Identification of Bitter Peptides in Lilium lancifolium Thunb.; Peptidomics, Computational Simulation and Cellular Functional Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Electronic Tongue Analysis and Sensory Evaluation
2.2.2. Peptide Extraction
2.2.3. Lc-Ms/Ms Analysis and Data Processing
2.2.4. Database Search and Quantification
2.2.5. Homology Modeling, Molecular Docking, and Md Simulation
2.2.6. Sensory Evaluation of Total Peptides and Four Candidate Bitter Peptides
2.2.7. Construction of Egfp-Tas2r14 Stable Cell Line and Peptide Ligand-Induced Fluorescence Imaging
2.2.8. Detection of Signaling Molecules Following Peptides Stimulation
2.2.9. Sequencing of the Transcriptome and Validation of Differentially Expressed Genes by RT-qPCR
3. Results
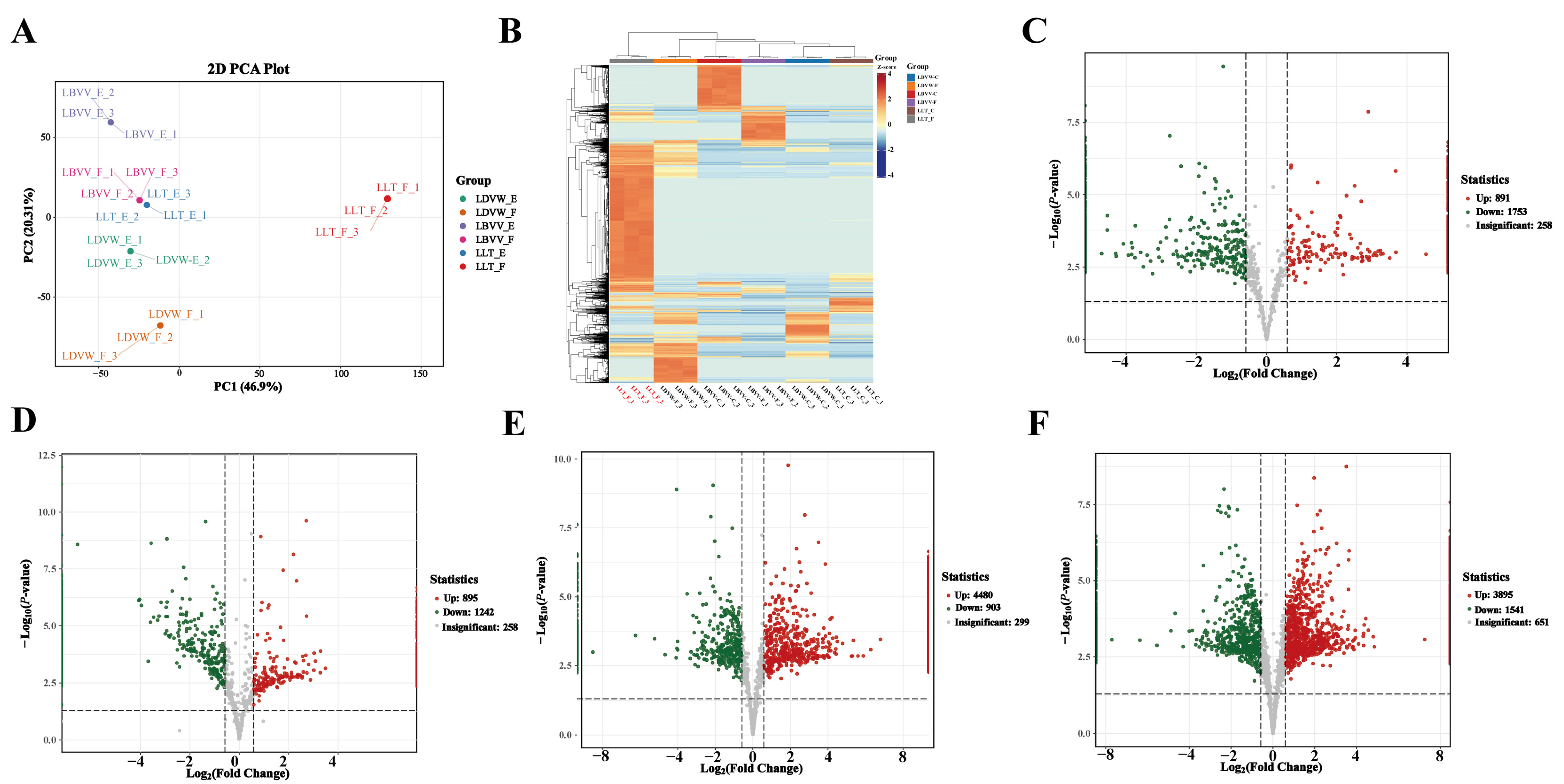
3.1. The Peptidomic Basis of Bitterness Formation in L. lancifolium and Electronic Tongue Validation
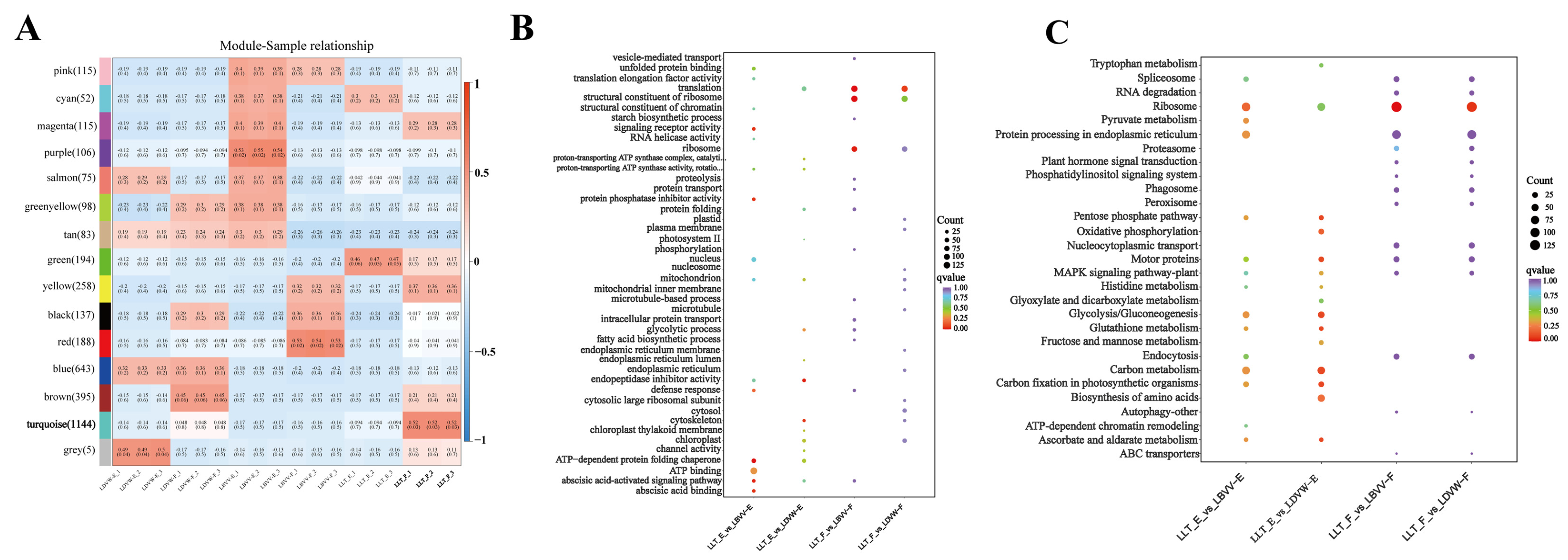
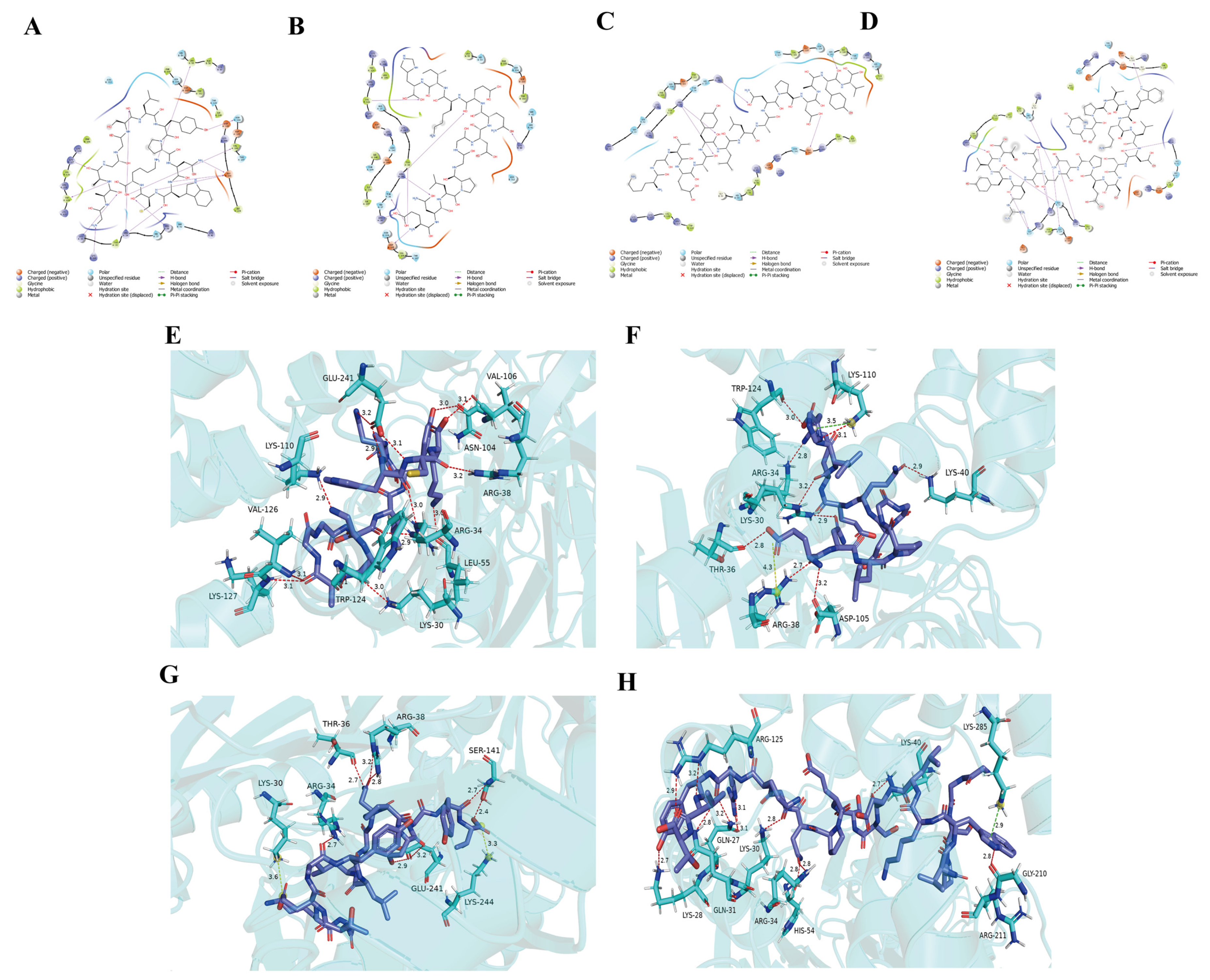
3.2. High-Throughput Screening of Bitter Peptides in L. lancifolium and Their Receptor Interaction Networks
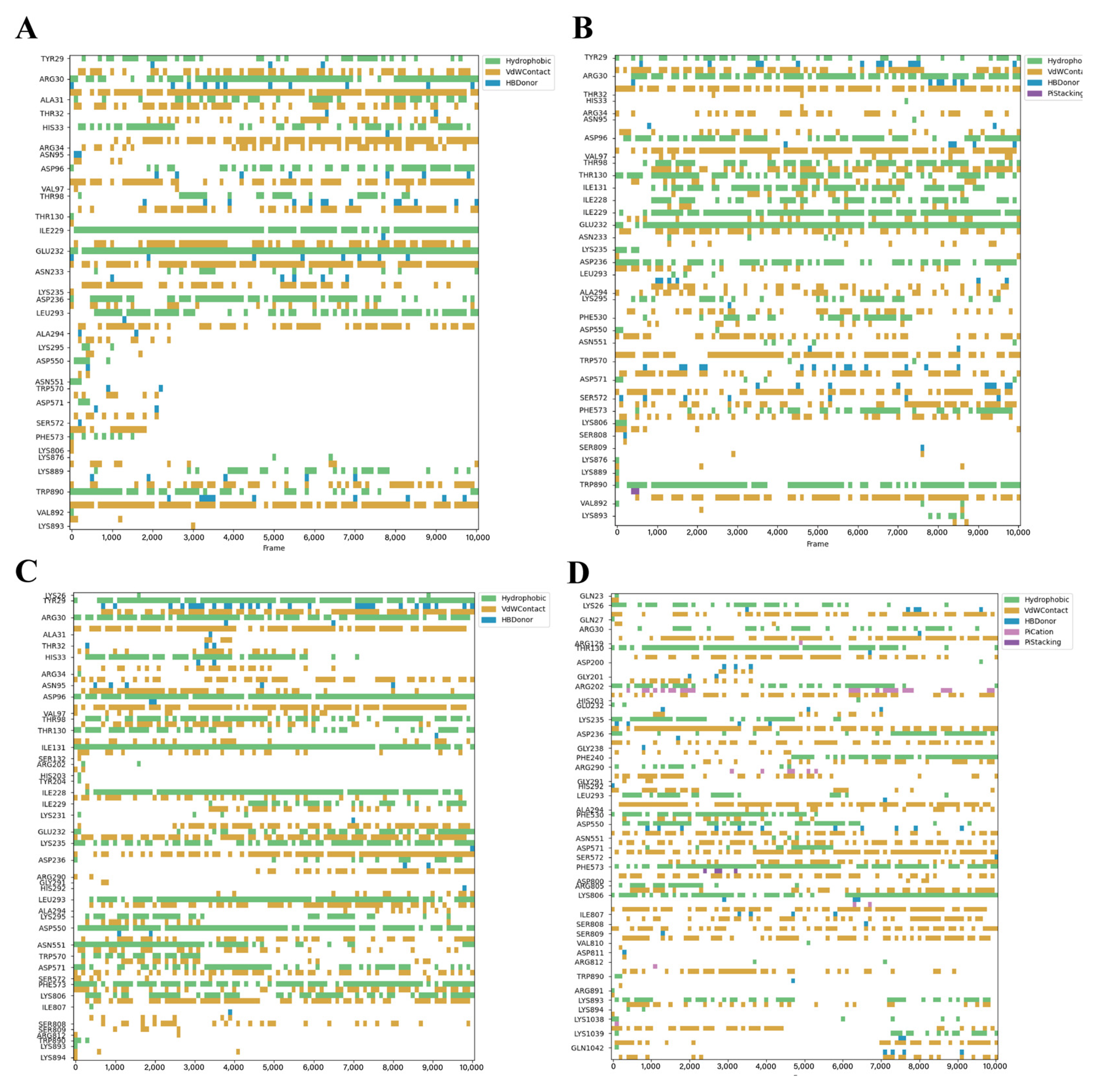
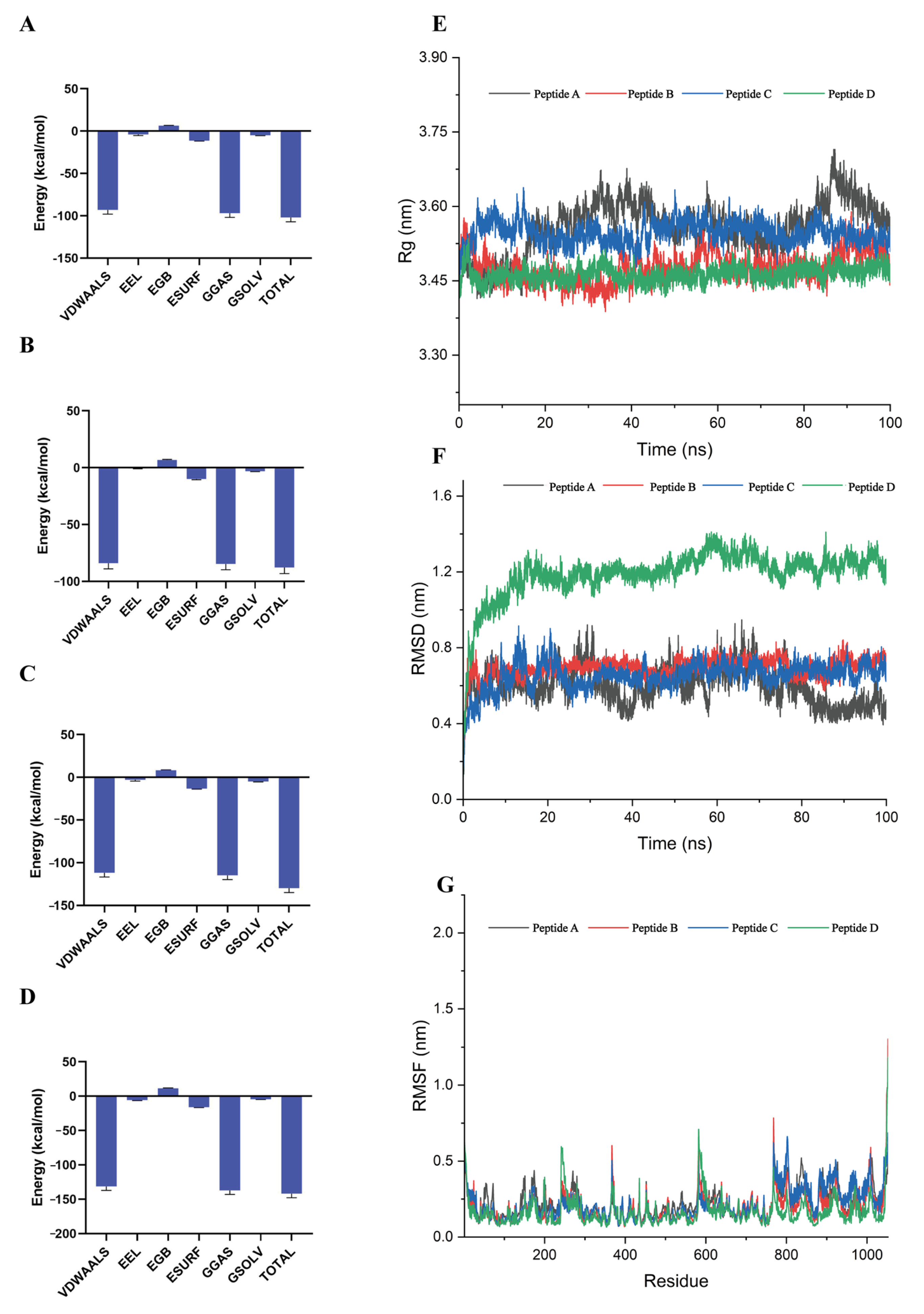
3.3. Dynamic Stability of Critical Binding Residues
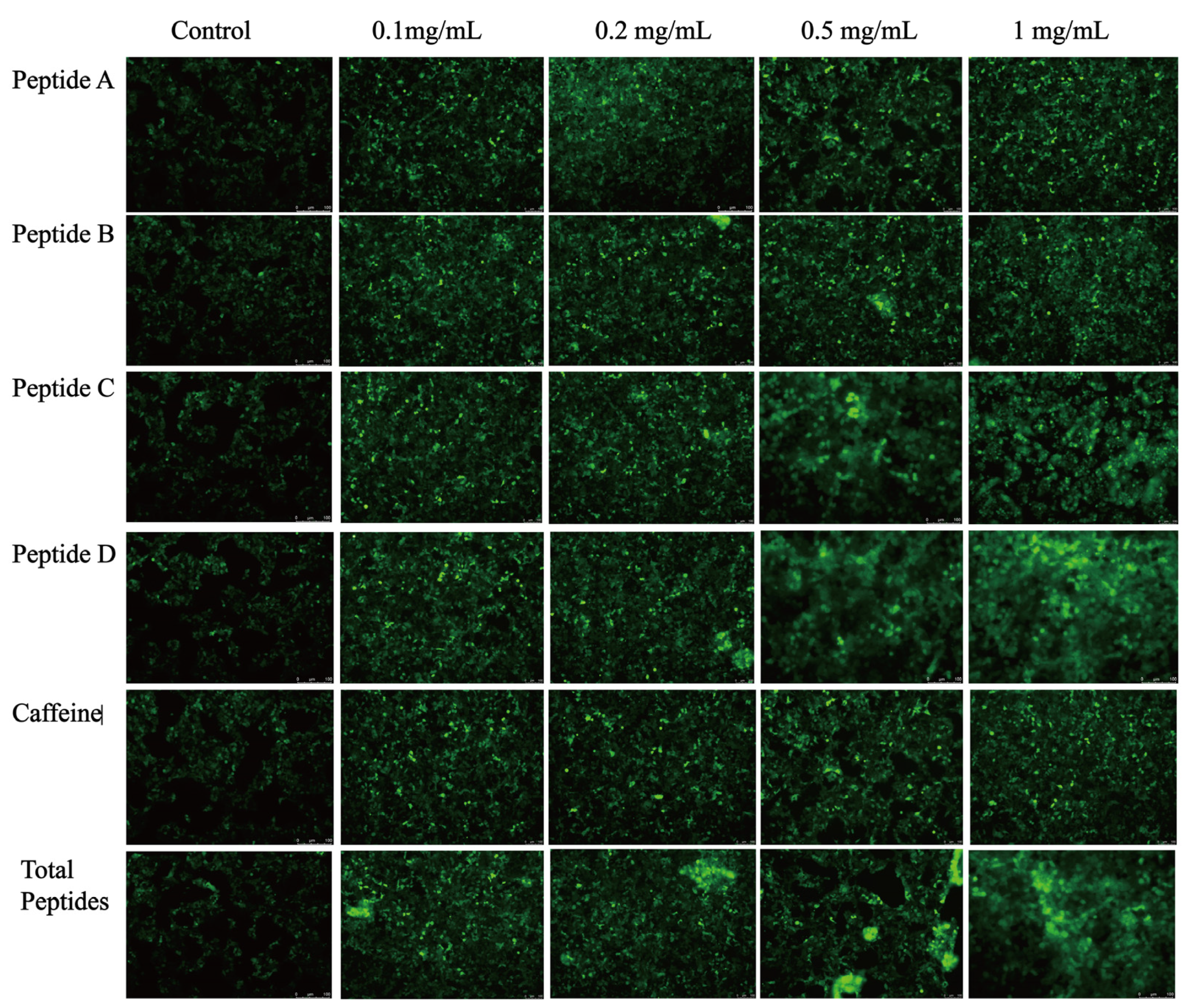
3.4. Sensory Evaluation and Validation of the Fluorescence Response
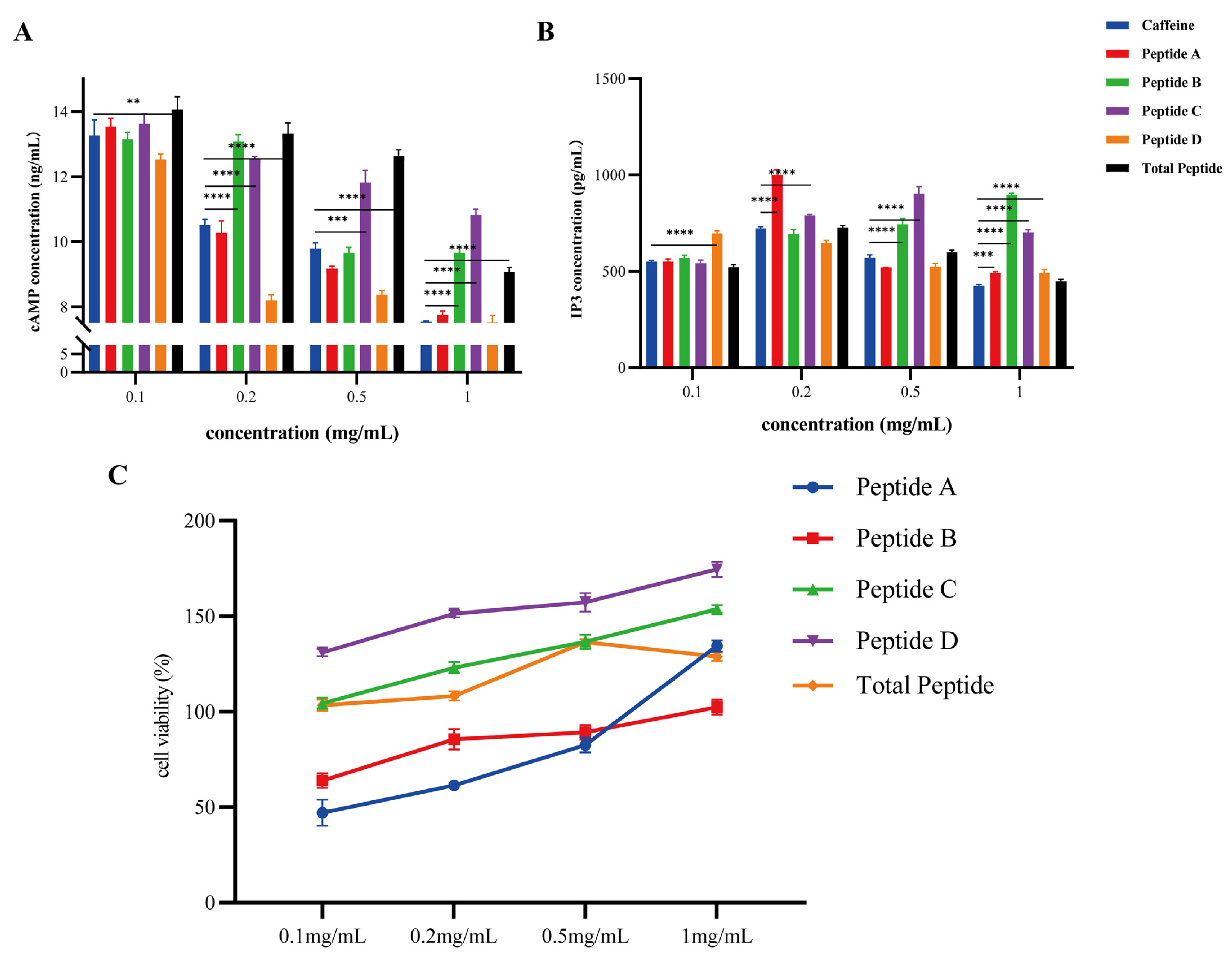
3.5. Plcb2/Ip3 Axis-Mediated Bitter Signal Transduction Mechanism
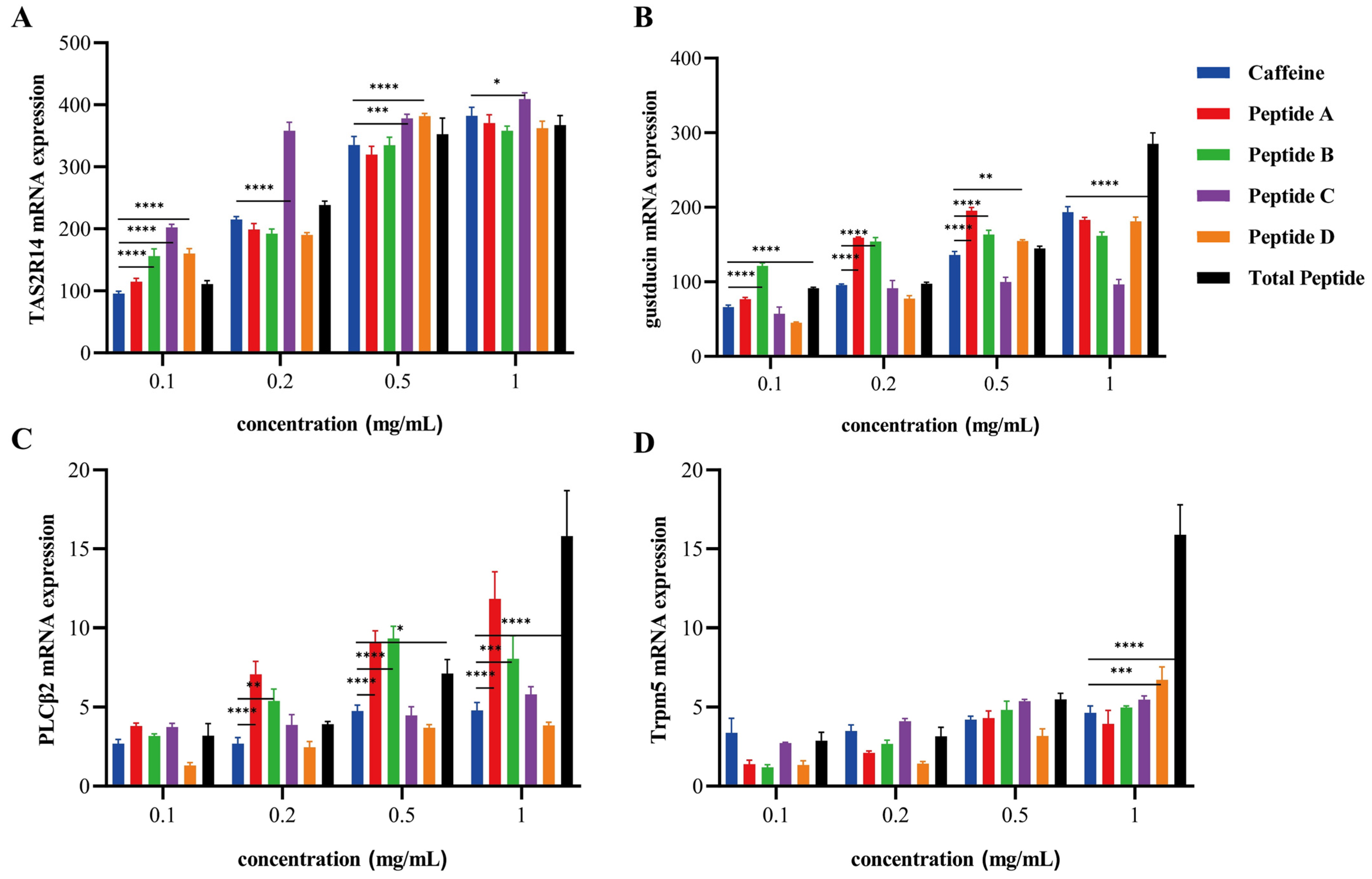
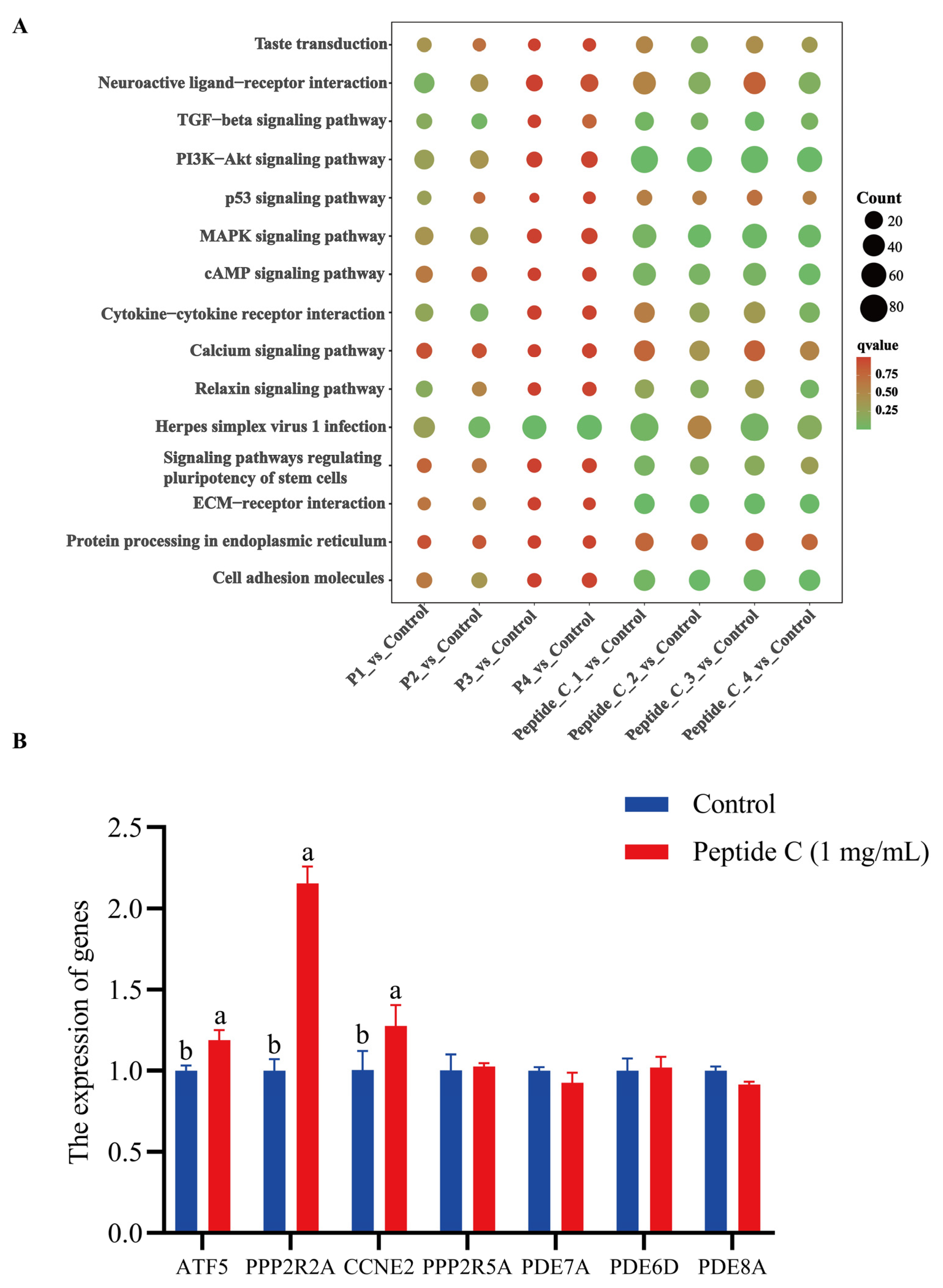
3.6. Transcriptomic Profiling Reveals Multi-Pathway Modulation of Bitter Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, J.; An, R.; Huang, X. Genus Lilium: A review on traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2021, 270, 113852. [Google Scholar] [CrossRef]
- Bakhshaie, M.; Khosravi, S.; Azadi, P.; Bagheri, H.; van Tuyl, J.M. Biotechnological advances in Lilium. Plant Cell Rep. 2016, 35, 1799–1826. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hou, J.; Zhang, Z.; Chen, L.; Ni, H.; Qian, Y.; Wu, W.; Long, H.; Zhang, L.; Li, F.; et al. In-depth exploration and comparison of chemical constituents from two Lilium species through offline two-dimensional liquid chromatography combined with multimode acquisition of high-resolution mass spectrometry. J. Chromatogr. A 2022, 1670, 462980. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wang, H.; Lang, L.; Dou, X.; Bai, J. Metabolome-based discrimination analysis of five Lilium bulbs associated with differences in secondary metabolites. Molecules 2021, 26, 1340. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tang, H.P.; Bai, Q.X.; Yu, A.Q.; Wang, S.; Wu, L.H.; Fu, L.; Wang, Z.B.; Kuang, H.X. Extraction, purification, structural characteristics, biological activities, and applications of polysaccharides from the genus Lilium: A review. Int. J. Biol. Macromol. 2024, 267, 131499. [Google Scholar] [CrossRef]
- Guo, J.; Lu, L.; Li, J.; Kang, S.; Li, G.; Li, S.; Yuan, M. Extraction, structure, pharmacological activity, and structural modification of Lilium polysaccharides. Fitoterapia 2024, 172, 105760. [Google Scholar] [CrossRef]
- Kan, J.; Hui, Y.; Xie, W.; Chen, C.; Liu, Y.; Jin, C. Lily bulbs’ polyphenols extract ameliorates oxidative stress and lipid accumulation in vitro and in vivo. J. Sci. Food Agric. 2021, 101, 5038–5048. [Google Scholar] [CrossRef]
- Han, S.Y.; Yi, Y.S.; Jeong, S.G.; Hong, Y.H.; Choi, K.J.; Hossain, M.A.; Hwang, H.; Rho, H.S.; Lee, J.; Kim, J.-H.; et al. Ethanol extract of lilium bulbs plays an anti-inflammatory role by targeting the IKK α/β-mediated NF-κ B pathway in macrophages. Am. J. Chin. Med. 2018, 46, 1281–1296. [Google Scholar] [CrossRef]
- Nanjing University of Chinese Medicine (Ed.) Zhong Yao Da Ci Dian, 2nd ed.; Shanghai Scientific & Technical Publishers: Shanghai, China, 2006. [Google Scholar]
- Editorial Board of Zhong Hua Ben Cao; State Administration of Traditional Chinese Medicine. Zhong Hua Ben Cao: Jing Xuan Ben; Shanghai Scientific & Technical Publishers: Shanghai, China, 1998. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press Co., Ltd.: Beijing, China, 2020; Volume 1, pp. 137–138. [Google Scholar]
- Richter, P.; Sebald, K.; Fischer, K.; Schnieke, A.; Jlilati, M.; Mittermeier-Klessinger, V.; Somoza, V. Gastric digestion of the sweet-tasting plant protein thaumatin releases bitter peptides that reduce H. pylori induced pro-inflammatory IL-17A release via the TAS2R16 bitter taste receptor. Food Chem. 2024, 448, 139157. [Google Scholar] [CrossRef]
- Gradl, K.; Richter, P.; Somoza, V. Bitter peptides formed during in-vitro gastric digestion induce mechanisms of gastric acid secretion and release satiating serotonin via bitter taste receptors TAS2R4 and TAS2R43 in human parietal cells in culture. Food Chem. 2025, 482, 144174. [Google Scholar] [CrossRef]
- Luo, T.; He, Y.; Jiang, L.; Yang, L.; Hou, X.; Shen, G.; Cui, Q.; Yu, J.; Ke, J.; Chen, S.; et al. Flavor perception and biological activities of bitter compounds in food. Food Chem. 2025, 477, 143532. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, X.; Yu, M.; Tian, J.; Chang, P.; Zhu, S. Bitter peptides in fermented soybean foods—A review. Plant Foods Hum. Nutr. 2023, 78, 261–269. [Google Scholar] [CrossRef]
- Peri, L.; Matzov, D.; Huxley, D.R.; Rainish, A.; Fierro, F.; Sapir, L.; Pfeiffer, T.; Waterloo, L.; Hübner, H.; Peleg, Y.; et al. A bitter anti-inflammatory drug binds at two distinct sites of a human bitter taste GPCR. Nat. Commun. 2024, 15, 9991. [Google Scholar] [CrossRef]
- Miller, Z.A.; Carey, R.M.; Lee, R.J. A deadly taste: Linking bitter taste receptors and apoptosis. Apoptosis 2025, 30, 674–692. [Google Scholar] [CrossRef]
- Matsushima, K.; Yang, D.; Oppenheim, J.J. Interleukin-8: An evolving chemokine. Cytokine 2022, 153, 155828. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Qian, W.; Subramaniyam, S.; Liu, S.; Xin, W. Denatonium enhanced the tone of denuded rat aorta via bitter taste receptor and phosphodiesterase activation. Eur. J. Pharmacol. 2020, 872, 172951. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, S.; Wang, B.; Wang, M.; Liao, P. Unraveling bitter peptides in wheat protein hydrolysates. Food Chem. Mol. Sci. 2025, 10, 100263. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Jiang, S.; Zhu, S.; Zhu, Y.; Chang, P.; Zheng, J. Identification of bitter peptides in sufu via homology modeling and molecular docking. J. Sci. Food Agric. 2025, 105, 8119–8126. [Google Scholar] [CrossRef]
- Dai, W.; Xiang, A.; Pan, D.; Xia, Q.; Sun, Y.; Wang, Y.; Wang, W.; Cao, J.; Zhou, C. Insights into the identification of bitter peptides from Jinhua ham and its taste mechanism by molecular docking and transcriptomics analysis. Food Res. Int. 2024, 189, 114534. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Zhao, W.; Li, J.; Shuian, D.; Liu, J. Identification of Oncorhynchus mykiss nebulin-derived peptides as bitter taste receptor TAS2R14 blockers by in silico screening and molecular docking. Food Chem. 2022, 368, 130839. [Google Scholar] [CrossRef]
- Giménez-Gómez, P.; Escudé-Pujol, R.; Capdevila, F.; Puig-Pujol, A.; Jiménez-Jorquera, C.; Gutiérrez-Capitán, M. Portable electronic tongue based on microsensors for the analysis of cava wines. Sensors 2016, 16, 1796. [Google Scholar] [CrossRef]
- Gomes, F.A.; Souza Junior, D.R.; Massafera, M.P.; Ronsein, G.E. Robust assessment of sample preparation protocols for proteomics of cells and tissues. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2024, 1872, 141030. [Google Scholar] [CrossRef] [PubMed]
- Subedi, P.; Schneider, M.; Philipp, J.; Azimzadeh, O.; Metzger, F.; Moertl, S.; Atkinson, M.J.; Tapio, S. Comparison of methods to isolate proteins from extracellular vesicles for mass spectrometry-based proteomic analyses. Anal. Biochem. 2019, 584, 113390. [Google Scholar] [CrossRef] [PubMed]
- Ochi, N.; Suzuki, T. Determination of lipophilic marine biotoxins (azaspiracids, brevetoxins, and okadaic acid group) and domoic acid in mussels by solid-phase extraction and reversed-phase liquid chromatography with tandem mass spectrometry. J. Chromatogr. A 2024, 1720, 464795. [Google Scholar] [CrossRef]
- Shams Nosrati, M.S.; Doustmohammadi, A.; Severino, M.; Romano, F.; Zafari, M.; Nemati, A.H.; Velmans, C.; Netzer, C.; Breuer, J.; Broekaert, I.J.; et al. Novel KIF26A variants associated with pediatric intestinal pseudo-obstruction (PIPO) and brain developmental defects. Clin. Genet. 2025, 107, 83–90. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Maestro, version 2020-4; Schrödinger, LLC: New York, NY, USA, 2020.
- Waterloo, L.; Hübner, H.; Fierro, F.; Pfeiffer, T.; Brox, R.; Löber, S.; Weikert, D.; Niv, M.Y.; Gmeiner, P. Discovery of 2-aminopyrimidines as potent agonists for the bitter taste receptor TAS2R14. J. Med. Chem. 2023, 66, 3499–3521. [Google Scholar] [CrossRef]
- Yang, L.; Dong, H.; Wang, J.; Dadmohammadi, Y.; Zhou, Y.; Lin, T.; Khongkomolsakul, W.; Meletharayil, G.; Kapoor, R.; Abbaspourrad, A. Fabrication and characterization of whey protein isolate-tryptophan nanoparticles by pH-shifting combined with heat treatment. Food Res. Int. 2024, 196, 115031. [Google Scholar] [CrossRef]
- Di Pizio, A.; Waterloo, L.A.W.; Brox, R.; Löber, S.; Weikert, D.; Behrens, M.; Gmeiner, P.; Niv, M.Y. Rational Design of Agonists for Bitter Taste Receptor TAS2R14: From Modeling to Bench and Back. Cell. Mol. Life Sci. 2020, 77, 531–542. [Google Scholar] [CrossRef]
- Qin, Y.; Jin, J.; Zhou, R.; Fang, L.; Liu, H.; Zhong, C.; Xie, Y.; Liu, P.; Qin, Y.; Zhang, S. Integrative analysis of metabolome and transcriptome provide new insights into the bitter components of Lilium lancifolium and Lilium brownii. J. Pharm. Biomed. Anal. 2022, 215, 114778. [Google Scholar] [CrossRef]
- Hu, X.; Ao, W.; Gao, M.; Wu, L.; Pei, Y.; Liu, S.; Wu, Y.; Zhao, F.; Sun, Q.; Liu, J.; et al. Bitter taste TAS2R14 activation by intracellular tastants and cholesterol. Nature 2024, 631, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, Y.-L.; Yang, Y.; Guo, S.; You, E.; Xie, X.; Jiang, Y.; Mao, C.; Xu, H.E.; Zhang, Y.; et al. Structural Basis of Tethered Agonism and G Protein Coupling of Protease-Activated Receptors. Cell Res. 2024, 34, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Hollenhorst, M.I.; Kumar, P.; Zimmer, M.; Salah, A.; Maxeiner, S.; Elhawy, M.I.; Evers, S.B.; Flockerzi, V.; Gudermann, T.; Chubanov, V. Taste receptor activation in tracheal brush cells by denatonium modulates ENaC channels via Ca2+, cAMP and ACh. Cells 2022, 11, 2411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tu, Z.; Wen, P.; Wang, H.; Hu, Y. Peptidomics screening and molecular docking with umami receptors T1R1/T1R3 of novel umami peptides from oyster (Crassostrea gigas) hydrolysates. J. Agric. Food Chem. 2023, 72, 634–646. [Google Scholar] [CrossRef]
- Zhao, J.; Liao, S.; Han, J.; Xie, Y.; Tang, J.; Zhao, J.; Shao, W.; Wang, Q.; Lin, H. Revealing the secret of umami taste of peptides derived from fermented broad bean paste. J. Agric. Food Chem. 2023, 71, 4706–4716. [Google Scholar] [CrossRef]
- Meng, T.K.; Han, R.L.; Ma, P.; Chen, S.X.; Qi, B.H.; Wang, Z.X.; Li, X.Y.; Deng, H.S. Microemulsion-based drug delivery system identifies pepper alkaloids as anti-obesity compounds. Acta Pharmacol. Sin. 2025, 46, 2310–2322. [Google Scholar] [CrossRef]
- Zhou, Y.-W.; Sun, J.; Wang, Y.; Chen, C.-P.; Tao, T.; Ma, M.; Chen, X.; Zhang, X.-N.; Yang, L.-Y.; Zhang, Z.-L.; et al. Tas2R activation relaxes airway smooth muscle by release of Gαt targeting on AChR signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2121513119. [Google Scholar] [CrossRef]
- Cherkashin, A.P.; Rogachevskaja, O.A.; Khokhlov, A.A.; Kabanova, N.V.; Bystrova, M.F.; Kolesnikov, S.S. Contribution of TRPC3-mediated Ca2+ entry to taste transduction. Pflügers Arch. Eur. J. Physiol. 2023, 475, 1009–1024. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Salido, G.M.; Rosado, J.A. Cross-talk between the adenylyl cyclase/cAMP pathway and Ca2+ homeostasis. Rev. Physiol. Biochem. Pharmacol. 2021, 179, 73–116. [Google Scholar] [CrossRef]
- Di Mise, A.; Tamma, G.; Ranieri, M.; Centrone, M.; van den Heuvel, L.; Mekahli, D.; Levtchenko, E.N.; Valenti, G. Activation of Calcium-Sensing Receptor increases intracellular calcium and decreases cAMP and mTOR in PKD1 deficient cells. Sci. Rep. 2018, 8, 5704. [Google Scholar] [CrossRef]
- Marosi, M.; Nenov, M.N.; Di Re, J.; Dvorak, N.M.; Alshammari, M.; Laezza, F. Inhibition of the Akt/PKB kinase increases Nav1.6-mediated currents and neuronal excitability in CA1 hippocampal pyramidal neurons. Int. J. Mol. Sci. 2022, 23, 1700. [Google Scholar] [CrossRef]
- Zhang, Y.; Smolen, P.D.; Cleary, L.J.; Byrne, J.H. Quantitative description of the interactions among kinase cascades underlying long-term plasticity of Aplysia sensory neurons. Sci. Rep. 2021, 11, 14931. [Google Scholar] [CrossRef]
- Jalsevac, F.; Descamps-Solà, M.; Vilalta, A.; Segú, H.; Blay, M.T.; Beltrán-Debón, R.; Rodríguez-Gallego, E.; Terra, X.; Ardévol, A.; Pinent, M. Subchronic modulation of bitter taste receptors (TAS2R) by procyanidins. Unravelling the complex interplay between stimulation and expression. J. Physiol. Biochem. 2025, 1–14. [Google Scholar] [CrossRef]
- Stephani, M.; Picchianti, L.; Gajic, A.; Beveridge, R.; Skarwan, E.; de Medina Hernandez, V.S.; Mohseni, A.; Clavel, M.; Zeng, Y.; Naumann, C.; et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 2020, 9, e58396. [Google Scholar] [CrossRef]
| Bitter Taste Receptor | Number of Effectively Bound Peptides | CDOCKER Interaction Energy (kcal/mol) |
|---|---|---|
| TAS2R3 | 2 | −73.38 |
| TAS2R4 | 3 | −65.18 |
| TAS2R7 | 2 | −49.82 |
| TAS2R10 | 1 | −167.33 |
| TAS2R14 | 76 | −119.73 |
| TAS2R39 | 1 | −139.65 |
| TAS2R41 | 44 | −98.12 |
| TAS2R46 | 121 | −114.88 |
| Energy (kcal/mol) | Peptide A | Peptide B | Peptide C | Peptide D |
|---|---|---|---|---|
| △Evdw | −93.05 | −83.88 | −111.74 | −131.4 |
| △Ele | −3.91 | −0.72 | −2.98 | −5.76 |
| △Esurf | 6.38 | 6.87 | 8.28 | 11.44 |
| △EGB | −11.32 | −9.99 | −13.25 | −15.96 |
| △EGas | −96.96 | −84.6 | −114.72 | −137.16 |
| △Esolv | −4.94 | −3.12 | −4.97 | −4.51 |
| △EBind | −101.9 | −87.72 | −129.69 | −141.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Zhong, X.; Sun, M.; Huang, P.; He, Y.; Gong, H.; Zhou, L.; Zeng, J.; Xiang, W. Identification of Bitter Peptides in Lilium lancifolium Thunb.; Peptidomics, Computational Simulation and Cellular Functional Assays. Foods 2025, 14, 4056. https://doi.org/10.3390/foods14234056
Dong Z, Zhong X, Sun M, Huang P, He Y, Gong H, Zhou L, Zeng J, Xiang W. Identification of Bitter Peptides in Lilium lancifolium Thunb.; Peptidomics, Computational Simulation and Cellular Functional Assays. Foods. 2025; 14(23):4056. https://doi.org/10.3390/foods14234056
Chicago/Turabian StyleDong, Zhuang, Xiaohong Zhong, Mengshan Sun, Peng Huang, Yuedong He, Haiyuan Gong, Li Zhou, Jianguo Zeng, and Wei Xiang. 2025. "Identification of Bitter Peptides in Lilium lancifolium Thunb.; Peptidomics, Computational Simulation and Cellular Functional Assays" Foods 14, no. 23: 4056. https://doi.org/10.3390/foods14234056
APA StyleDong, Z., Zhong, X., Sun, M., Huang, P., He, Y., Gong, H., Zhou, L., Zeng, J., & Xiang, W. (2025). Identification of Bitter Peptides in Lilium lancifolium Thunb.; Peptidomics, Computational Simulation and Cellular Functional Assays. Foods, 14(23), 4056. https://doi.org/10.3390/foods14234056






