Seafood Labeling in Croatia: Molecular Evidence and Regulatory Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Ethics Approval
2.3. DNA Extraction from Tissue and Sequencing Identification
2.4. Mislabeling Determination
2.5. Statistical Analysis
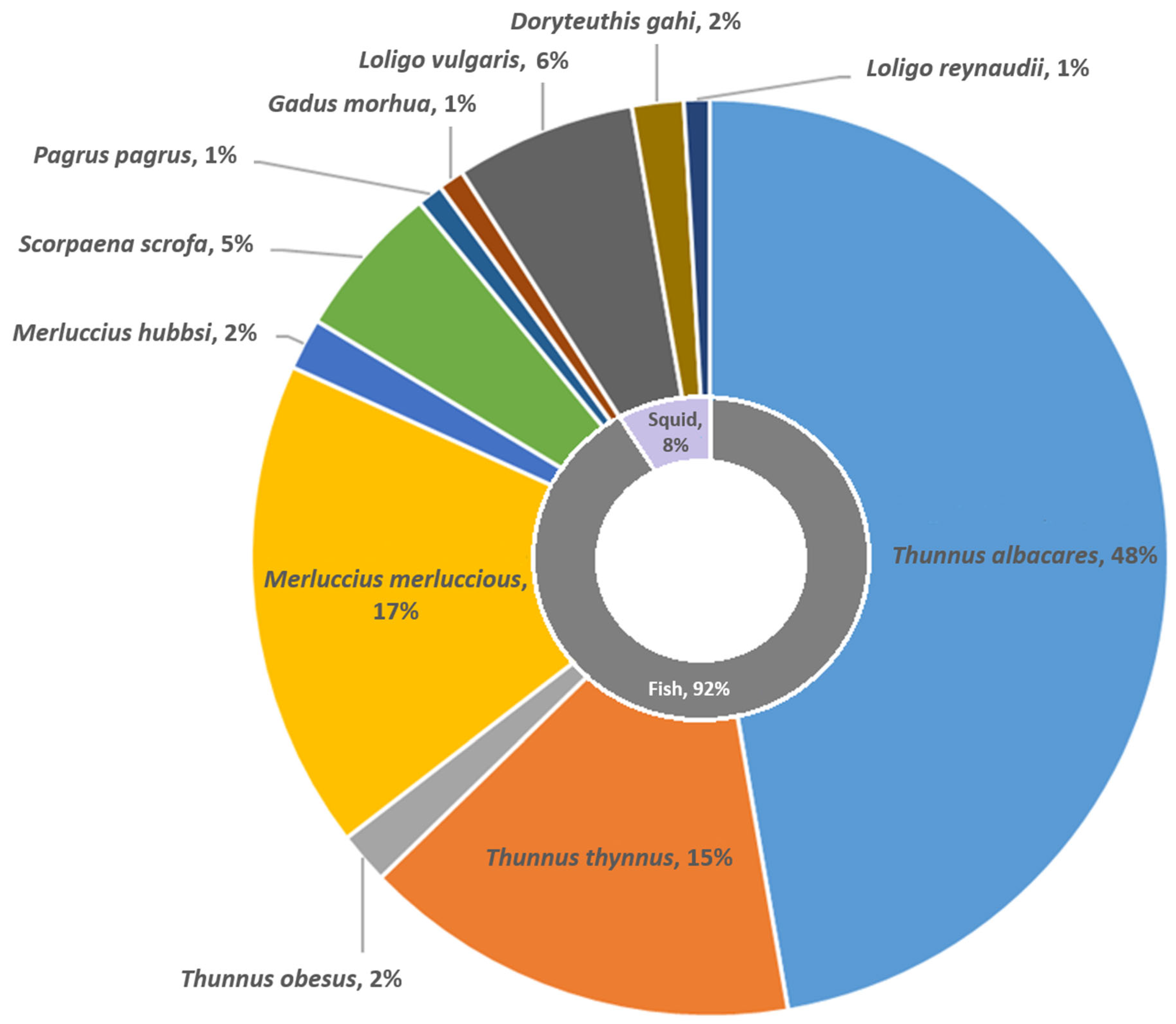
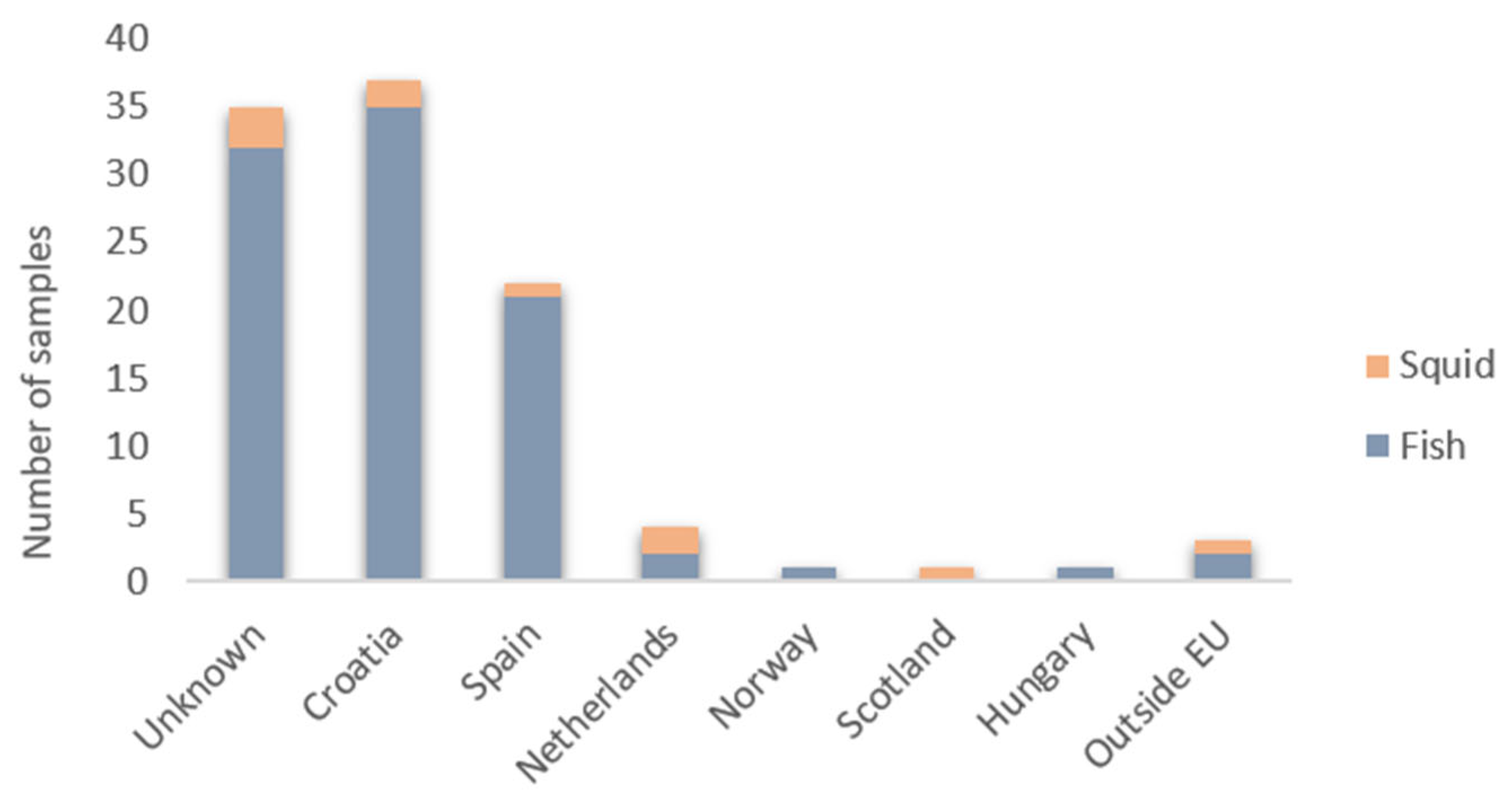
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture 2024; Food and Agriculture Organization of the United Nations: Rome, Italy, 2024. [Google Scholar]
- Ministry of Agriculture, Forestry and Fisheries. Annual Report on Marine Fisheries—Data for 2023. Available online: https://podaci.ribarstvo.hr/web-arhiva/2024/12/20/godisnji-izvjestaj-o-morskom-ribarstvu-prelim2023/ (accessed on 2 July 2024).
- Eurofish. Croatia. Eurofish International Organisation, 2022. Available online: https://eurofish.dk/member-countries/croatia/ (accessed on 31 July 2024).
- Spink, J.; Moyer, D.C. Defining the Public Health Threat of Food Fraud. J. Food Sci. 2011, 76, R157–R163. [Google Scholar] [CrossRef]
- Horreo, J.L.; Fitze, P.S.; Jiménez-Valverde, A.; Noriega, J.A.; Pelaez, M.L. Amplification of 16S rDNA Reveals Important Fish Mislabelling in Madrid Restaurants. Food Control 2019, 96, 146–150. [Google Scholar] [CrossRef]
- Crego-Prieto, V.; Campo, D.; Perez, J.; Martinez, J.L.; Garcia-Vazquez, E.; Roca, A. Inaccurate Labelling Detected at Landings and Markets: The Case of European Megrims. Fish. Res. 2012, 129, 106–109. [Google Scholar] [CrossRef]
- Doukakis, P.; Pikitch, E.K.; Rothschild, A.; DeSalle, R.; Amato, G.; Kolokotronis, S.O. Testing the Effectiveness of an International Conservation Agreement: Marketplace Forensics and CITES Caviar Trade Regulation. PLoS ONE 2012, 7, e40907. [Google Scholar] [CrossRef]
- Palmeira, C.A.M.; Rodrigues-Filho, L.F.S.; Sales, J.B.L.; Vallinoto, M.; Schneider, H.; Sampaio, I. Commercialization of a Critically Endangered Species (Largetooth Sawfish, Pristis perotteti) in Fish Markets of Northern Brazil: Authenticity by DNA Analysis. Food Control 2013, 34, 249–252. [Google Scholar] [CrossRef]
- Di Pinto, A.; Marchetti, P.; Mottola, A.; Bozzo, G.; Bonerba, E.; Ceci, E.; Bottaro, M.; Tantillo, G. Species Identification in Fish Fillet Products Using DNA Barcoding. Fish. Res. 2015, 170, 9–13. [Google Scholar] [CrossRef]
- Harris, D.J.; Rosado, D.; Xavier, R. DNA Barcoding Reveals Extensive Mislabeling in Seafood Sold in Portuguese Supermarkets. J. Aquat. Food Prod. Technol. 2016, 25, 1375–1380. [Google Scholar] [CrossRef]
- Ward, R.D.; Hanner, R.; Hebert, P.D. The Campaign to DNA Barcode All Fishes, FISH-BOL. J. Fish Biol. 2009, 74, 329–356. [Google Scholar] [CrossRef]
- Holmes, L., Jr.; Thompson, J. Environmental Sciences in Public Health. In Basics of Public Health Core Competencies; Jones & Bartlett Publishers: Burlington, MA, USA, 2009; p. 49. [Google Scholar]
- Regulation (EU) No 1379/2013 of the European Parliament and of the Council of 11 December 2013 on the Common Organisation of the Markets in Fishery and Aquaculture Products, Amending Council Regulations (EC) No 1184/2006 and (EC) No 1224/2009 and Repealing Council Regulation (EC) No 104/2000. Off. J. Eur. Union 2013, 56, L354.
- European Commission. EU Fisheries Control System Gets a Major Revamp. Blue Economy Observatory, 15 February 2024. Available online: https://blue-economy-observatory.ec.europa.eu/news/eu-fisheries-control-system-gets-major-revamp-2024-02-15_en (accessed on 2 August 2024).
- Griffiths, A.M.; Sotelo, C.G.; Mendes, R.; Pérez-Martín, R.I.; Schröder, U.; Shorten, M.; Silva, H.A.; Verrez-Bagnis, V.; Mariani, S. Current methods for seafood authenticity testing in Europe: Is there a need for harmonisation? Food Control 2014, 45, 95–100. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. Lond. Ser. B 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Galimberti, A.; De Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA Barcoding as a New Tool for Food Traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
- European Parliament. European Parliament Resolution of 14 January 2014 on the Food Crisis, Fraud in the Food Chain and the Control Thereof (2013/2091(INI)). Off. J. Eur. Union 2014, 59, C482. [Google Scholar]
- Committee on the Environment. Draft Report on the Food Crisis, Fraud in the Food Chain and the Control Thereof. In Public Health and Food Safety from European Parliament; EU Parliament Publisher: Strasbourg, France, 2013; pp. 1–9. [Google Scholar]
- Guardone, L.; Tinacci, L.; Costanzo, F.; Azzarelli, D.; D’Amico, P.; Tasselli, G.; Magni, A.; Guidi, A.; Nucera, D.; Armani, A. DNA Barcoding as a Tool for Detecting Mislabeling of Fishery Products Imported from Third Countries: An Official Survey Conducted at the Border Inspection Post of Livorno-Pisa (Italy). Food Control 2017, 80, 204–216. [Google Scholar] [CrossRef]
- Stern, D.B.; Nallar, E.C.; Rathod, J.; Crandall, K.A. DNA Barcoding Analysis of Seafood Accuracy in Washington, DC Restaurants. PeerJ 2017, 5, e3234. [Google Scholar] [CrossRef]
- Christiansen, H.; Fournier, N.; Hellemans, B.; Volckaert, F.A. Seafood Substitution and Mislabeling in Brussels’ Restaurants and Canteens. Food Control 2018, 85, 66–75. [Google Scholar] [CrossRef]
- Armani, A.; Tinacci, L.; Lorenzetti, R.; Benvenuti, A.; Susini, F.; Gasperetti, L.; Ricci, E.; Guarducci, C.; Guidi, A. Is Raw Better? A Multiple DNA Barcoding Approach (Full and Mini) Based on Mitochondrial and Nuclear Markers Reveals Low Rates of Misdescription in Sushi Products Sold on the Italian Market. Food Control 2017, 79, 126–133. [Google Scholar] [CrossRef]
- Espiñeira, M.; González-Lavín, N.; Vieites, J.M.; Santaclara, F.J. Development of a Method for the Genetic Identification of Flatfish Species on the Basis of Mitochondrial DNA Sequences. J. Agric. Food Chem. 2008, 56, 8954–8961. [Google Scholar] [CrossRef]
- Barbuto, M.; Galimberti, A.; Ferri, E.; Labra, M.; Malandra, R.; Galli, P.; Casiraghi, M. DNA Barcoding Reveals Fraudulent Substitutions in Shark Seafood Products: The Italian Case of “Palombo” (Mustelus spp.). Food Res. Int. 2010, 43, 376–381. [Google Scholar] [CrossRef]
- Garcia-Vazquez, E.; Perez, J.; Martinez, J.L.; Pardinas, A.F.; Lopez, B.; Karaiskou, N.; Triantafyllidis, A. High Level of Mislabeling in Spanish and Greek Hake Markets Suggests the Fraudulent Introduction of African Species. J. Agric. Food Chem. 2011, 59, 475–480. [Google Scholar] [CrossRef]
- Bartlett, S.E.; Davidson, W.S. Identification of Thunnus Tuna Species by the Polymerase Chain Reaction and Direct Sequence Analysis of Their Mitochondrial Cytochrome b Genes. Can. J. Fish. Aquat. Sci. 1991, 48, 309–317. [Google Scholar] [CrossRef]
- Logan, C.A.; Alter, S.E.; Haupt, A.J.; Tomalty, K.; Palumbi, S.R. An Impediment to Consumer Choice: Overfished Species Are Sold as Pacific Red Snapper. Biol. Conserv. 2008, 141, 1591–1599. [Google Scholar] [CrossRef]
- Mariani, S.; Griffiths, A.M.; Velasco, A.; Kappel, K.; Jérôme, M.; Perez-Martin, R.I.; Schröder, U.; Verrez-Bagnis, V.; Silva, H.; Vandamme, S.G.; et al. Low Mislabeling Rates Indicate Marked Improvements in European Seafood Market Operations. Front. Ecol. Environ. 2015, 13, 536–540. [Google Scholar] [CrossRef]
- Xiong, X.; D’Amico, P.; Guardone, L.; Castigliego, L.; Guidi, A.; Gianfaldoni, D.; Armani, A. The Uncertainty of Seafood Labeling in China: A Case Study on Cod, Salmon and Tuna. Mar. Policy 2016, 68, 123–135. [Google Scholar] [CrossRef]
- Marko, P.B.; Lee, S.C.; Rice, A.M.; Gramling, J.M.; Fitzhenry, T.M.; McAlister, J.S.; Harper, G.R.; Moran, A.L. Fisheries: Mislabeling of a Depleted Reef Fish. Nature 2004, 430, 309–310. [Google Scholar] [CrossRef]
- Warner, K.; Timme, W.; Lowell, B.; Hirschfield, M. Oceana Study Reveals Seafood Fraud Nationwide; Oceana: Washington, DC, USA, 2013; pp. 1–69. [Google Scholar]
- Oceana. Oceana 2016 Annual Report. Available online: https://oceana.org/wp-content/uploads/sites/18/2016_annual_report.pdf (accessed on 21 December 2024).
- Nagalakshmi, K.; Annam, P.K.; Venkateshwarlu, G.; Pathakota, G.B.; Lakra, W.S. Mislabeling in Indian Seafood: An Investigation Using DNA Barcoding. Food Control 2016, 59, 196–200. [Google Scholar] [CrossRef]
- Senathipathi, D.N.; Benjakul, S.; Sukkapat, P.; Detcharoen, M.; Moorthy, G.; Saetang, J. DNA Barcoding Revealed Mislabeling of Imported Seafood Products in Thailand. Fishes 2024, 9, 215. [Google Scholar] [CrossRef]
- Tommasi, R.B.; Lamia, S.M.; Van, D.; Estrada, I.; Kuchler, Z.L.; Ramsey, D.; Tamang, J.; Kaneko, G.; Ehsan, H. DNA Barcoding Revealed a High Percentage of Mislabelling in Commercial Fish Products: The First Empirical Survey in South Texas. bioRxiv 2023, 2023, 08. [Google Scholar] [CrossRef]
- Carvalho, D.C.; Palhares, R.M.; Drummond, M.G.; Frigo, T.B. DNA Barcoding Identification of Commercialized Seafood in South Brazil: A Governmental Regulatory Forensic Program. Food Control 2015, 50, 784–788. [Google Scholar] [CrossRef]
- Munguia-Vega, A.; Terrazas-Tapia, R.; Dominguez-Contreras, J.F.; Reyna-Fabian, M.; Zapata-Morales, P. DNA Barcoding Reveals Global and Local Influences on Patterns of Mislabeling and Substitution in the Trade of Fish in Mexico. PLoS ONE 2022, 17, e0265960. [Google Scholar] [CrossRef]
- Cawthorn, D.M.; Steinman, H.A.; Witthuhn, R.C. DNA Barcoding Reveals a High Incidence of Fish Species Misrepresentation and Substitution on the South African Market. Food Res. Int. 2012, 46, 30–40. [Google Scholar] [CrossRef]
- Anonymous. Act on Animal Protection in Research Purposes. Off. Gaz. 2013, 55, 1129. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Asensio, L.; Samaniego, L.; Pavón, M.A.; Gonzalez, I.; Garcia, T.; Martin, R. Detection of Grouper Mislabelling in the Fish Market by an Immunostick Colorimetric ELISA Assay. Food Agric. Immunol. 2008, 19, 141–147. [Google Scholar] [CrossRef]
- Miller, D.D.; Mariani, S. Smoke, Mirrors, and Mislabeled Cod: Poor Transparency in the European Seafood Industry. Front. Ecol. Environ. 2010, 8, 517–521. [Google Scholar] [CrossRef]
- Filonzi, L.; Chiesa, S.; Vaghi, M.; Marzano, F.N. Molecular Barcoding Reveals Mislabelling of Commercial Fish Products in Italy. Food Res. Int. 2010, 43, 1383–1388. [Google Scholar] [CrossRef]
- Pardo, M.Á.; Jiménez, E.; Pérez-Villarreal, B. Misdescription Incidents in the Seafood Sector. Food Control 2016, 62, 277–283. [Google Scholar] [CrossRef]
- Filonzi, L.; Vaghi, M.; Ardenghi, A.; Rontani, P.M.; Voccia, A.; Nonnis Marzano, F. Efficiency of DNA Mini-Barcoding to Assess Mislabeling in Commercial Fish Products in Italy: An Overview of the Last Decade. Foods 2021, 10, 1449. [Google Scholar] [CrossRef]
- Minoudi, S.; Karaiskou, N.; Avgeris, M.; Gkagkavouzis, K.; Tarantili, P.; Triantafyllidou, D.; Kafetzopoulos, D.; Triantafyllidis, A. Seafood Mislabeling in Greek Market Using DNA Barcoding. Food Control 2020, 113, 107213. [Google Scholar] [CrossRef]
- Pazartzi, T.; Siaperopoulou, S.; Gubili, C.; Maradidou, S.; Loukovitis, D.; Chatzispyrou, A.; Tsikliras, A.C.; Batargias, C.; Mylonas, C.C.; Kotoulas, G. High Levels of Mislabeling in Shark Meat—Investigating Patterns of Species Utilization with DNA Barcoding in Greek Retailers. Food Control 2019, 98, 179–186. [Google Scholar] [CrossRef]
- Bénard-Capelle, J.; Guillonneau, V.; Nouvian, C.; Fournier, N.; Le Loët, K.; Dettai, A. Fish Mislabelling in France: Substitution Rates and Retail Types. PeerJ 2015, 2, e714. [Google Scholar] [CrossRef] [PubMed]
- Paolacci, S.; Mendes, R.; Klapper, R.; Velasco, A.; Ramilo-Fernandez, G.; Munoz-Colmenero, M.; Sotelo, C.G. Labels on Seafood Products in Different European Countries and Their Compliance to EU Legislation. Mar. Policy 2021, 134, 104810. [Google Scholar] [CrossRef]
- Velez-Zuazo, X.; Alfaro-Shigueto, J.; Rosas-Puchuri, U.; Guidino, C.; Pasara-Polack, A.; Riveros, J.C.; Mangel, J.C. High Incidence of Mislabeling and a Hint of Fraud in the Ceviche and Sushi Business. Food Control 2021, 129, 108224. [Google Scholar] [CrossRef]
- Pascual, C.Y.; Reche, M.; Fiandor, A.; Valbuena, T.; Cuevas, T.; Esteban, M.M. Fish Allergy in Childhood. Pediatr. Allergy Immunol. 2008, 19, 573–579. [Google Scholar] [CrossRef]
- Pascual, C.; Esteban, M.M.; Crespo, J.F. Fish Allergy: Evaluation of the Importance of Cross-Reactivity. J. Pediatr. 1992, 121, S29–S34. [Google Scholar] [CrossRef]
- Koyama, H.; Kakami, M.; Kawamura, M.; Tokuda, R.; Kondo, Y.; Tsuge, I.; Urisu, A. Grades of 43 Fish Species in Japan Based on IgE-Binding Activity. Allergol. Int. 2006, 55, 311–316. [Google Scholar] [CrossRef]
- Van Do, T.; Elsayed, S.; Florvaag, E.; Hordvik, I.; Endresen, C. Allergy to Fish Parvalbumins: Studies on the Cross-Reactivity of Allergens from 9 Commonly Consumed Fish. J. Allergy Clin. Immunol. 2005, 116, 1314–1320. [Google Scholar] [CrossRef]
- Machado-Schiaffino, G.; Martinez, J.L.; Garcia-Vazquez, E. Detection of Mislabeling in Hake Seafood Employing mtSNPs-Based Methodology with Identification of Eleven Hake Species of the Genus Merluccius. J. Agric. Food Chem. 2008, 56, 5091–5095. [Google Scholar] [CrossRef]
- Muñoz-Colmenero, M.; Blanco, O.; Arias, V.; Martinez, J.L.; Garcia-Vazquez, E. DNA Authentication of Fish Products Reveals Mislabeling Associated with Seafood Processing. Fisheries 2016, 41, 128–138. [Google Scholar] [CrossRef]
- Kappel, K.; Schröder, U. Substitution of High-Priced Fish with Low-Priced Species: Adulteration of Common Sole in German Restaurants. Food Control 2016, 59, 478–486. [Google Scholar] [CrossRef]
- Giagkazoglou, Z.; Loukovitis, D.; Gubili, C.; Chatziplis, D.; Symeonidis, A.; Imsiridou, A. Untangling the Cephalopod Market: Authentication of Seafood Products in Greece with DNA-Barcoding. Food Control 2024, 163, 110523. [Google Scholar] [CrossRef]
- Rosa, R.; Pereira, J.; Nunes, M.L. Biochemical Composition of Cephalopods with Different Life Strategies, with Special Reference to a Giant Squid, Architeuthis sp. Mar. Biol. 2005, 146, 739–751. [Google Scholar] [CrossRef]
- Benetti, D.D.; Partridge, G.J.; Stieglitz, J. Overview on Status and Technological Advances in Tuna Aquaculture Around the World. Adv. Tuna Aquac. 2016, 1, 1–19. [Google Scholar]
- Sotelo, C.G.; Velasco, A.; Perez-Martin, R.I.; Kappel, K.; Schröder, U.; Verrez-Bagnis, V.; Griffiths, A. Tuna Labels Matter in Europe: Mislabeling Rates in Different Tuna Products. PLoS ONE 2018, 13, e0196641. [Google Scholar] [CrossRef]
- Servusova, E.; Piskata, Z. Identification of Selected Tuna Species in Commercial Products. Molecules 2021, 26, 1137. [Google Scholar] [CrossRef]

| Primer | Sequence 5′-3′ | Reference |
|---|---|---|
| FishF1 | TCAACCAACCACAAAGACATTGGCAC | Ward et al., (2009) [11] |
| FishF2 | TCGACTAATCATAAAGATATCGGCAC | Ward et al., (2009) [11] |
| FishR1 | TAGACTTCTGGGTGGCCAAAGAATCA | Ward et al., (2009) [11] |
| FishR2 | ACTTCAGGGTGACCGAAGAATCAGAA | Ward et al., (2009) [11] |
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | Folmer et al., (1994) [41] |
| HC02198 | TAAACTTCAGGGTGACCAAAAAATCA | Folmer et al., (1994) [41] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grbin, D.; Zrnčić, S.; Oraić, D.; Alfier, M.; Cindrić, M.; Jović, L.; Sučec, I.; Zupičić, I.G. Seafood Labeling in Croatia: Molecular Evidence and Regulatory Insights. Foods 2025, 14, 917. https://doi.org/10.3390/foods14060917
Grbin D, Zrnčić S, Oraić D, Alfier M, Cindrić M, Jović L, Sučec I, Zupičić IG. Seafood Labeling in Croatia: Molecular Evidence and Regulatory Insights. Foods. 2025; 14(6):917. https://doi.org/10.3390/foods14060917
Chicago/Turabian StyleGrbin, Dorotea, Snježana Zrnčić, Dražen Oraić, Matea Alfier, Mario Cindrić, Lucija Jović, Ivica Sučec, and Ivana Giovanna Zupičić. 2025. "Seafood Labeling in Croatia: Molecular Evidence and Regulatory Insights" Foods 14, no. 6: 917. https://doi.org/10.3390/foods14060917
APA StyleGrbin, D., Zrnčić, S., Oraić, D., Alfier, M., Cindrić, M., Jović, L., Sučec, I., & Zupičić, I. G. (2025). Seafood Labeling in Croatia: Molecular Evidence and Regulatory Insights. Foods, 14(6), 917. https://doi.org/10.3390/foods14060917









