRef]
- Allard, M.W.; Strain, E.; Rand, H.; Melka, D.; Correll, W.A.; Hintz, L.; Stevens, E.; Timme, R.; Lomonaco, S.; Chen, Y.; et al. Whole genome sequencing uses for foodborne contamination and compliance: Discovery of an emerging contamination event in an ice cream facility using whole genome sequencing. Infect. Genet. Evol. 2019, 73, 214–220. [Google Scholar] [CrossRef]
- Tong, S.; Ma, L.; Ronholm, J.; Hsiao, W.; Lu, X. Whole genome sequencing of Campylobacter in agri-food surveillance. Curr. Opin. Food Sci. 2021, 39, 130–139. [Google Scholar] [CrossRef]
- Lakicevic, B.; Jankovic, V.; Pietzka, A.; Ruppitsch, W. Wholegenome sequencing as the gold standard approach for control of Listeria monocytogenes in the food chain. J. Food Prot. 2023, 86, 100003. [Google Scholar] [CrossRef]
- Smith, A.M.; Tau, N.P.; Ngomane, H.M.; Sekwadi, P.; Ramalwa, N.; Moodley, K.; Govind, C.; Khan, S.; Archary, M.; Thomas, J. Whole-genome sequencing to investigate two concurrent outbreaks of Salmonella Enteritidis in South Africa, 2018. J. Med. Microbiol. 2020, 69, 1303–1307. [Google Scholar] [CrossRef]
- Ramadan, A.A. Bacterial typing methods from past to present: A comprehensive overview. Gene Rep. 2022, 29, 101675. [Google Scholar] [CrossRef]
- Song, D.; Su, Q.; Jia, A.; Fu, S.; Ma, X.; Li, T.; Man, C.; Yang, X.; Jiang, Y. A Method to Directly Identify Cronobacter sakazakii in Liquid Medium by MALDI-TOF MS. Foods 2023, 12, 1981. [Google Scholar] [CrossRef]
- Rossen, J.W.A.; Friedrich, A.W.; Moran-Gilad, J. Practical Issues in Implementing Whole-Genome-Sequencing in Routine Diagnostic Microbiology. Clin. Microbiol. Infect. 2017, 24, 355–360. [Google Scholar] [CrossRef]
- Yan, S.; Jiang, Z.; Zhang, W.; Liu, Z.; Dong, X.; Li, D.; Liu, Z.; Li, C.; Liu, X.; Zhu, L. Genomes-based MLST, cgMLST, wgMLST and SNP analysis of Salmonella Typhimurium from animals and humans. Comp. Immunol. Microb. 2023, 96, 101973. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef]
- Sabat, A.; Budimir, A.; Nashev, D.; Sa-Leao, R.; Friedrich, A. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance 2013, 18, 20380. [Google Scholar] [CrossRef] [PubMed]
- Goering, R.V. Pulsed field gel electrophoresis: A review of application and interpretation in the molecular epidemiology of infectious disease. Infect. Genet. Evol. 2010, 10, 866–875. [Google Scholar] [CrossRef]
- Tran, A.; Rowlinson, M.-C. Application of Next-Generation Sequencing in Public Health Epidemiology and Outbreak Investigation. Adv. Mol. Pathol. 2019, 2, 89–97. [Google Scholar] [CrossRef]
- Park, S.-H.; Jung, J.-H.; Seo, D.-H.; Lee, H.-L.; Kim, G.-W.; Park, S.-Y.; Shin, W.-C.; Hong, S.; Park, C.-S. Differentiation of lactic acid bacteria based on RFLP analysis of the tuf gene. Food Sci. Biotechnol. 2012, 21, 911–915. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Parisi, A.; Latorre, L.; Normanno, G.; Miccolupo, A.; Fraccalvieri, R.; Lorusso, V.; Santagada, G. Amplified Fragment Length Polymorphism and Multi-Locus Sequence Typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol. 2010, 27, 101–108. [Google Scholar] [CrossRef]
- Ranjbar, R.; Karami, A.; Farshad, S.; Giammanco, G.M.; Mammina, C. Typing methods used in the molecular epidemiology of microbial pathogens: A how-to guide. New Microbiol. 2014, 37, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lanini, S.; D’Arezzo, S.; Puro, V.; Martini, L.; Imperi, F.; Piselli, P.; Montanaro, M.; Paoletti, S.; Visca, P.; Ippolito, G. Molecular Epidemiology of a Pseudomonas aeruginosa Hospital Outbreak Driven by a Contaminated Disinfectant-Soap Dispenser. PLoS ONE 2011, 6, e17064. [Google Scholar] [CrossRef]
- Zhao, J.; Li, T.; Xu, Z.; Wang, Z.; Yang, S.; Chen, A. AFLP markers for meat traceability of cattle in the Chinese market. Food Control 2018, 91, 421–426. [Google Scholar] [CrossRef]
- Mir, I.A.; Kashyap, S.K.; Maherchandani, S. Isolation, serotype diversity and antibiogram of Salmonella enterica isolated from different species of poultry in India. Asian Pac. Trop. Biomed. 2015, 5, 561–567. [Google Scholar] [CrossRef]
- Boxrud, D.; Pederson-Gulrud, K.; Wotton, J.; Medus, C.; Lyszkowicz, E.; Besser, J.; Bartkus, J.M. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 2007, 45, 536–543. [Google Scholar] [CrossRef]
- Yang, B.; Qiao, L.; Zhang, X.; Cui, Y.; Xia, X.; Cui, S.; Wang, X.; Meng, X.; Ge, W.; Shi, X. Serotyping, antimicrobial susceptibility, pulse field gel electrophoresis analysis of Salmonella isolates from retail foods in Henan Province, China. Food Control 2013, 32, 228–235. [Google Scholar] [CrossRef]
- Ziebell, K.; Chui, L.; King, R.; Johnson, S.; Boerlin, P.; Johnson, R.P. Subtyping of Canadian isolates of Salmonella enteritidis using Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) alone and in combination with Pulsed-Field Gel Electrophoresis (PFGE) and phage typing. J. Microbiol. 2017, 139, 29–36. [Google Scholar] [CrossRef]
- Tourasse, N.J.; Jolley, K.A.; Kolstø, A.-B.; Økstad, O.A. Core genome multilocus sequence typing scheme for Bacillus cereus group bacteria. Res. Microbiol. 2023, 174, 104050. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; He, Q.; Zhang, W.; Zhang, H.; Sun, T. Single molecule, real-time sequencing technology improves the sensitivity for detecting bacteria in koumiss, a traditional fermented mare milk product. Sci. Bull. 2020, 65, 2065–2067. [Google Scholar] [CrossRef]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef]
- Allard, M.W.; Strain, E.; Melka, D.; Bunning, K.; Musser, S.M.; Brown, E.W.; Timme, R. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 2016, 54, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, G.; Agarwal, S.; Marshall, A.; Jones, D.H.; Sulaiman, I.M.; Sur, S.; Banerjee, P. Application of advanced genomic tools in food safety rapid diagnostics: Challenges and opportunities. Curr. Opin. Food Sci. 2022, 47, 100886. [Google Scholar] [CrossRef]
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 2018, 49, 224–229. [Google Scholar] [CrossRef]
- Joensen, K.G.; Kiil, K.; Gantzhorn, M.R.; Nauerby, B.; Engberg, J.; Holt, H.M.; Nielsen, H.L.; Petersen, A.M.; Kuhn, K.G.; Sandø, G. Whole-genome sequencing to detect numerous Campylobacter jejuni outbreaks and match patient isolates to sources, Denmark, 2015-2017. Emerg. Infect. Dis. 2020, 26, 523. [Google Scholar] [CrossRef] [PubMed]
- Stasiewicz, M.J.; Oliver, H.F.; Wiedmann, M.; den Bakker, H.C. Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl. Environ. Microbiol. 2015, 81, 6024–6037. [Google Scholar] [CrossRef]
- Thépault, A.; Méric, G.; Rivoal, K.; Pascoe, B.; Mageiros, L.; Touzain, F.; Rose, V.; Béven, V.; Chemaly, M.; Sheppard, S.K. Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl. Environ. Microbiol. 2017, 83, 03085–03116. [Google Scholar] [CrossRef]
- Franz, E.; Gras, L.M.; Dallman, T. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr. Opin. Food Sci. 2016, 8, 74–79. [Google Scholar] [CrossRef]
- Pightling, A.W.; Pettengill, J.B.; Luo, Y.; Baugher, J.D.; Rand, H.; Strain, E. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol. 2018, 9, 1482. [Google Scholar] [CrossRef]
- Crowe, S.J.; Green, A.; Hernandez, K.; Peralta, V.; Bottichio, L.; Defibaugh-Chavez, S.; Douris, A.; Gieraltowski, L.; Hise, K.; La-Pham, K.; et al. Utility of Combining Whole Genome Sequencing with Traditional Investigational Methods to Solve Foodborne Outbreaks of Salmonella Infections Associated with Chicken: A New Tool for Tackling This Challenging Food Vehicle. J. Food Prot. 2017, 80, 654–660. [Google Scholar] [CrossRef]
- Kase, J.A.; Zhang, G.; Chen, Y. Recent foodborne outbreaks in the United States linked to atypical vehicles-lessons learned. Curr. Opin. Food Sci. 2017, 18, 56–63. [Google Scholar] [CrossRef]
- Sachs, J.D.; Karim, S.S.A.; Aknin, L.; Allen, J.; Brosbøl, K.; Colombo, F.; Barron, G.C.; Espinosa, M.F.; Gaspar, V.; Gaviria, A.; et al. The Lancet Commission on lessons for the future from the COVID-19 pandemic. Lancet 2022, 400, 1224–1280. [Google Scholar] [CrossRef] [PubMed]
- Olenslager, K.; Yim, J.; Dickey, L.; Bueno, E.; Tifrea, D.F.; Crumpler, M.; Huang, S.; Gohil, S.K. Whole Genome Sequencing (WGS) for COVID-19 Outbreak Evaluations—How Much Does It Add to Bootstrap Epidemiology & Contact Tracing? Am. J. Infect. Control 2022, 50, S25. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Lei, C.-W.; Liu, S.-Y.; Chen, X.; Gao, Y.-F.; Zhang, Y.; Tang, Y.; Zhang, A.; Yang, X.; Wang, H.-N. Tracking Salmonella enterica by whole genome sequencing of isolates recovered from broiler chickens in a poultry production system. Int. J. Food Microbiol. 2021, 350, 109246. [Google Scholar] [CrossRef]
- Salaheen, S.; Chowdhury, N.; Hanning, I.; Biswas, D. Zoonotic bacterial pathogens and mixed crop-livestock farming. Poult. Sci. 2015, 94, 1398–1410. [Google Scholar] [CrossRef]
- Doyle, M.P.; Erickson, M.C. Reducing the Carriage of Foodborne Pathogens in Livestock and Poultry. Poult. Sci. 2006, 85, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ye, K.; Zhu, Y.; Huang, Y.; Wang, G.; Wang, H.; Zhou, G. Prevalence, antimicrobial resistance and genetic diversity of Listeria monocytogenes isolated from chilled pork in Nanjing, China. LWT Food Sci. Technol. 2015, 64, 905–910. [Google Scholar] [CrossRef]
- Morales-Partera, A.M.; Cardoso-Toset, F.; Luque, I.; Astorga, R.J.; Maldonado, A.; Herrera-León, S.; Hernández, M.; Gómez-Laguna, J.; Tarradas, C. Prevalence and diversity of Salmonella spp., Campylobacter spp., and Listeria monocytogenes in two free-range pig slaughterhouses. Food Control 2018, 92, 208–215. [Google Scholar] [CrossRef]
- Choi, Y.M.; Park, H.J.; Jang, H.I.; Kim, S.A.; Imm, J.Y.; Hwang, I.G.; Rhee, M.S. Changes in microbial contamination levels of porcine carcasses and fresh pork in slaughterhouses, processing lines, retail outlets, and local markets by commercial distribution. Res. Vet. Sci. 2013, 94, 413–418. [Google Scholar] [CrossRef]
- Arguello, H.; Sørensen, G.; Carvajal, A.; Baggesen, D.L.; Rubio, P.; Pedersen, K. Prevalence, serotypes and resistance patterns of Salmonella in Danish pig production. Res. Vet. Sci. 2013, 95, 334–342. [Google Scholar] [CrossRef]
- Hellstrom, S.; Laukkanen, R.; Siekkinen, K.-M.; Ranta, J.; Mauala, R.; Korkeala, H. Listeria monocytogenes Contamination in Pork Can Originate from Farms. J. Food Prot. 2010, 73, 641–648. [Google Scholar] [CrossRef]
- Hernández, M.; Gómez-Laguna, J.; Luque, I.; Herrera-León, S.; Maldonado, A.; Reguillo, L.; Astorga, R.J. Salmonella prevalence and characterization in a free-range pig processing plant: Tracking in trucks, lairage, slaughter line and quartering. Int. J. Food Microbiol. 2013, 162, 48–54. [Google Scholar] [CrossRef]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abdela, W.; Adesiyun, A.A. Antimicrobial Resistance of Salmonella Isolates Recovered from Chickens Sold at Retail Outlets in Trinidad. J. Food Prot. 2018, 81, 1880–1889. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abebe, W.; Adesiyun, A.A. Occurrence, Risk Factors, Serotypes, and Antimicrobial Resistance of Salmonella Strains Isolated from Imported Fertile Hatching Eggs, Hatcheries, and Broiler Farms in Trinidad and Tobago. J. Food Prot. 2022, 85, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Xiao, R.; Wu, C.; Zheng, Z.; Deng, Y.; Chen, K.; Xiang, Y.; Xu, C.; Zou, L.; Liao, M.; et al. Characterization of the prevalence of Salmonella in different retail chicken supply modes using genome-wide and machine-learning analyses. Food Res. Int. 2024, 191, 114654. [Google Scholar] [CrossRef]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Wang, P.; Robertson, I.; Abraham, S.; Sahibzada, S.; Habib, I. Antimicrobial resistance and genomic characterisation of Escherichia coli isolated from caged and non-caged retail table eggs in Western Australia. Int. J. Food Microbiol. 2021, 340, 109054. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, R.; Carraro, L.; Pauletto, M.; Gallo, M.; Andreani, N.A.; Weiss, G.; Tessaro, C.; Babbucci, M.; Cardazzo, B. Molecular typing and genome sequencing allow the identification of persistent Listeria monocytogenes strains and the tracking of the contamination source in food environments. Int. J. Food Microbiol. 2023, 386, 110025. [Google Scholar] [CrossRef]
- Wakabayashi, Y.; Takemoto, K.; Iwasaki, S.; Yajima, T.; Kido, A.; Yamauchi, A.; Kuroiwa, K.; Kumai, Y.; Yoshihara, S.; Tokumoto, H.; et al. Isolation and characterization of Staphylococcus argenteus strains from retail foods and slaughterhouses in Japan. Int. J. Food Microbiol. 2022, 363, 109503. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Baltazar, A.; Godínez-Oviedo, A.; Segura-García, L.E.; Hernández-Pérez, C.F.; Hernández-Iturriaga, M.; Cabrera-Díaz, E. Genomic diversity of Salmonella enterica isolated from raw chicken at retail establishments in Mexico. Int. J. Food Microbiol. 2024, 411, 110526. [Google Scholar] [CrossRef]
- Atlaw, N.A.; Keelara, S.; Correa, M.; Foster, D.; Gebreyes, W.; Aidara-Kane, A.; Harden, L.; Thakur, S.; Fedorka-Cray, P.J. Evidence of sheep and abattoir environment as important reservoirs of multidrug resistant Salmonella and extended-spectrum beta-lactamase Escherichia coli. Int. J. Food Microbiol. 2022, 363, 109516. [Google Scholar] [CrossRef]
- Palacios-Gorba, C.; Moura, A.; Markovich, Y.; Tessaud-Rita, N.; Gómez-Martín, Á.; Bracq-Dieye, H.; Gomis, J.; Vales, G.; Pastor-Martín, M.; Thouvenot, P.; et al. Genomic characterization of Listeria spp. isolated from tonsils, udder and feces of domestic dairy ruminants in Spain. Microbes Infect. 2023, 25, 105079. [Google Scholar] [CrossRef]
- Molina, A.; Thye, T.; Muñoz-Vargas, L.; Zamora-Sanabria, R.; Chercos, D.H.; Hernández-Rojas, R.; Robles, N.; Aguilar, D.; May, J.; Dekker, D. Molecular characterization of antibiotic resistant Salmonella enterica across the poultry production chain in Costa Rica: A cross-sectional study. Int. J. Food Microbiol. 2024, 416, 110663. [Google Scholar] [CrossRef]
- Rossler, E.; Olivero, C.; Soto, L.P.; Frizzo, L.S.; Zimmermann, J.; Rosmini, M.R.; Sequeira, G.J.; Signorini, M.L.; Zbrun, M.V. Prevalence, genotypic diversity and detection of virulence genes in thermotolerant Campylobacter at different stages of the poultry meat supply chain. Int. J. Food Microbiol. 2020, 326, 108641. [Google Scholar] [CrossRef]
- Ricke, S.; Dawoud, T.; Kwon, Y. Application of Molecular Methods for Traceability of Foodborne Pathogens in Food Safety Systems. In Food Safety; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar] [CrossRef]
- Mylius, S.D.; Nauta, M.J.; Havelaar, A.H. Cross-Contamination During Food Preparation: A Mechanistic Model Applied to Chicken-borne Campylobacter. Risk Anal. 2007, 27, 803–813. [Google Scholar] [CrossRef]
- Lai, H.; Tang, Y.; Wang, Z.; Ren, F.; Kong, L.; Jiao, X.; Huang, J. Handling practice as a critical point influencing the transmission route of campylobacter throughout a commercial restaurant kitchen in China. Food Control 2022, 139, 109056. [Google Scholar] [CrossRef]
- Habib, I.; Harb, A.; Hansson, I.; Vågsholm, I.; Osama, W.; Adnan, S.; Anwar, M.; Agamy, N.; Boqvist, S. Challenges and opportunities towards the development of risk assessment at the consumer phase in developing countries—The case of Campylobacter cross-contamination during handling of raw chicken in two middle eastern countries. Pathogens 2020, 9, 62. [Google Scholar] [CrossRef]
- Cardoso, M.J.; Ferreira, V.; Truninger, M.; Maia, R.; Teixeira, P. Cross-contamination events of Campylobacter spp. in domestic kitchens associated with consumer handling practices of raw poultry. Int. J. Food Microbiol. 2021, 338, 108984. [Google Scholar] [CrossRef]
- Gplden, C.E.; Rothrock, M.J.; Mishra, A. Mapping foodborne pathogen contamination throughout the conventional and alternative poultry supply chains. Poult. Sci. 2021, 100, 101157. [Google Scholar] [CrossRef]
- Duchez, R.; Vingadassalon, N.; Merda, D.; Van Nieuwenhuysen, T.; Byrne, B.; Kourtis, C.; Nia, Y.; Hennekinne, J.-A.; Cavaiuolo, M. Genetic relatedness of Staphylococcus aureus isolates within food outbreaks by single nucleotide polymorphisms. Int. J. Food Microbiol. 2025, 433, 111115. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Chen, Q.; Liu, Y.; Qiao, Z.; Sang, S.; Zhang, J.; Zhan, S.; Wu, Z.; Liu, L. Research advances of advanced glycation end products in milk and dairy products: Formation, determination, control strategy and immunometabolism via gut microbiota. Food Chem. 2023, 417, 135861. [Google Scholar] [CrossRef]
- Oliver, S.P.; Jayarao, B.M.; Almeida, R.A. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathog. Dis. 2005, 2, 115–129. [Google Scholar] [CrossRef] [PubMed]
- da Costa Krewer, C.; Santos Amanso, E.; Veneroni Gouveia, G.; de Lima Souza, R.; da Costa, M.M.; Aparecido Mota, R. Resistance to antimicrobials and biofilm formation in Staphylococcus spp. isolated from bovine mastitis in the Northeast of Brazil. Trop. Anim. Health Prod. 2015, 47, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Jiang, Y.; Gobius, K.S. Key pathogenic bacteria associated with dairy foods: On-farm ecology and products associated with foodborne pathogen transmission. Int. Dairy J. 2018, 84, 28–35. [Google Scholar] [CrossRef]
- Hunt, K.; Butler, F.; Jordan, K. Factors affecting staphylococcal enterotoxin Cbovine production in milk. Int. Dairy J. 2014, 39, 41–46. [Google Scholar] [CrossRef]
- Tracy, S.; Kock, M.M.; Ehlers, M.M. Molecular Characterization of Staphylococcus aureus Isolated from Bovine Mastitis and Close Human Contacts in South African Dairy Herds: Genetic Diversity and Inter-Species Host Transmission. Front. Microbiol. 2017, 8, 511. [Google Scholar] [CrossRef]
- Solenne, C.; Luis, E.; Huybert, G.; Zagmutt, F.J. Outbreak-Related Disease Burden Associated with Consumption of Unpasteurized Cow’s Milk and Cheese, United States, 2009–2014. Emerg. Infect. Dis. 2017, 23, 957–964. [Google Scholar] [CrossRef]
- Wenberg, M.F.; Porretta, M. Foodborne Diseases Active Surveillance Network (FoodNet). J. Am. Diet. Assoc. 1998, 98, A101. [Google Scholar] [CrossRef]
- Nancy, M.; Pratik, B.; Steven, S.; Khalil, K.; Irshad, S. Molecular Surveillance of Cronobacter spp. Isolated from a Wide Variety of Foods from 44 Different Countries by Sequence Typing of 16S rRNA, rpoB and O-Antigen Genes. Foods 2017, 6, 36. [Google Scholar] [CrossRef]
- Yi, M.; He, P.; Li, J.; Zhang, J.; Lin, L.; Wang, L.; Zhao, L. A portable toolbox based on time-resolved fluoroimmunoassay and immunomagnetic separation for Cronobacter sakazakii on-site detection in dairy. Int. Dairy J. 2022, 133, 105425. [Google Scholar] [CrossRef]
- Phiri, B.S.J.; Hang’Ombe, B.M.; Mulenga, E.; Mubanga, M.; Maurischat, S.; Wichmann-Schauer, H.; Schaarschmidt, S.; Fetsch, A. Prevalence and diversity of Staphylococcus aureus in the Zambian dairy value chain: A public health concern. Int. J. Food Microbiol. 2022, 375, 109737. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef]
- Wang, S.; Weller, D.; Falardeau, J.; Strawn, L.K.; Mardones, F.O.; Adell, A.D.; Moreno Switt, A.I. Food safety trends: From globalization of whole genome sequencing to application of new tools to prevent foodborne diseases. Trends Food Sci. Technol. 2016, 57, 188–198. [Google Scholar] [CrossRef]
- Jackson, B.R.; Tarr, C.; Strain, E.; Jackson, K.A.; Conrad, A.; Carleton, H.; Katz, L.S.; Stroika, S.; Gould, L.H.; Mody, R.K. Implementation of Nationwide Real-time Whole-genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Rev. Infect. Dis. 2016, 63, 380–386. [Google Scholar] [CrossRef]
- Gilmour, M.W.; Graham, M.; Domselaar, G.V.; Tyler, S.; Kent, H.; Trout-Yakel, K.M.; Larios, O.; Allen, V.; Lee, B.; Nadon, C. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genom. 2010, 11, 120. [Google Scholar] [CrossRef]
- Toro, M.; Retamal, P.; Ayers, S.; Barreto, M.; Allard, M.; Brown, E.W.; Gonzalez-Escalona, N.; Dudley, E.G. Whole genome sequencing analysis of Salmonella enteritidis isolated in Chile provides insights about possible transmission between gulls, poultry and humans. Appl. Environ. Microbiol. 2016, 82, 6223–6232. [Google Scholar] [CrossRef] [PubMed]
- Samarasekera, U. Salmonella Typhimurium outbreak linked to chocolate. Lancet Infect. Dis. 2022, 22, 947. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Melzer, F.; Sayour, A.E.; Shell, W.S.; Linde, J.; Abdel-Glil, M.; El-Soally, S.A.; Elschner, M.C.; Sayour, H.E.; Ramadan, E.S. Whole-genome sequencing for tracing the genetic diversity of Brucella abortus and Brucella melitensis isolated from livestock in Egypt. Pathogens 2021, 10, 759. [Google Scholar] [CrossRef]
- Brocke, V.; Fritz, J.; Holder, C.; Eichner, M.; Brockmann, S. WHO European strategic action plan on antibiotic resistance: How to preserve antibiotics. J. Pediatr. Infect. Dis. 2014, 9, 127–134. [Google Scholar] [CrossRef]
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M.A.P. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- Erickson, V.I.; Alfifi, A.; Hounmanou, Y.G.M.; Sana, M.J.; Christensen, J.P.; Dalsgaard, A. Genomic traits of Aeromonas veronii isolated from slaughtered Danish broilers. Vet. Microbiol. 2023, 283, 109772. [Google Scholar] [CrossRef]
- Baker, K.S.; Campos, J.; Pichel, M.; Della Gaspera, A.; Duarte-Martínez, F.; Campos-Chacón, E.; Bolaños-Acuña, H.M.; Guzmán-Verri, C.; Mather, A.E.; Velasco, S.D.; et al. Whole genome sequencing of Shigella sonnei through PulseNet Latin America and Caribbean: Advancing global surveillance of foodborne illnesses. Clin. Microbiol. Infect. 2017, 23, 845–853. [Google Scholar] [CrossRef]
- Martins, B.T.F.; Meirelles, J.L.d.; Omori, W.P.; Oliveira, R.R.d.; Yamatogi, R.S.; Call, D.R.; Nero, L.A. Comparative genomics and antibiotic resistance of Yersinia enterocolitica obtained from a pork production chain and human clinical cases in Brazil. Food Res. Int. 2022, 152, 110917. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2018, 47, 687–692. [Google Scholar] [CrossRef]
- Marco, B.G.; Fratamico, P.M.; Jayanthi, G.; Isha, P.; Bagi, L.K.; Sabine, D.; Patrick, F.; Federica, B.; Aniello, A.; Tiziana, P. Characterization of Shiga Toxin Subtypes and Virulence Genes in Porcine Shiga Toxin-Producing Escherichia coli. Front. Microbiol. 2016, 7, 574. [Google Scholar] [CrossRef]
- Billington, C.; Kingsbury, J.M.; Rivas, L. Metagenomics Approaches for Improving Food Safety: A Review. J. Food Prot. 2022, 85, 448–464. [Google Scholar] [CrossRef]

| Scope of Application | Website | |
|---|---|---|
| Bowtie/ Bowtie2 (2.5.4) | A tool for efficiently comparing high-throughput sequencing data, especially for short-read-length sequencing data. | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml (accessed on 16 May 2024) |
| BWA (MEM) | Tool for comparing sequencing data, supporting both short- and long-read length sequencing data. | https://bio-bwa.sourceforge.net/ (accessed on 28 February 2010) |
| SAMtools (1.2) | Tools for processing and analyzing comparison results, with the possibility of sorting, filtering, indexing, converting, etc. | https://www.htslib.org/doc/1.2/samtools.html (accessed on 15 December 2015) |
| GATK (3.7) | Tools for analyzing high-throughput sequencing data, including variant detection, splice variant detection, RNA-seq analysis and other features. | https://wiki.rc.usf.edu/index.php/Genome_Analysis_ToolKit_(GATK) (accessed on 13 March 2023) |
| Picard (3.4.0) | Toolset for processing and analyzing sequencing data, including de-duplication, sorting, and format conversion. | https://broadinstitute.github.io/picard/ (accessed on 13 April 2024) |
| BLAST (1.4.0) | Tools for comparing and recognizing biological sequences, widely used for sequence similarity searching and annotation. | https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 17 March 2025) |
| Years | Region | Sample | Location | Pathogens | Positive Rate | References |
|---|---|---|---|---|---|---|
| 2015 | Nanjing, China | Pork | Open-air markets | Listeria monocytogenes | 6.9% | [56] |
| 2018 | Andalusia, Spain | Free-range pig | Slaughterhouses | Salmonella | 12.93% | [57] |
| Campylobacter | 17.17% | |||||
| L. monocytogenes | 9.37% | |||||
| 2013 | South Korea | Pork | Slaughterhouse, processing line retail outlet local market | Bacillus cereus | 4.41% | [58] |
| Escherichia coli O157:H7 | ND | |||||
| L. monocytogenes | 5.89% | |||||
| Salmonella | 1.20% | |||||
| S. Aureus | 0.83% | |||||
| Y. enterocolitica | ND | |||||
| 2013 | Danish | Pig | Farms | Salmonella | 40.9% | [59] |
| Slaughterhouses | 7.4% | |||||
| 2010 | Finland | Pig | feed and litter | L. monocytogenes | 11% | [60] |
| rectal swabs | 1% | |||||
| intestinal contents | 1% | |||||
| tonsils | 24% | |||||
| pluck sets | 5% | |||||
| carcasses | 1% | |||||
| meat cuts | 4% | |||||
| 2013 | Spain | Pig | pre-scalding (slaughter line) | Salmonella | 36.25% | [61] |
| trucks | 23.21% | |||||
| cecal contents (slaughter line) | 21.25% | |||||
| Tonsils (slaughter line) | 17.50% | |||||
| ileocecal lymph nodes | 16.25% | |||||
| lairage | 14.06% | |||||
| 2021 | Sichuan, China | Chicken | defeathering | Salmonella enterica | 50% | [53] |
| evisceration | 36.67% | |||||
| disinfection and pre-cooling | 15% | |||||
| segmentation | 6.67% | |||||
| refrigeration | 3.33% | |||||
| 2018 | Trinidad | Chicken | cottage poultry processors | Salmonella | 20.5% | [62] |
| supermarkets | 8.3% | |||||
| 2022 | Trinidad and Tobago | Chicken | hatcheries | Salmonella | 7.6% | [63] |
| broiler farms | 2.8% | |||||
| 2024 | Guangzhou, China | Retail chicken meat | live poultry | Salmonella | 67.5% | [64] |
| frozen | 50% | |||||
| chilled | 43.3% | |||||
| 2022 | Jiangsu, China | Ducks | hatchery samples | Salmonella | 35.7% | [9] |
| market samples | 29.2% | |||||
| farm samples | 23.6% | |||||
| slaughterhouse samples | 9.4% | |||||
| 2019 | Southern China | Duck, Fish | Integrated fishery | Escherichia coli | 55.17% | [65] |
| Slaughter house | 56% | |||||
| Market | 40.32% | |||||
| 2021 | Australia | Egg | Cage | Escherichia coli | 20.3% | [66] |
| Barn-laid | 20% | |||||
| Free-range | 19.5% | |||||
| 2023 | Italy | Food processing environment | Meat | Listeria monocytogenes | 17.57% | [67] |
| Dairy | 4.47% | |||||
| Fish product | 0.96% | |||||
| RTE | 1.6% | |||||
| Breeding farms | 0.32% | |||||
| Large retail | 0.16% | |||||
| 2022 | Japan | Raw food products for retailing | Feather | Staphylococcus aureus complex | 52.63% | [68] |
| Feces | 12.07% | |||||
| Chiller water | 9.8% | |||||
| Slaughterhouse environment | 17.19% | |||||
| Carcass | 64.04% | |||||
| 2024 | Mexico | Raw chicken | fresh markets | Salmonella enterica | 23.56% | [69] |
| supermarkets | 28.64% | |||||
| butcher shops | 29.68% | |||||
| 2022 | North Carolina, USA | Sheep | Feces | Escherichia coli | 27.3% | [70] |
| Cecal contents | 21.9% | |||||
| Carcass swab | 10.2% | |||||
| Abattoir resting area feces | 20.0% | |||||
| Environmental samples | Soil samples | 58.9% | ||||
| Lairage swabs | 65.8% | |||||
| Animal feed | 30.4% | |||||
| Water | 18.8% | |||||
| 2023 | Spain | Cow | Feces | Listeria ivanovii | 0.5% | [71] |
| Tonsils | 1.1% | |||||
| Udder | 7.1% | |||||
| 2024 | Costa Rica, USA | Chicken | chicken meat | Salmonella enterica | 58.5% | [72] |
| chicken caecal | 38.0% | |||||
| 2020 | Argentina | Poultry meat supply chain | Poultry | Thermotolerant Campylobacter | 33% | [73] |
| Wild-living birds | 24% | |||||
| Darkling beetles | 20% | |||||
| Farm workers boots | 17% | |||||
| Darkling beetle larvae | 10% | |||||
| Flies | 5% | |||||
| Litter | 5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, K.; Song, D.; Li, S.; Wang, X.; Dai, L.; Pei, X.; Yang, X.; Jiang, Y. Significance of Whole-Genome Sequencing for the Traceability of Foodborne Pathogens: During the Processing of Meat and Dairy Products. Foods 2025, 14, 1410. https://doi.org/10.3390/foods14081410
Dong K, Song D, Li S, Wang X, Dai L, Pei X, Yang X, Jiang Y. Significance of Whole-Genome Sequencing for the Traceability of Foodborne Pathogens: During the Processing of Meat and Dairy Products. Foods. 2025; 14(8):1410. https://doi.org/10.3390/foods14081410
Chicago/Turabian StyleDong, Kai, Danliangmin Song, Shihang Li, Xu Wang, Lina Dai, Xiaoyan Pei, Xinyan Yang, and Yujun Jiang. 2025. "Significance of Whole-Genome Sequencing for the Traceability of Foodborne Pathogens: During the Processing of Meat and Dairy Products" Foods 14, no. 8: 1410. https://doi.org/10.3390/foods14081410
APA StyleDong, K., Song, D., Li, S., Wang, X., Dai, L., Pei, X., Yang, X., & Jiang, Y. (2025). Significance of Whole-Genome Sequencing for the Traceability of Foodborne Pathogens: During the Processing of Meat and Dairy Products. Foods, 14(8), 1410. https://doi.org/10.3390/foods14081410





