Beluga Optimization Algorithm for Near-Infrared Spectral Variable Selection of Complex Samples
Abstract
1. Introduction
2. Theory and Algorithm
2.1. Beluga Whale Optimization
2.2. Chemometric Methods
2.3. Discretized BWO-PLS
- The algorithm initializes a population of beluga whales.
- For the position of each beluga whale, use the corresponding spectral variables and target vector to construct a PLS model and calculate the fitness.
- Beluga whale positions are updated in accordance with BWO. Subsequently, a transfer function is applied to convert these updated continuous positions into new binary vectors.
- The fitness of each beluga whale is re-evaluated based on its new binary position vector after each update. This process continues until the fitness values converge.
- The binary position vector of the whale with the highest fitness is output as the optimal variable combination. A final PLS model is constructed from this optimal variable subset for predicting the target property.
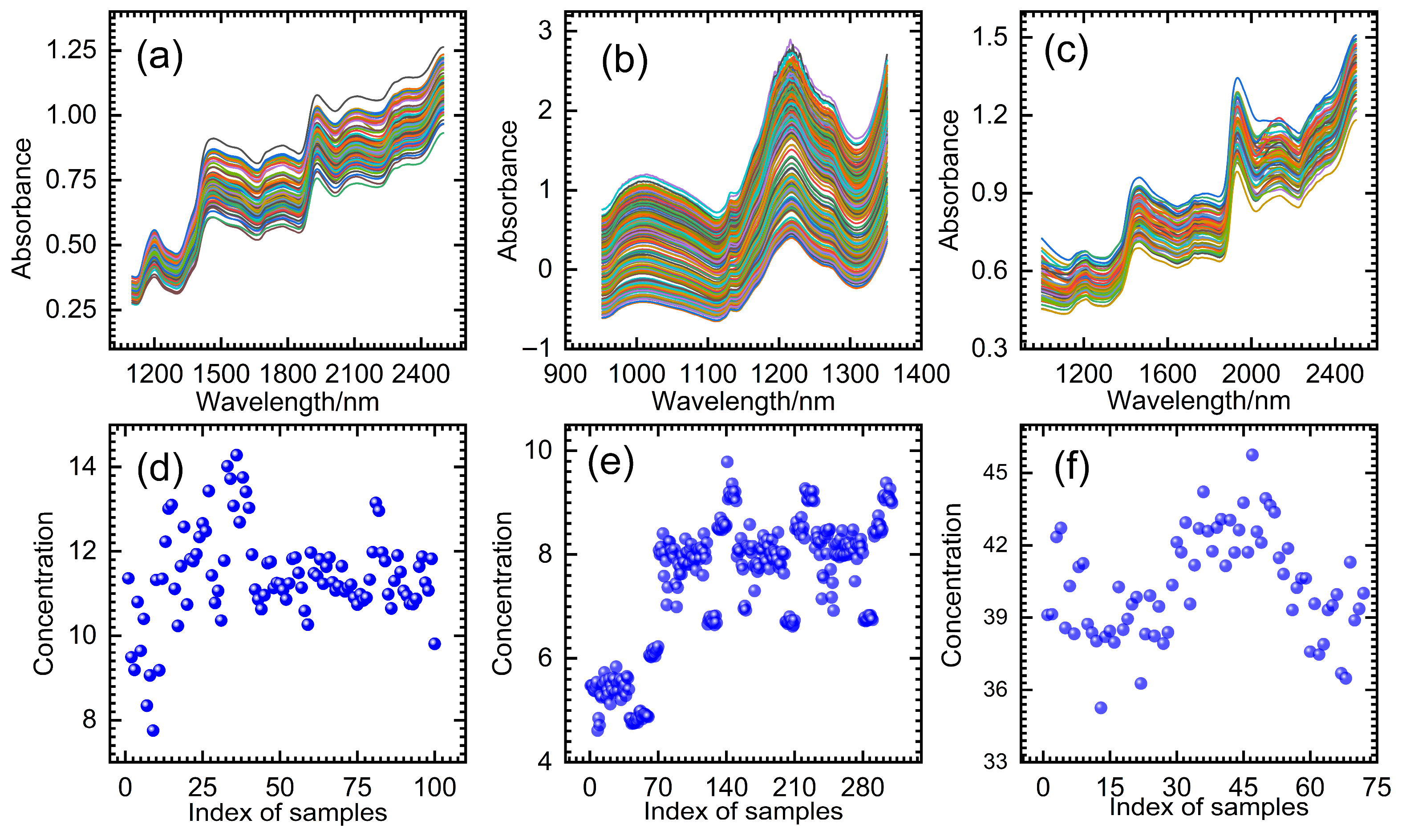
3. Experiments
4. Results and Discussion
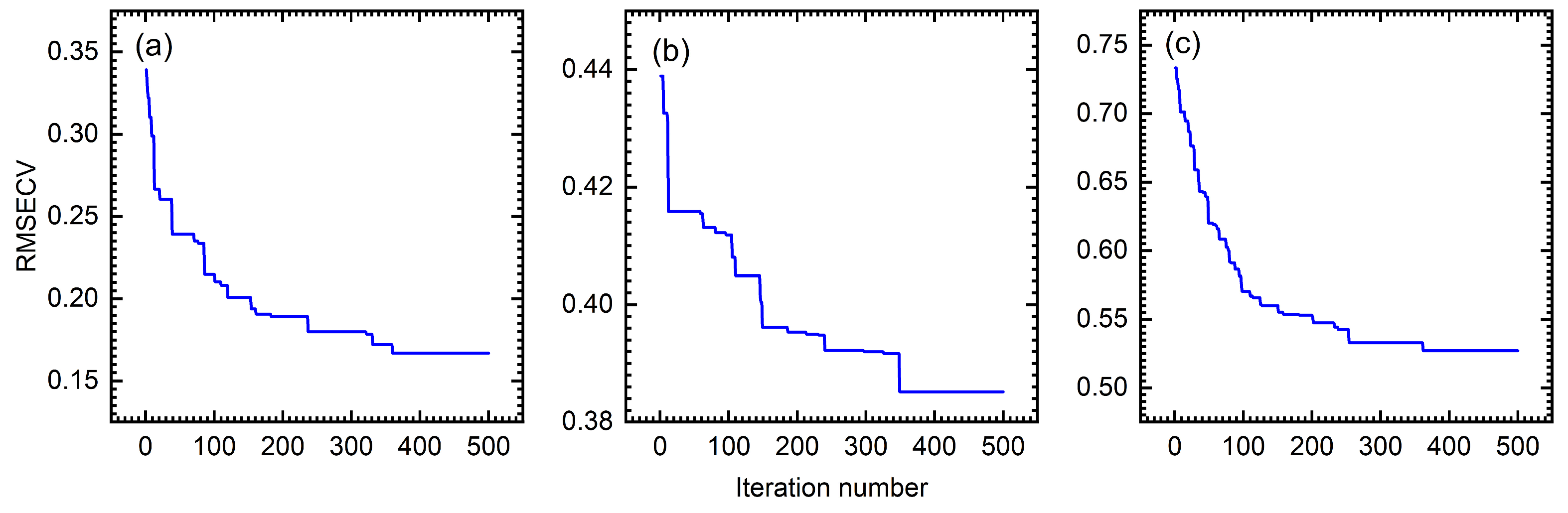
4.1. Iteration Number of BWO
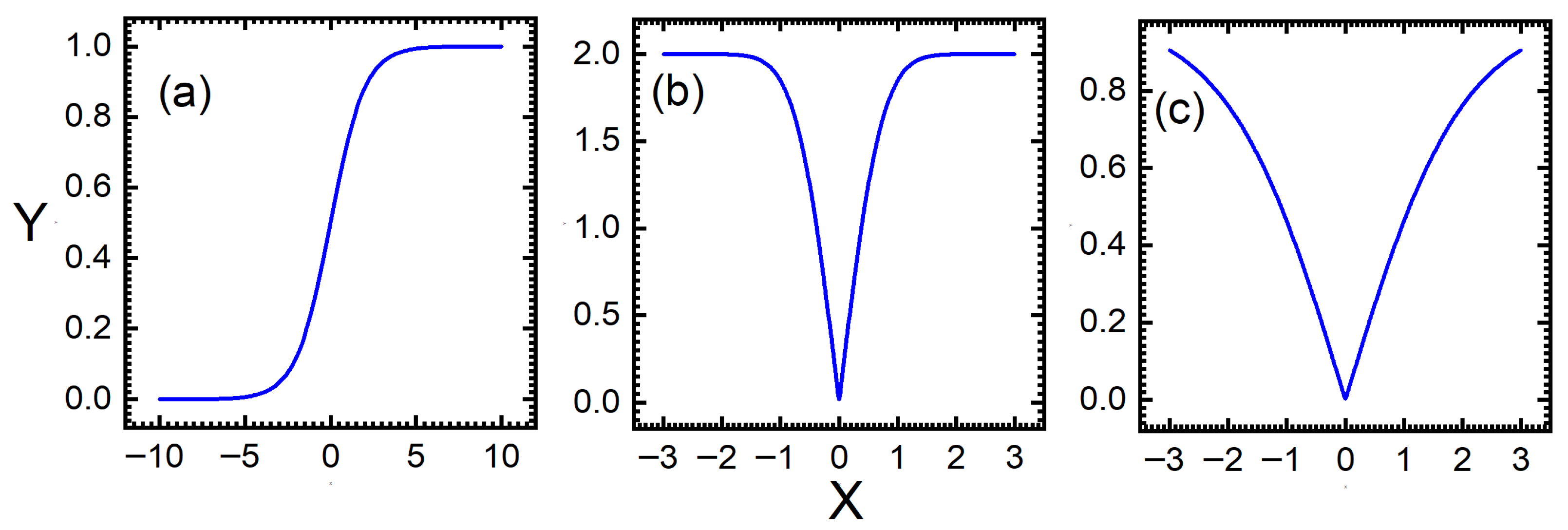
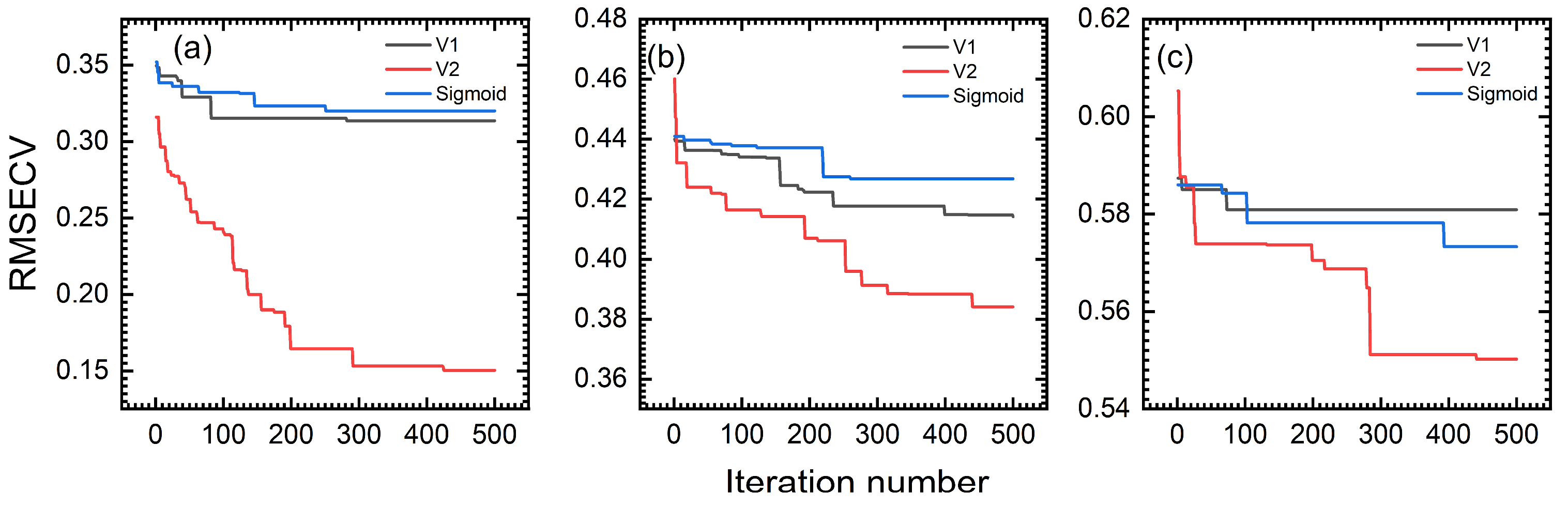
4.2. Discretization Function
4.3. Prediction Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NIR | Near-infrared |
| BWO | Beluga whale optimization |
| DBWO | Discretized BWO |
| PLS | Partial least squares |
| RT | Randomization test |
| UVE | Uninformative variable elimination |
| MC-UVE | Monte Carlo uninformative variable elimination |
| ANN | Artificial neural network |
| PCR | Principal component regression |
| SVR | Support vector regression |
| GPR | Gaussian process regression |
| ELM | Extreme learning machine |
| SI | Swarm intelligence |
| LVs | Latent variables |
| RMSEP | Root mean square error of prediction |
| R2 | Determination coefficients |
| KS | Kennard–Stone |
| RMSECV | Root mean square error of cross validation |
References
- Liu, Y.X.; Sun, H.; Zhao, C.Y.; Ai, C.K.; Bian, X.H. Extreme Learning Machine Combined with Whale Optimization Algorithm for Spectral Quantitative Analysis of Complex Samples. J. Chemom. 2024, 38, e3590. [Google Scholar] [CrossRef]
- Wu, D.Y.; Johnson, J.B.; Zhang, K.; Guo, Y.G.; Liu, D.; Wang, Z.G.; Bian, X.H. Variational mode decomposition unfolded partial least squares regression for ultraviolet–visible spectral analysis of edible oil blend, fuel oil and aqueous samples. Microchem. J. 2024, 196, 109587. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.M.; Wang, R.Q.; Waterhouse, G.I.N.; Xu, Z.X. SERS-tag technology in food safety and detection: Sensing from the “fingerprint” region to the “biological-silent” region. J. Future Foods 2024, 4, 309–323. [Google Scholar] [CrossRef]
- Feng, X.Y.; Chen, Z.G.; Yi, S.J.; Wang, P.H. A three-stage wavelength selection algorithm for near-infrared spectroscopy calibration. Spectrochim. Acta A 2025, 324, 125029. [Google Scholar] [CrossRef]
- Jiao, X.B.; Guo, D.L.; Zhang, X.J.; Su, Y.P.; Ma, R.; Chen, L.W.; Tian, K.; Su, J.Y.; Sahati, T.; Aierkenjiang, X.; et al. The Application of Near-Infrared Spectroscopy Combined with Chemometrics in the Determination of the Nutrient Composition in Chinese Cyperus esculentus L. Foods 2025, 14, 366. [Google Scholar] [CrossRef]
- Sem, V. Interpretability of selected variables and performance comparison of variable selection methods in a polyethylene and polypropylene NIR classification task. Spectrochim. Acta A 2021, 258, 119850. [Google Scholar] [CrossRef]
- Yun, T. Quantitative Analysis of Near-Infrared Spectroscopy of Blended Fabrics Based on Convolutional Neural Network. IEEE Access 2023, 11, 46644–46652. [Google Scholar] [CrossRef]
- Sun, J.Q.; Zhang, Y.Y.; Zhang, Y.M.; Zhao, H.G.; Han, G.T.; Via, B.K.; Jiang, W. Determination of natural turmeric dyes using near-infrared spectroscopy. Ind. Crops Prod. 2024, 222, 119817. [Google Scholar] [CrossRef]
- Long, T.Z.; Yi, H.; Kang, Y.T.; Qiao, Y.; Guan, Y.; Chen, C. Study on bionics-based swarm intelligence optimization algorithms for wavelength selection in near-infrared spectroscopy. Infrared Phys. Technol. 2024, 143, 105594. [Google Scholar] [CrossRef]
- Yun, Y.H.; Li, H.D.; Deng, B.C.; Cao, D.S. An overview of variable selection methods in multivariate analysis of near-infrared spectra. Trends Anal. Chem. 2019, 113, 102–115. [Google Scholar] [CrossRef]
- Wang, H.P.; Chen, P.; Dai, J.W.; Liu, D.; Li, J.Y.; Xu, Y.P.; Chu, X.L. Recent advances of chemometric calibration methods in modern spectroscopy: Algorithms, strategy, and related issues. Trends Anal. Chem. 2022, 153, 116648. [Google Scholar] [CrossRef]
- Firouz, M.S.; Omid, M.; Babaei, M.; Rashvand, M. Dielectric spectroscopy coupled with artificial neural network for classification and quantification of sesame oil adulteration. Inf. Process. Agric. 2022, 9, 233–242. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Shi, S.J.; Zhang, K.D.; Li, L.; Jiang, Y. Application of two near-infrared instruments combined with chemometrics and key variable selection algorithms for the determination of anthocyanins content in black rice. Microchem. J. 2024, 207, 112034. [Google Scholar] [CrossRef]
- Wang, Z.J.; Zuo, C.Z.; Chen, M.; Song, J.; Tu, K.; Lan, W.J.; Li, C.Y.; Pan, L.Q. A Novel Variable Selection Method Based on Ordered Predictors Selection and Successive Projections Algorithm for Predicting Gastrodin Content in Fresh Gastrodia elata Using Fourier Transform Near-Infrared Spectroscopy and Chemometrics. Foods 2023, 12, 4435. [Google Scholar] [CrossRef]
- Yu, Y.; Chai, Y.H.; Li, Z.T.; Li, Z.M.; Ren, Z.Y.; Dong, H.; Chen, L. Quantitative predictions of protein and total flavonoids content in Tartary and common buckwheat using near-infrared spectroscopy and chemometrics. Food Chem. 2025, 462, 141033. [Google Scholar] [CrossRef]
- Cui, C.H.; Caporaso, N.; Chen, J.W.; Fearn, T. Farinograph characteristics of wheat flour predicted by near infrared spectroscopy with an ensemble modelling method. J. Food Eng. 2023, 359, 111689. [Google Scholar] [CrossRef]
- Li, H.H.; Nunekpeku, X.; Zhang, W.; Adade, S.Y.S.S.; Ahmad, W.; Sheng, W.; Chen, Q.S. Quantitative prediction of minced chicken gel strength under ultrasonic treatment by NIR spectroscopy coupled with nonlinear chemometric tools evaluated using APaRPs. Food Chem. 2025, 463, 141373. [Google Scholar] [CrossRef]
- Liu, J.J.; Xu, Y.F.; Liu, S.K.; Yu, S.X.; Yu, Z.R.; Low, S.S. Application and Progress of Chemometrics in Voltammetric Biosensing. Biosensors 2022, 12, 494. [Google Scholar] [CrossRef]
- de Paulo, E.H.; Rech, A.M.; Weiler, F.H.; Nascimento, M.H.C.; Filgueiras, P.R.; Ferrao, M.F. Determination of total protein and sugar content in soy-based beverages using variable selection methods applied to ATR-FTIR spectroscopy. J. Food Compos. Anal. 2024, 135, 106639. [Google Scholar] [CrossRef]
- Zhu, J.J.; Jiang, X.; Wang, Q.J.; Wu, J.Z.; Wu, S.D.; Chen, X.J.; Chen, Q.S. A novel hybrid variable selection strategy with application to molecular spectroscopic analysis. Chemom. Intell. Lab. Syst. 2023, 236, 104795. [Google Scholar] [CrossRef]
- Abrantes, G.; Almeida, V.; Maia, A.J.; Nascimento, R.; Nascimento, C.; Silva, Y.; Silva, Y.; Veras, G. Comparison between Variable-Selection Algorithms in PLS Regression with Near-Infrared Spectroscopy to Predict Selected Metals in Soil. Molecules 2023, 28, 6959. [Google Scholar] [CrossRef] [PubMed]
- Moro, M.K.; dos Santos, F.D.; Folli, G.S.; Romao, W.; Filgueiras, P.R. A review of chemometrics models to predict crude oil properties from nuclear magnetic resonance and infrared spectroscopy. Fuel 2021, 303, 121283. [Google Scholar] [CrossRef]
- Wang, H.H.; Zhang, M.J.; Xiong, Z.X.; Yang, W.Y.; Wu, T.; Du, Y.P. Flexible optimization of variables based on exponential and linear attenuation elimination-binary dragonfly algorithm in near infrared spectroscopic analysis. Infrared Phys. Technol. 2024, 140, 105374. [Google Scholar] [CrossRef]
- Bian, X.H.; Zhao, Z.Z.; Liu, J.W.; Liu, P.; Shi, H.B.; Tan, X.Y. Discretized butterfly optimization algorithm for variable selection in the rapid determination of cholesterol by near-infrared spectroscopy. Anal. Methods 2023, 15, 5190–5198. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Kalita, D.; Ahmed, T.M.; ElMasry, G.; Makino, Y. Effect of variable selection algorithms on model performance for predicting moisture content in biological materials using spectral data. Anal. Chim. Acta 2021, 1202, 339390. [Google Scholar] [CrossRef]
- Morelli, J.B.; Mcgoverin, C.; Nieuwoudt, M.; Holroyd, S.E.; Pilkington, L.I. Chemometric techniques for the prediction of milk composition from MIR spectral data: A review. Food Chem. 2024, 469, 142465. [Google Scholar] [CrossRef]
- Chen, T.; Wang, B. Bayesian variable selection for gaussian process regression: Application to chemometric calibration of spectrometers. Neurocomputing 2010, 73, 2718–2726. [Google Scholar] [CrossRef]
- Bian, X.H.; Zhang, R.L.; Liu, P.; Xiang, Y.; Wang, S.Y.; Tan, X.Y. Near infrared spectroscopic variable selection by a novel swarm intelligence algorithm for rapid quantification of high order edible blend oil. Spectrochim. Acta A 2022, 284, 121788. [Google Scholar] [CrossRef]
- Su, Y.; He, K.; Liu, W.Z.; Li, J.; Hou, K.Y.; Lv, S.Y.; He, X.W. Detection of soluble solid content in table grapes during storage based on visible-near-infrared spectroscopy. Food Innov. Adv. 2025, 4, 10–18. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, Q.S. Determination of Adulteration Content in Extra Virgin Olive Oil Using FT-NIR Spectroscopy Combined with the BOSS-PLS Algorithm. Molecules 2019, 24, 2134. [Google Scholar] [CrossRef]
- Brezocnik, L.; Fister, I.; Podgorelec, V. Swarm Intelligence Algorithms for Feature Selection: A Review. Appl. Sci. 2018, 8, 1521. [Google Scholar] [CrossRef]
- Zhong, C.T.; Li, G.; Meng, Z. Beluga whale optimization: A novel nature-inspired metaheuristic algorithm. Knowl.-Based Syst. 2022, 251, 109215. [Google Scholar] [CrossRef]
- Sun, X.X.; Zhu, L.K.; Liu, D.Y. Blueberry bruise non-destructive detection based on hyperspectral information fusion combined with multi-strategy improved Beluga Whale Optimization algorithm. Front. Plant Sci. 2024, 115, 1411485. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Zhang, M.J.; Yang, M.; Wang, D.G. NHBBWO: A novel hybrid butterfly-beluga whale optimization algorithm with the dynamic strategy for WSN coverage optimization. Peer-to-Peer Netw. Appl. 2025, 18, 80. [Google Scholar] [CrossRef]
- Esfahani, K.K.; Zade, B.M.H.; Mansouri, N. Multi-objective feature selection algorithm using Beluga Whale Optimization. Chemom. Intell. Lab. Syst. 2025, 257, 105295. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Deng, J.H.; Ding, Z.D.; Jiang, H. Quantification of heavy metal Cd in peanut oil using near-infrared spectroscopy combined with chemometrics: Analysis and comparison of variable selection methods. Infrared Phys. Technol. 2024, 141, 105447. [Google Scholar] [CrossRef]
- Kalivas, J.H. Two data sets of near infrared spectra. Chemom. Intell. Lab. Syst. 1997, 37, 255–259. [Google Scholar] [CrossRef]
- Dyrby, M.; Engelsen, S.B.; Norgaard, L.; Bruhn, M.; Lundsberg-Nielsen, L. Chemometric quantitation of the active substance in a pharmaceutical tablet using near-infrared (NIR) transmittance and NIR FT-Raman spectra. Appl. Spectrosc. 2002, 56, 579–585. [Google Scholar] [CrossRef]
- Agussabti; Rahmaddiansyah; Satriyo, P.; Munawar, A.A. Data analysis on near infrared spectroscopy as a part of technology adoption for cocoa farmer in Aceh Province, Indonesia. Data Brief 2020, 29, 105251. [Google Scholar] [CrossRef]

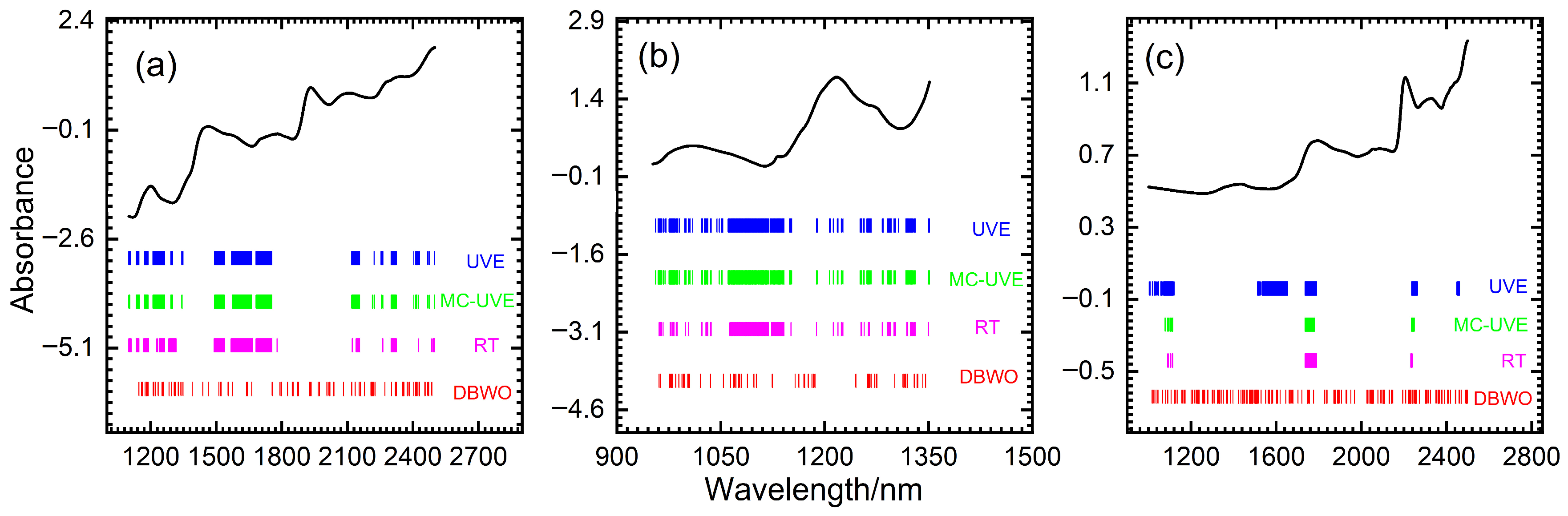
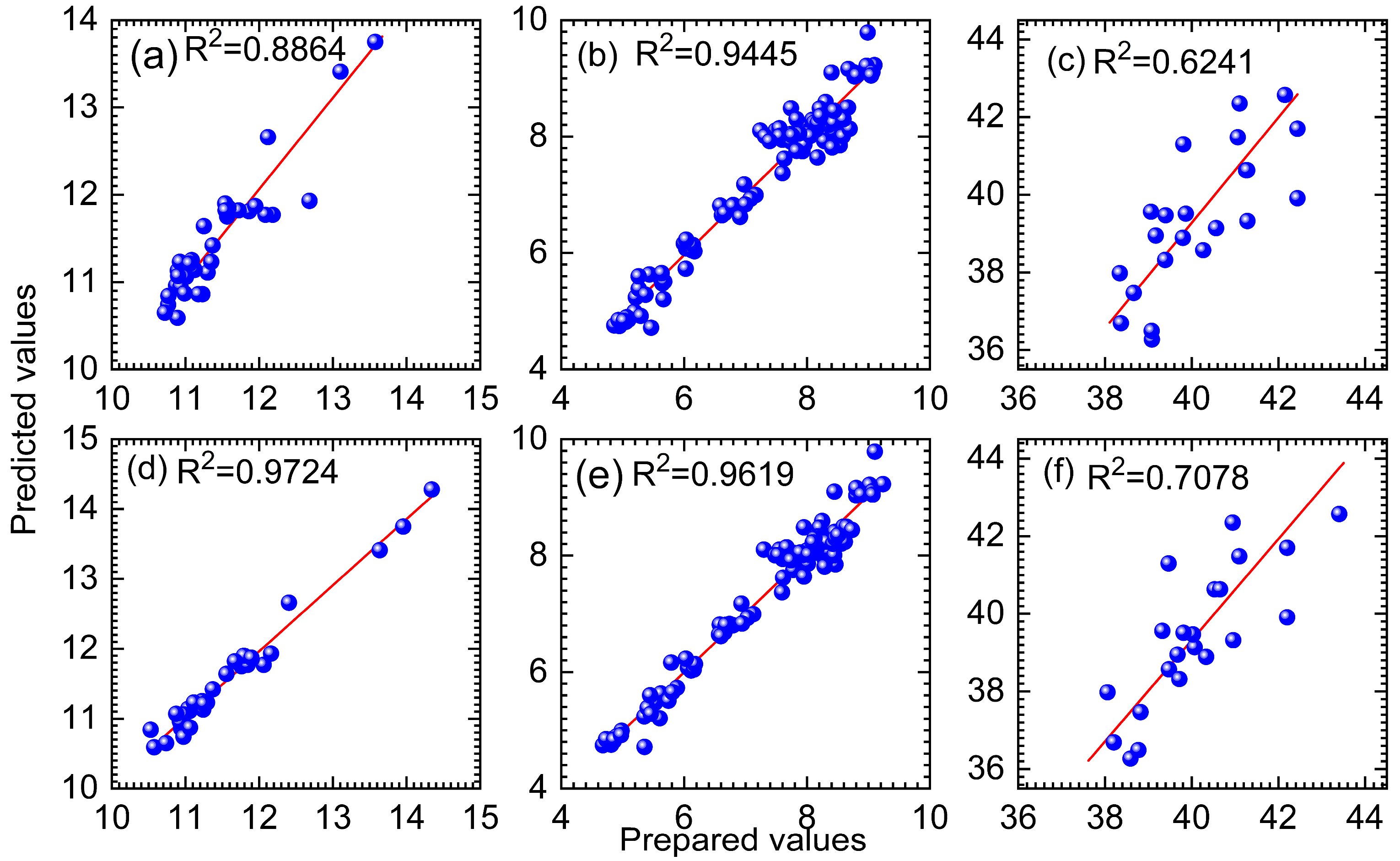
| Dataset | Methods | Variable Number | RMSEP | R2 |
|---|---|---|---|---|
| Wheat | PLS | 701 | 0.2846 | 0.8864 |
| UVE-PLS | 210 | 0.3138 | 0.8619 | |
| MC-UVE-PLS | 205 | 0.3209 | 0.8556 | |
| RT-PLS | 200 | 0.2975 | 0.8759 | |
| DBWO-PLS | 87 | 0.1419 | 0.9724 | |
| Tablets | PLS | 404 | 0.3119 | 0.9445 |
| UVE-PLS | 190 | 0.3081 | 0.9458 | |
| MC-UVE-PLS | 185 | 0.3019 | 0.9455 | |
| RT-PLS | 135 | 0.3023 | 0.9478 | |
| DBWO-PLS | 55 | 0.2585 | 0.9619 | |
| Cocoa beans | PLS | 1557 | 1.6215 | 0.6241 |
| UVE-PLS | 290 | 1.5602 | 0.6389 | |
| MC-UVE-PLS | 75 | 1.6384 | 0.6368 | |
| RT-PLS | 70 | 1.5962 | 0.6357 | |
| DBWO-PLS | 167 | 1.3781 | 0.7078 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kousar, J.; Yang, L.; Xiang, J.; Mao, Q.; Bian, X. Beluga Optimization Algorithm for Near-Infrared Spectral Variable Selection of Complex Samples. Foods 2025, 14, 4266. https://doi.org/10.3390/foods14244266
Kousar J, Yang L, Xiang J, Mao Q, Bian X. Beluga Optimization Algorithm for Near-Infrared Spectral Variable Selection of Complex Samples. Foods. 2025; 14(24):4266. https://doi.org/10.3390/foods14244266
Chicago/Turabian StyleKousar, Javaria, Liping Yang, Jiale Xiang, Qingwei Mao, and Xihui Bian. 2025. "Beluga Optimization Algorithm for Near-Infrared Spectral Variable Selection of Complex Samples" Foods 14, no. 24: 4266. https://doi.org/10.3390/foods14244266
APA StyleKousar, J., Yang, L., Xiang, J., Mao, Q., & Bian, X. (2025). Beluga Optimization Algorithm for Near-Infrared Spectral Variable Selection of Complex Samples. Foods, 14(24), 4266. https://doi.org/10.3390/foods14244266







