Dynamic Modelling of Listeria monocytogenes Growth in a Milk Model Medium as Affected by pH and Selected Lactic Acid Bacteria Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Preparation
2.2. Inoculation of the Milk Model Medium
2.3. Microbiological and Physicochemical Analysis
2.4. Modelling of L. monocytogenes and LAB in Monoculture and Coculture in the Milk Model Medium
2.5. Estimation of Parameters
2.6. External Validation
3. Results and Discussion
3.1. Growth of L. monocytogenes and LAB Strains in the Milk Model as Affected by Initial pH
3.1.1. Growth of L. monocytogenes and LAB Strains in Monoculture
3.1.2. Growth of L. monocytogenes in Coculture with LAB Strains
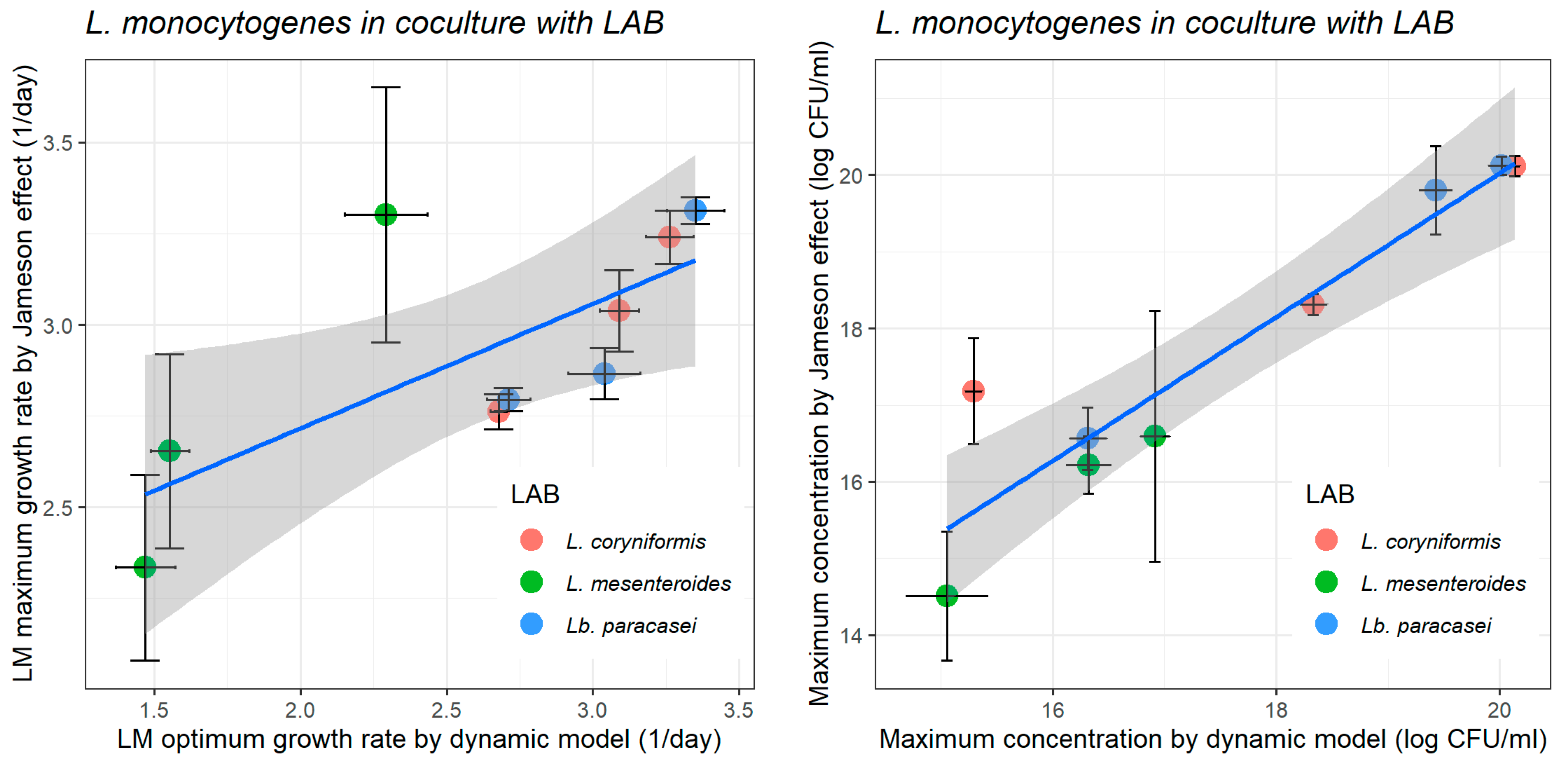
3.2. A Comparison Between the Kinetic Parameters of L. monocytogenes in the Milk Model Obtained by the pH-Driven Dynamic Model and the Jameson-Effect Competition Model
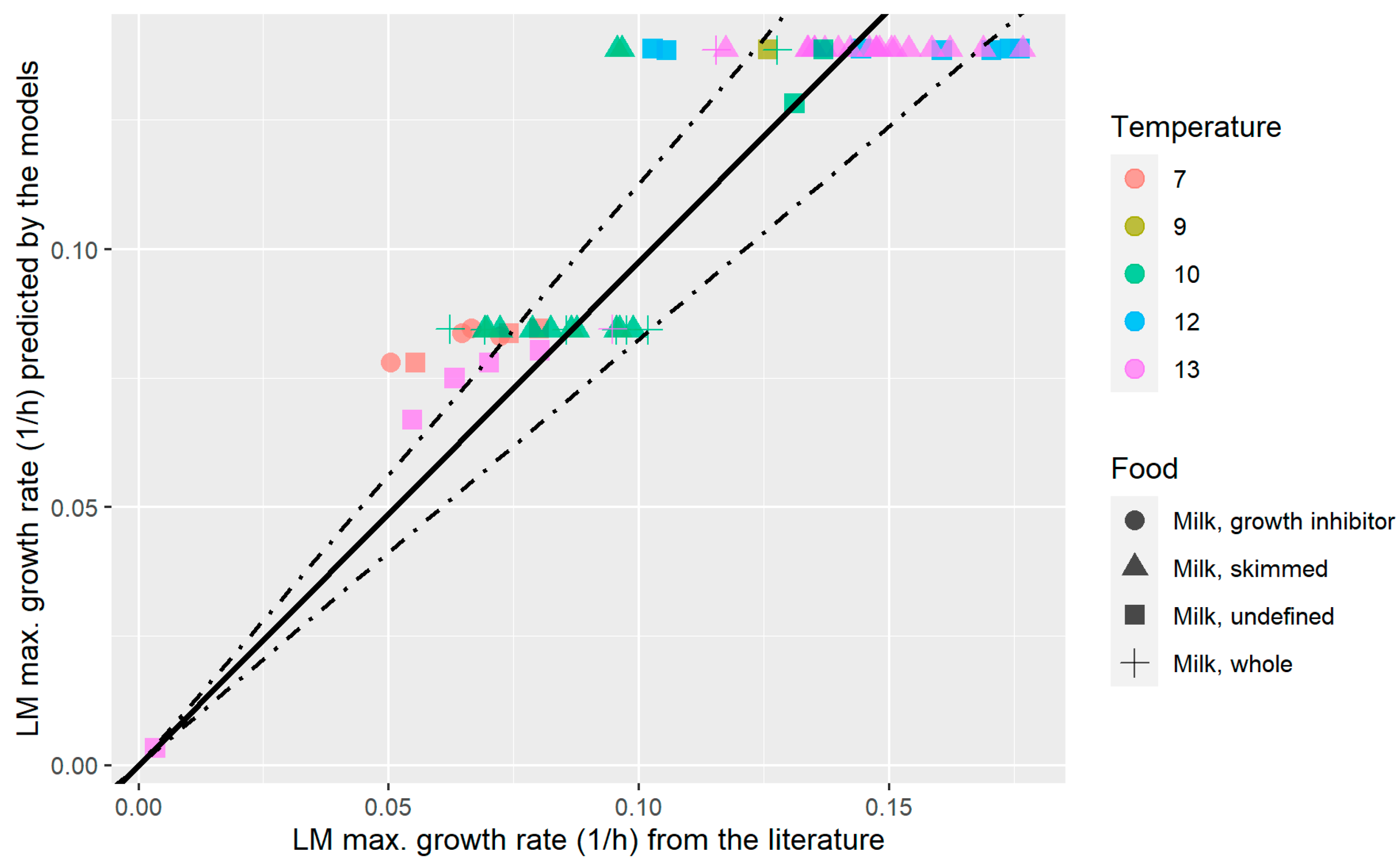
3.3. External Validation of the pH-Driven Dynamic Model of L. monocytogenes in the Milk Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sibanda, T.; Buys, E.M. Listeria monocytogenes Pathogenesis: The Role of Stress Adaptation. Microorganisms 2022, 10, 1522. [Google Scholar] [CrossRef] [PubMed]
- Warriner, K.; Namvar, A. What Is the Hysteria with Listeria? Trends Food Sci. Technol. 2009, 20, 245–254. [Google Scholar] [CrossRef]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Listeria monocytogenes in Cheese and the Dairy Environment Remains a Food Safety Challenge: The Role of Stress Responses. Food Res. Int. 2015, 67, 75–90. [Google Scholar] [CrossRef]
- Pyz-Łukasik, R.; Gondek, M.; Winiarczyk, D.; Michalak, K.; Paszkiewicz, W.; Piróg-Komorowska, A.; Policht, A.; Ziomek, M. Occurrence of Listeria monocytogenes in Artisanal Cheeses from Poland and Its Identification by MALDI-TOF MS. Pathogens 2021, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Allerberger, F.; Wagner, M. Listeriosis: A Resurgent Foodborne Infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Saraoui, T.; Leroi, F.; Chevalier, F.; Cappelier, J.-M.; Passerini, D.; Pilet, M.-F. Bioprotective Effect of Lactococcus piscium CNCM I-4031 Against Listeria monocytogenes Growth and Virulence. Front. Microbiol. 2018, 9, 1564. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC) The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [CrossRef]
- Osek, J.; Wieczorek, K. Listeria monocytogenes—How This Pathogen Uses Its Virulence Mechanisms to Infect the Hosts. Pathogens 2022, 11, 1491. [Google Scholar] [CrossRef]
- Fotopoulou, E.T.; Jenkins, C.; Painset, A.; Amar, C. Listeria monocytogenes: The Silent Assassin. J. Med. Microbiol. 2024, 73, 001800. [Google Scholar] [CrossRef]
- EFSA Dashboard on Listeria monocytogenes (European, Food Safety Authority) 2024. Available online: https://www.efsa.europa.eu/en/microstrategy/listeria-dashboard (accessed on 5 September 2025).
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Escamez, P.S.F.; Girones, R.; Herman, L.; Koutsoumanis, K.; Norrung, B.; et al. Listeria monocytogenes Contamination of Ready-to-Eat Foods and the Risk for Human Health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Chan, Y.C.; Wiedmann, M. Physiology and Genetics of Listeria monocytogenes Survival and Growth at Cold Temperatures. Crit. Rev. Food Sci. Nutr. 2009, 49, 237–253. [Google Scholar] [CrossRef]
- Lomonaco, S.; Decastelli, L.; Nucera, D.; Gallina, S.; Manila Bianchi, D.; Civera, T. Listeria monocytogenes in Gorgonzola: Subtypes, Diversity and Persistence over Time. Int. J. Food Microbiol. 2009, 128, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, B.; Szymczak, M.; Trafiałek, J. Prevalence of Listeria species and L. monocytogenes in Ready-to-Eat Foods in the West Pomeranian Region of Poland: Correlations between the Contamination Level, Serogroups, Ingredients, and Producers. Food Microbiol. 2020, 91, 103532. [Google Scholar] [CrossRef] [PubMed]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef] [PubMed]
- Morandi, S.; Silvetti, T.; Battelli, G.; Brasca, M. Can Lactic Acid Bacteria Be an Efficient Tool for Controlling Listeria monocytogenes Contamination on Cheese Surface? The Case of Gorgonzola Cheese. Food Control 2019, 96, 499–507. [Google Scholar] [CrossRef]
- Saraoui, T.; Fall, P.A.; Leroi, F.; Antignac, J.-P.; Chéreau, S.; Pilet, M.F. Inhibition Mechanism of Listeria monocytogenes by a Bioprotective Bacteria Lactococcus Piscium CNCM I-4031. Food Microbiol. 2016, 53, 70–78. [Google Scholar] [CrossRef]
- Gonzales-Barron, U.; Cadavez, V.; Guillier, L.; Sanaa, M. A Critical Review of Risk Assessment Models for Listeria monocytogenes in Dairy Products. Foods 2023, 12, 4436. [Google Scholar] [CrossRef]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Castellano, P.; Pérez Ibarreche, M.; Blanco Massani, M.; Fontana, C.; Vignolo, G.M. Strategies for Pathogen Biocontrol Using Lactic Acid Bacteria and Their Metabolites: A Focus on Meat Ecosystems and Industrial Environments. Microorganisms 2017, 5, 38. [Google Scholar] [CrossRef]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocins: Novel Solutions to Age Old Spore-Related Problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef]
- Webb, L.; Ma, L.; Lu, X. Impact of Lactic Acid Bacteria on the Control of Listeria monocytogenes in Ready-to-Eat Foods. Food Qual. Saf. 2022, 6, fyac045. [Google Scholar] [CrossRef]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef]
- Pessione, E. Lactic Acid Bacteria Contribution to Gut Microbiota Complexity: Lights and Shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef]
- Topisirovic, L.; Kojic, M.; Fira, D.; Golic, N.; Strahinic, I.; Lozo, J. Potential of Lactic Acid Bacteria Isolated from Specific Natural Niches in Food Production and Preservation. Int. J. Food Microbiol. 2006, 112, 230–235. [Google Scholar] [CrossRef]
- Silva, B.N.; Fernandes, N.; Carvalho, L.; Faria, A.S.; Teixeira, J.A.; Rodrigues, C.; Gonzales-Barron, U.; Cadavez, V. Lactic Acid Bacteria from Artisanal Raw Goat Milk Cheeses: Technological Properties and Antimicrobial Potential. Ital. J. Food Saf. 2023, 12, 11559. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.N.; Faria, A.S.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Technological Potential of Lactic Acid Bacteria Isolated from Portuguese Goat’s Raw Milk Cheeses. Biol. Life Sci. Forum 2021, 6, 9. [Google Scholar] [CrossRef]
- Henderson, L.O.; Cabrera-Villamizar, L.A.; Skeens, J.; Kent, D.; Murphy, S.; Wiedmann, M.; Guariglia-Oropeza, V. Environmental Conditions and Serotype Affect Listeria monocytogenes Susceptibility to Phage Treatment in a Laboratory Cheese Model. J. Dairy Sci. 2019, 102, 9674–9688. [Google Scholar] [CrossRef] [PubMed]
- Schvartzman, M.S.; Belessi, C.; Butler, F.; Skandamis, P.N.; Jordan, K.N. Effect of pH and Water Activity on the Growth Limits of Listeria monocytogenes in a Cheese Matrix at Two Contamination Levels. J. Food Prot. 2011, 74, 1805–1813. [Google Scholar] [CrossRef]
- Engstrom, S.K.; Cheng, C.; Seman, D.; Glass, K.A. Growth of Listeria monocytogenes in a Model High-Moisture Cheese on the Basis of pH, Moisture, and Acid Type. J. Food Prot. 2020, 83, 1335–1344. [Google Scholar] [CrossRef]
- Schvartzman, M.S.; Gonzalez-Barron, U.; Butler, F.; Jordan, K. Modeling the Growth of Listeria monocytogenes on the Surface of Smear- or Mold-Ripened Cheese. Front. Cell. Infect. Microbiol. 2014, 4, 90. [Google Scholar] [CrossRef]
- Tirloni, E.; Stella, S.; Bernardi, C.; Dalgaard, P.; Rosshaug, P.S. Predicting Growth of Listeria monocytogenes in Fresh Ricotta. Food Microbiol. 2019, 78, 123–133. [Google Scholar] [CrossRef]
- Leong, W.M.; Geier, R.; Engstrom, S.; Ingham, S.; Ingham, B.; Smukowski, M. Growth of Listeria monocytogenes, Salmonella spp., Escherichia coli O157:H7, and Staphylococcus aureus on Cheese during Extended Storage at 25 Degrees C. J. Food Prot. 2014, 77, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Campagnollo, F.B.; Margalho, L.P.; Kamimura, B.A.; Feliciano, M.D.; Freire, L.; Lopes, L.S.; Alvarenga, V.O.; Cadavez, V.A.P.; Gonzales-Barron, U.; Schaffner, D.W.; et al. Selection of Indigenous Lactic Acid Bacteria Presenting Anti-Listerial Activity, and Their Role in Reducing the Maturation Period and Assuring the Safety of Traditional Brazilian Cheeses. Food Microbiol. 2018, 73, 288–297. [Google Scholar] [CrossRef]
- Meloni, M.P.; Piras, F.; Siddi, G.; Cabras, D.; Comassi, E.; Lai, R.; McAuliffe, O.; De Santis, E.P.L.; Scarano, C. Comparison of Activity of Commercial Protective Cultures and Thermophilic Lactic Acid Bacteria against Listeria monocytogenes: A New Perspective to Improve the Safety of Sardinian PDO Cheeses. Foods 2023, 12, 1182. [Google Scholar] [CrossRef]
- Borges, D.O.; Matsuo, M.M.; Bogsan, C.S.B.; da Silva, T.F.; Casarotti, S.N.; Penna, A.L.B. Leuconostoc mesenteroides subsp. mesenteroides SJRP55 Reduces Listeria monocytogenes Growth and Impacts on Fatty Acids Profile and Conjugated Linoleic Acid Content in Fermented Cream. LWT 2019, 107, 264–271. [Google Scholar] [CrossRef]
- Gonzales-Barron, U.; Campagnollo, F.B.; Schaffner, D.W.; Sant’Ana, A.S.; Cadavez, V.A.P. Behavior of Listeria monocytogenes in the Presence or Not of Intentionally-Added Lactic Acid Bacteria during Ripening of Artisanal Minas Semi-Hard Cheese. Food Microbiol. 2020, 91, 103545. [Google Scholar] [CrossRef] [PubMed]
- Ananou, S.; Garriga, M.; Hugas, M.; Maqueda, M.; Martínez-Bueno, M.; Gálvez, A.; Valdivia, E. Control of Listeria monocytogenes in Model Sausages by Enterocin AS-48. Int. J. Food Microbiol. 2005, 103, 179–190. [Google Scholar] [CrossRef]
- Serrano, S.; Morais, S.; Semedo-Lemsaddek, T. Tradition Unveiled: A Comprehensive Review of Microbiological Studies on Portuguese Traditional Cheeses, Merging Conventional and OMICs Analyses. Front. Ind. Microbiol. 2024, 2, 1420042. [Google Scholar] [CrossRef]
- Settanni, L.; Moschetti, G. Non-Starter Lactic Acid Bacteria Used to Improve Cheese Quality and Provide Health Benefits. Food Microbiol. 2010, 27, 691–697. [Google Scholar] [CrossRef]
- Morgan, F.; Bonnin, V.; Mallereau, M.-P.; Perrin, G. Survival of Listeria Monocytogenes during Manufacture, Ripening and Storage of Soft Lactic Cheese Made from Raw Goat Milk. Int. J. Food Microbiol. 2001, 64, 217–221. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; ISO 11290-1: Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria Spp.—Part 1: Detection Method. International Organization for Standardization (ISO): Geneva, Switzerland, 2017.
- ISO 15214:1998; ISO 15214: Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Emumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 °C. International Organization for Standardization (ISO): Genève, Switzerland, 1998.
- Nunes Silva, B.; Cadavez, V.; Teixeira, J.A.; Ellouze, M.; Gonzales-Barron, U. Cardinal Parameter Meta-Regression Models Describing Listeria monocytogenes Growth in Broth. Food Res. Int. Ott. Ont 2020, 136, 109476. [Google Scholar] [CrossRef]
- Charles, S. Predictive Modelling of the Growth Kinetics of a Population of Listeria monocytogenes. Ph.D. Thesis, Claude Bernard University Lyon, Lyon France, 1996. [Google Scholar]
- Donnelly, C.W.; Briggs, E.H. Psychrotrophic Growth and Thermal Inactivation of Listeria monocytogenes as a Function of Milk Composition. J. Food Prot. 1986, 49, 994–999. [Google Scholar] [CrossRef] [PubMed]
- El-Shenawy, M.A.; Marth, E.H. Behavior of Listeria monocytogenes in the Presence of Gluconic Acid and During Preparation of Cottage Cheese Curd Using Gluconic Acid. J. Dairy Sci. 1990, 73, 1429–1438. [Google Scholar] [CrossRef]
- ComBase: A Web Resource for Quantitative and Predictive Food Microbiology. Data from the FSA Food Standards Agency Funded Data Generated at Champden and Chorleywood Food Research Association, UK. Available online: https://combase.errc.ars.usda.gov (accessed on 6 August 2025).
- Jia, Z.; Huang, L.; Wei, Z.; Yao, Y.; Fang, T.; Li, C. Dynamic Kinetic Analysis of Growth of Listeria monocytogenes in Pasteurized Cow Milk. J. Dairy Sci. 2021, 104, 2654–2667. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.L.; Schmidt, R.H. Growth of Listeria monocytogenes at 10 °C in Milk Preincubated with Selected Pseudomonads1. J. Food Prot. 1988, 51, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, F.; Michel, M.; Lebrihi, A.; Lefebvre, G. Effect of the Bacteriocin Carnocin CP5 and of the Producing Strain Carnobacterium piscicola CP5 on the Viability of Listeria monocytogenes ATCC 15313 in Salt Solution, Broth and Skimmed Milk, at Various Incubation Temperatures. Int. J. Food Microbiol. 1994, 22, 155–172. [Google Scholar] [CrossRef]
- Pearson, L.J.; Marth, E.H. Behavior of Listeria monocytogenes in the Presence of Cocoa, Carrageenan, and sugar in a Milk Medium Incubated With and Without Agitation. J. Food Prot. 1990, 53, 30–37. [Google Scholar] [CrossRef]
- Rajkowski, K.T.; Calderone, S.M.; Jones, E. Effect of Polyphosphate and Sodium Chloride on the Growth of Listeria monocytogenes and Staphylococcus aureus in Ultra-High Temperature Milk. J. Dairy Sci. 1994, 77, 1503–1508. [Google Scholar] [CrossRef]
- Rosenow, E.M.; Marth, E.H. Addition of Cocoa Powder, Cane Sugar, and Carrageenan to Milk Enhances Growth of Listeria monocytogenes. J. Food Prot. 1987, 50, 726–730. [Google Scholar] [CrossRef]
- Walker, S.J.; Archer, P.; Banks, J.G. Growth of Listeria monocytogenes at Refrigeration Temperatures. J. Appl. Bacteriol. 1990, 68, 157–162. [Google Scholar] [CrossRef]
- Rosso, L.; Lobry, J.R.; Bajard, S.; Flandrois, J.P. Convenient Model To Describe the Combined Effects of Temperature and pH on Microbial Growth. Appl. Environ. Microbiol. 1995, 61, 610–616. [Google Scholar] [CrossRef]
- Martinez-Rios, V.; Gkogka, E.; Dalgaard, P. New Term to Quantify the Effect of Temperature on pHmin-Values Used in Cardinal Parameter Growth Models for Listeria monocytogenes. Front. Microbiol. 2019, 10, 1510. [Google Scholar] [CrossRef]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria Monocytogenes—How This Pathogen Survives in Food-Production Environments? Front. Microbiol. 2022, 13, 866462. [Google Scholar] [CrossRef]
- Mishra, C.; Lambert, J. Production of Anti-Microbial Substances by Probiotics. Asia Pac. J. Clin. Nutr. 1996, 5, 20–24. [Google Scholar]
- Barbosa, J.; Borges, S.; Teixeira, P. Influence of Sub-Lethal Stresses on the Survival of Lactic Acid Bacteria after Spray-Drying in Orange Juice. Food Microbiol. 2015, 52, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Ono, H.; Kitagawa, H.; Ito, H.; Mu, F.; Sawa, N.; Zendo, T.; Sonomoto, K. Identification and Characterization of Leucocyclicin Q, a Novel Cyclic Bacteriocin Produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 2011, 77, 8164–8170. [Google Scholar] [CrossRef] [PubMed]
- Adamberg, K.; Kask, S.; Laht, T.-M.; Paalme, T. The Effect of Temperature and pH on the Growth of Lactic Acid Bacteria: A pH-Auxostat Study. Int. J. Food Microbiol. 2003, 85, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Kirk, M.F. pH as a Primary Control in Environmental Microbiology: 1. Thermodynamic Perspective. Front. Environ. Sci. 2018, 6, 101. [Google Scholar] [CrossRef]
- Vera-Peña, M.Y.; Rodriguez-Rodriguez, W.L. Effect of pH on the Growth of Three Lactic Acid Bacteria Strains Isolated from Sour Cream. Univ. Sci. 2020, 25, 341–358. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic Acid Bacteria as Starter Cultures: An Update in Their Metabolism and Genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- M’hamed, A.C.; Ncib, K.; Merghni, A.; Migaou, M.; Lazreg, H.; Snoussi, M.; Noumi, E.; Mansour, M.B.; Maaroufi, R.M. Characterization of Probiotic Properties of Lacticaseibacillus paracasei L2 Isolated from a Traditional Fermented Food “Lben. ” Life 2022, 13, 21. [Google Scholar] [CrossRef]
- Safi, E.; Haddad, M.; Hasan, M.; Al-Dalain, S.Y.; Proestos, C.; Siddiqui, S.A. Characterization of Potential Probiotic Activity of Lactic Acid Bacteria Isolated from Camel Colostrum by Biochemical and Molecular Methods. Vet. Med. Int. 2023, 2023, 8334152. [Google Scholar] [CrossRef]
- Kondybayev, A.; Konuspayeva, G.; Strub, C.; Loiseau, G.; Mestres, C.; Grabulos, J.; Manzano, M.; Akhmetsadykova, S.; Achir, N. Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage. Fermentation 2022, 8, 367. [Google Scholar] [CrossRef]
- Cadavez, V.A.P.; Campagnollo, F.B.; Silva, R.A.; Duffner, C.M.; Schaffner, D.W.; Sant’Ana, A.S.; Gonzales-Barron, U. A Comparison of Dynamic Tertiary and Competition Models for Describing the Fate of Listeria monocytogenes in Minas Fresh Cheese during Refrigerated Storage. Food Microbiol. 2019, 79, 48–60. [Google Scholar] [CrossRef]
- Costa, H.H.S.; Souza, M.R.; Acúrcio, L.B.; Cunha, A.F.; Resende, M.F.S.; Nunes, Á.C. Probiotic Potential of Lactic Acid Bacteria Isolated from Minas Artisanal Cheese from Serra da Canastra, MG. Arq. Bras. Med. Veterinária E Zootec. 2013, 65, 1858–1866. [Google Scholar] [CrossRef]
- Mejlholm, O.; Dalgaard, P. Modelling and Predicting the Simultaneous Growth of Listeria monocytogenes and Psychrotolerant Lactic Acid Bacteria in Processed Seafood and Mayonnaise-Based Seafood Salads. Food Microbiol. 2015, 46, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mellefont, L.A.; McMeekin, T.A.; Ross, T. Effect of Relative Inoculum Concentration on Listeria monocytogenes Growth in Co-Culture. Int. J. Food Microbiol. 2008, 121, 157–168. [Google Scholar] [CrossRef]
- Østergaard, N.B.; Eklöw, A.; Dalgaard, P. Modelling the Effect of Lactic Acid Bacteria from Starter- and Aroma Culture on Growth of Listeria monocytogenes in Cottage Cheese. Int. J. Food Microbiol. 2014, 188, 15–25. [Google Scholar] [CrossRef]
- Costa, J.C.C.P.; Bolívar, A.; Valero, A.; Carrasco, E.; Zurera, G.; Pérez-Rodríguez, F. Evaluation of the Effect of Lactobacillus sakei strain L115 on Listeria monocytogenes at Different Conditions of Temperature by Using Predictive Interaction Models. Food Res. Int. 2020, 131, 108928. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.C.R.; Staliano, C.D.; Vieira, A.D.S.; Villarreal, M.L.M.; Todorov, S.D.; Saad, S.M.I.; Franco, B.D.G.d.M. Bacteriocin Production and Inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. Sakei 2a in a Potentially Synbiotic Cheese Spread. Food Microbiol. 2015, 48, 143–152. [Google Scholar] [CrossRef]
- Panebianco, F.; Giarratana, F.; Caridi, A.; Sidari, R.; De Bruno, A.; Giuffrida, A. Lactic Acid Bacteria Isolated from Traditional Italian Dairy Products: Activity against Listeria monocytogenes and Modelling of Microbial Competition in Soft Cheese. LWT 2021, 137, 110446. [Google Scholar] [CrossRef]
- Vera Pingitore, E.; Todorov, S.D.; Sesma, F.; Gombossy de Melo Franco, B.D. Application of Bacteriocinogenic Enterococcus Mundtii CRL35 and Enterococcus faecium ST88Ch in the Control of Listeria Monocytogenes in Fresh Minas Cheese. Food Microbiol. 2012, 32, 38–47. [Google Scholar] [CrossRef]
- Blanco-Lizarazo, C.M.; Sotelo-Díaz, I.; Llorente-Bousquets, A. In Vitro Modelling of Simultaneous Interactions of Listeria monocytogenes, Lactobacillus sakei, and Staphylococcus carnosus. Food Sci. Biotechnol. 2016, 25, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Guillier, L.; Stahl, V.; Hezard, B.; Notz, E.; Briandet, R. Modelling the Competitive Growth between Listeria monocytogenes and Biofilm Microflora of Smear Cheese Wooden Shelves. Int. J. Food Microbiol. 2008, 128, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Bagi, L.K. Microbial Competition: Effect of Culture Conditions on the Suppression of Listeria monocytogenes Scott A by Carnobacterium piscicola†. J. Food Prot. 1997, 60, 254–261. [Google Scholar] [CrossRef]
- Castro, R.D.; Oliveira, L.G.; Sant’Anna, F.M.; Luiz, L.M.P.; Sandes, S.H.C.; Silva, C.I.F.; Silva, A.M.; Nunes, A.C.; Penna, C.F.A.M.; Souza, M.R. Lactic Acid Microbiota Identification in Water, Raw Milk, Endogenous Starter Culture, and Fresh Minas Artisanal Cheese from the Campo Das Vertentes Region of Brazil during the Dry and Rainy Seasons. J. Dairy Sci. 2016, 99, 6086–6096. [Google Scholar] [CrossRef]
- Daba, H.; Pandian, S.; Gosselin, J.F.; Simard, R.E.; Huang, J.; Lacroix, C. Detection and Activity of a Bacteriocin Produced by Leuconostoc mesenteroides. Appl. Environ. Microbiol. 1991, 57, 3450–3455. [Google Scholar] [CrossRef]
- Huang, Y.; Ye, K.; Yu, K.; Wang, K.; Zhou, G. The Potential Influence of Two Enterococcus faecium on the Growth of Listeria monocytogenes. Food Control 2016, 67, 18–24. [Google Scholar] [CrossRef]
- Parente, E.; Ricciardi, A. Production, Recovery and Purification of Bacteriocins from Lactic Acid Bacteria. Appl. Microbiol. Biotechnol. 1999, 52, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Quinto, E.J.; Marín, J.M.; Schaffner, D.W. Effect of the Competitive Growth of Lactobacillus sakei MN on the Growth Kinetics of Listeria monocytogenes Scott A in Model Meat Gravy. Food Control 2016, 63, 34–45. [Google Scholar] [CrossRef]
- Elli, M.; Zink, R.; Rytz, A.; Reniero, R.; Morelli, L. Iron Requirement of Lactobacillus spp. in Completely Chemically Defined Growth Media. J. Appl. Microbiol. 2000, 88, 695–703. [Google Scholar] [CrossRef]
- Héchard, Y.; Dérijard, B.; Letellier, F.; Cenatiempo, Y. Characterization and Purification of Mesentericin Y105, an Anti-Listeria Bacteriocin from Leuconostoc mesenteroides. Microbiology 1992, 138, 2725–2731. [Google Scholar] [CrossRef]
- Wulijideligen; Asahina, T.; Hara, K.; Arakawa, K.; Nakano, H.; Miyamoto, T. Production of Bacteriocin by Leuconostoc mesenteroides 406 Isolated from Mongolian Fermented Mare’s Milk, Airag. Anim. Sci. J. 2012, 83, 704–711. [Google Scholar] [CrossRef]
- Papadopoulou, O.S.; Argyri, A.A.; Varzakis, E.; Sidira, M.; Kourkoutas, Y.; Galanis, A.; Tassou, C.; Chorianopoulos, N.G. Use of Lactobacilli strains with Probiotic Potential in Traditional Fermented Milk and Their Impact on Quality and Safety Related to Listeria monocytogenes. Int. Dairy J. 2019, 98, 44–53. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Gutiérrez-Chocoza, M.A.; López-Romero, J.C.; García-Galaz, A.; González-Ríos, H.; Peña-Ramos, A.; Juneja, V.K.; Pérez-Báez, A.J.; Valenzuela-Melendres, M. Modeling the Effects of Temperature and pH on Listeria monocytogenes Growth in Mexican-Style Pork Chorizo. Appl. Food Res. 2023, 3, 100336. [Google Scholar] [CrossRef]
- Le Marc, Y.; Huchet, V.; Bourgeois, C.M.; Guyonnet, J.P.; Mafart, P.; Thuault, D. Modelling the Growth Kinetics of Listeria as a Function of Temperature, pH and Organic Acid Concentration. Int. J. Food Microbiol. 2002, 73, 219–237. [Google Scholar] [CrossRef]
- Martín-Miguélez, J.M.; Robledo, J.; Martín, I.; Castaño, C.; Delgado, J.; Córdoba, J.J. Biocontrol of L. monocytogenes with Selected Autochthonous Lactic Acid Bacteria in Raw Milk Soft-Ripened Cheese under Different Water Activity Conditions. Foods 2024, 13, 172. [Google Scholar] [CrossRef]
- Rosshaug, P.S.; Detmer, A.; Ingmer, H.; Larsen, M.H. Modeling the Growth of Listeria monocytogenes in Soft Blue-White Cheese. Appl. Environ. Microbiol. 2012, 78, 8508–8514. [Google Scholar] [CrossRef] [PubMed]
- Ross, T. Indices for Performance Evaluation of Predictive Models in Food Microbiology. J. Appl. Bacteriol. 1996, 81, 501–508. [Google Scholar] [CrossRef]
- Mejlholm, O.; Gunvig, A.; Borggaard, C.; Blom-Hanssen, J.; Mellefont, L.; Ross, T.; Leroi, F.; Else, T.; Visser, D.; Dalgaard, P. Predicting Growth Rates and Growth Boundary of Listeria monocytogenes—An International Validation Study with Focus on Processed and Ready-to-Eat Meat and Seafood. Int. J. Food Microbiol. 2010, 141, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.; Dalgaard, P.; Tienungoon, S. Predictive Modelling of the Growth and Survival of Listeria in Fishery Products. Int. J. Food Microbiol. 2000, 62, 231–245. [Google Scholar] [CrossRef]
- Mejlholm, O.; Dalgaard, P. Development and Validation of an Extensive Growth and Growth Boundary Model for Psychrotolerant Lactobacillus spp. in Seafood and Meat Products. Int. J. Food Microbiol. 2013, 167, 244–260. [Google Scholar] [CrossRef]
- Lobacz, A.; Kowalik, J. A Predictive Model for Listeria monocytogenes in UHT Dairy Products with Various Fat Content during Cold Storage. J. Food Saf. 2015, 35, 119–127. [Google Scholar] [CrossRef]
- Nyhan, L.; Begley, M.; Mutel, A.; Qu, Y.; Johnson, N.; Callanan, M. Predicting the Combinatorial Effects of Water Activity, pH and Organic Acids on Listeria Growth in Media and Complex Food Matrices. Food Microbiol. 2018, 74, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Noviyanti, F.; Shimizu, S.; Hosotani, Y.; Koseki, S.; Inatsu, Y.; Kawasaki, S. Predictive Growth Model of Listeria monocytogenes Under Fluctuating Temperature Conditions in Pasteurized Milk by Using Real-Time Polymerase Chain Reaction. Foodborne Pathog. Dis. 2020, 17, 693–700. [Google Scholar] [CrossRef] [PubMed]
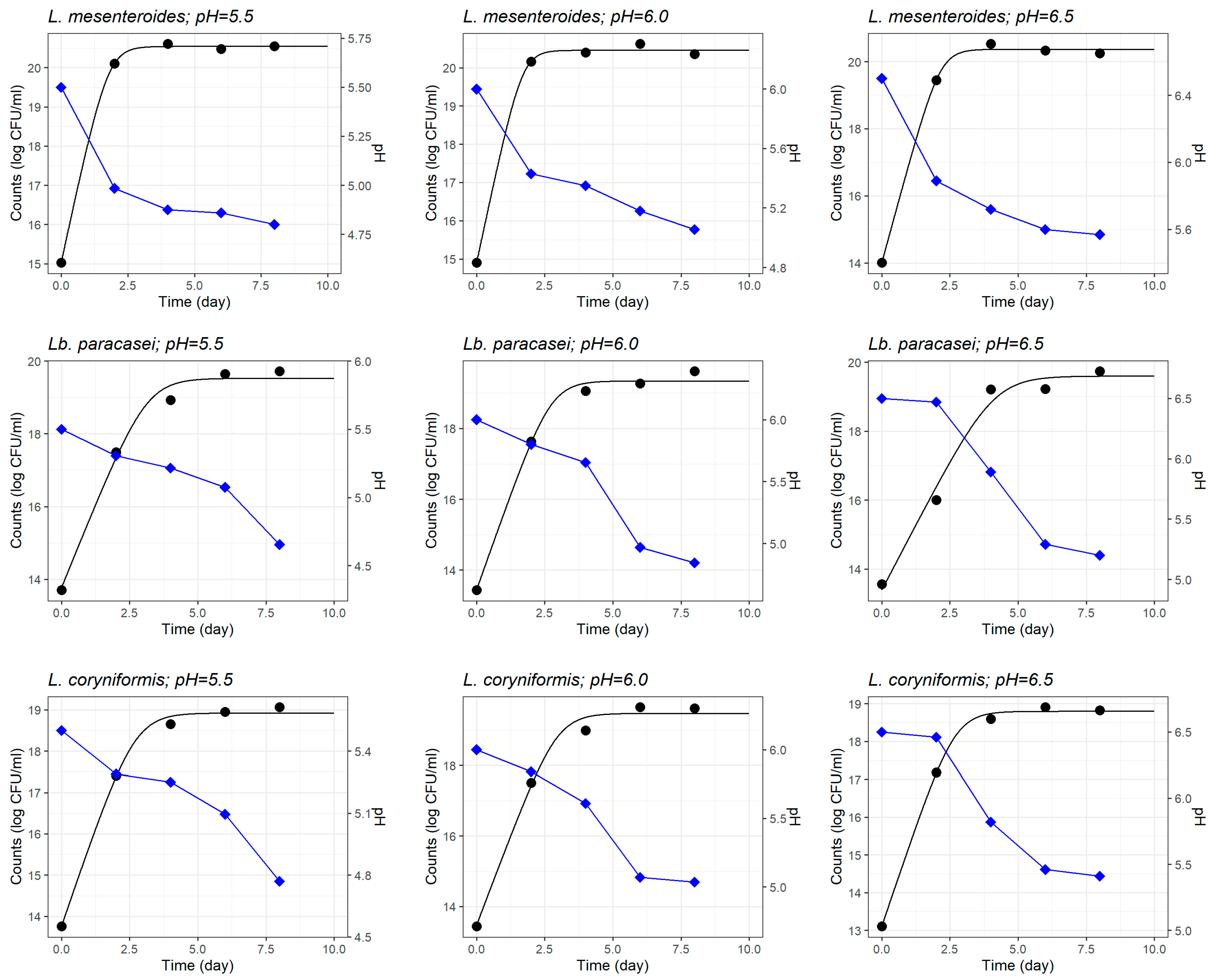
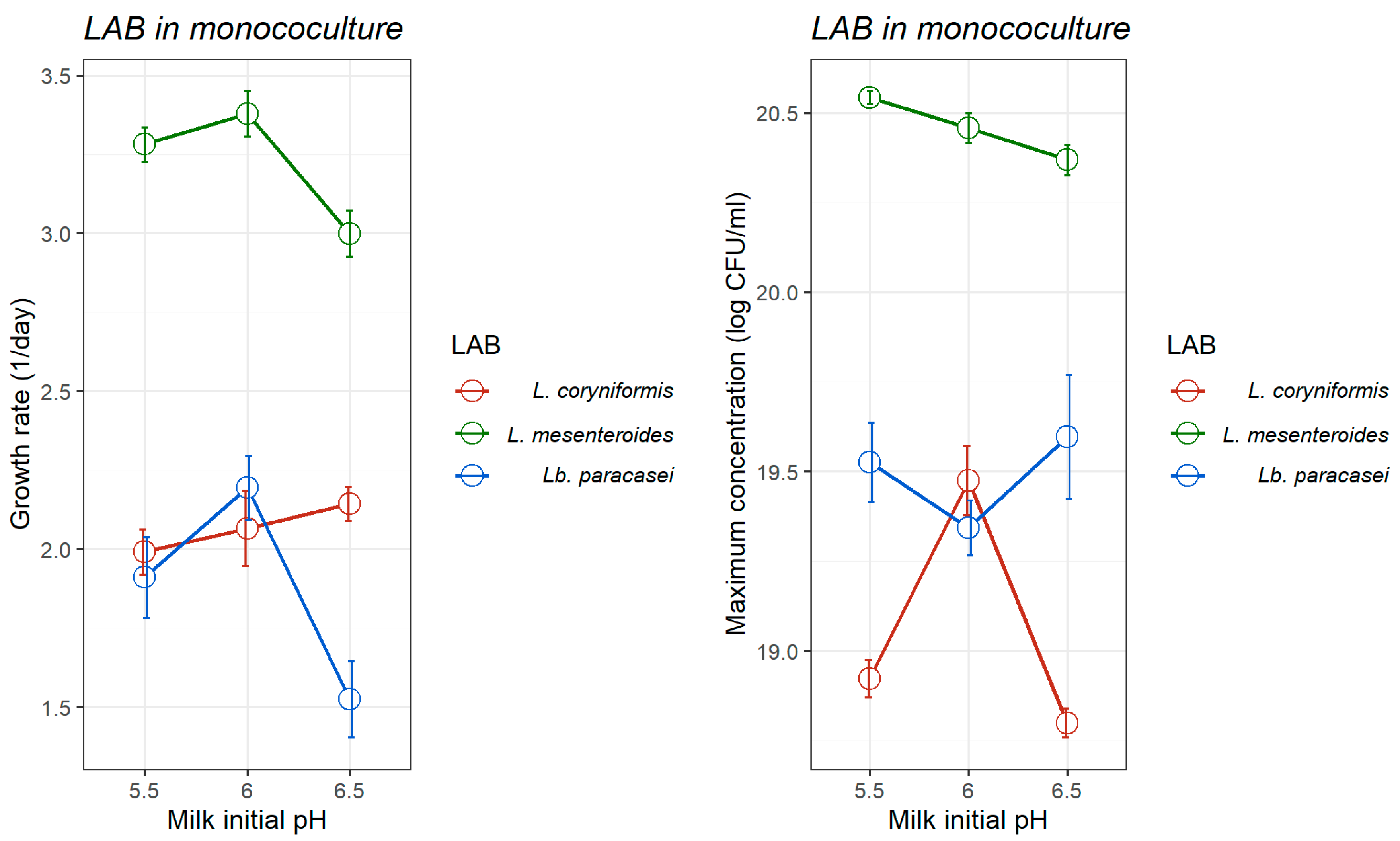
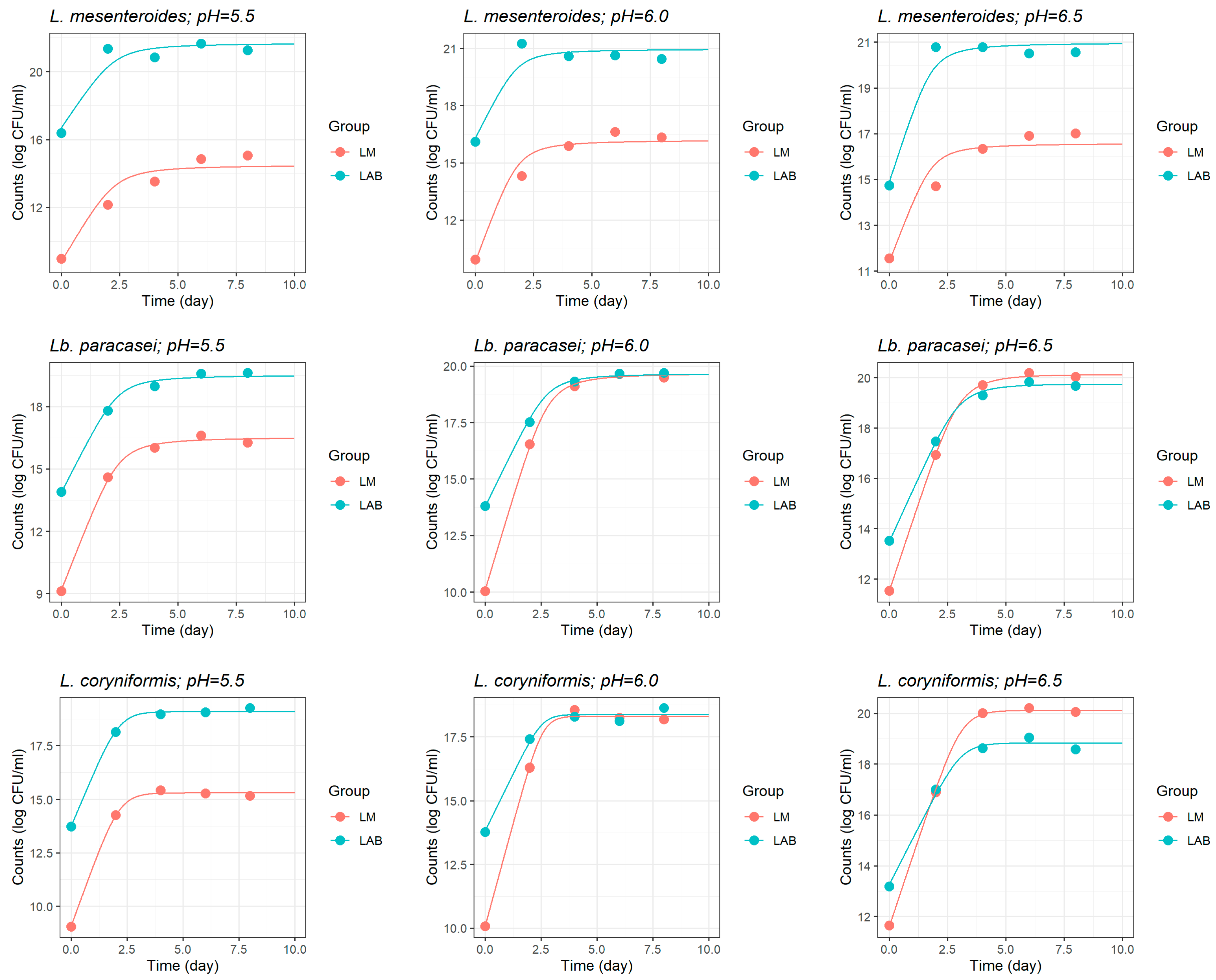
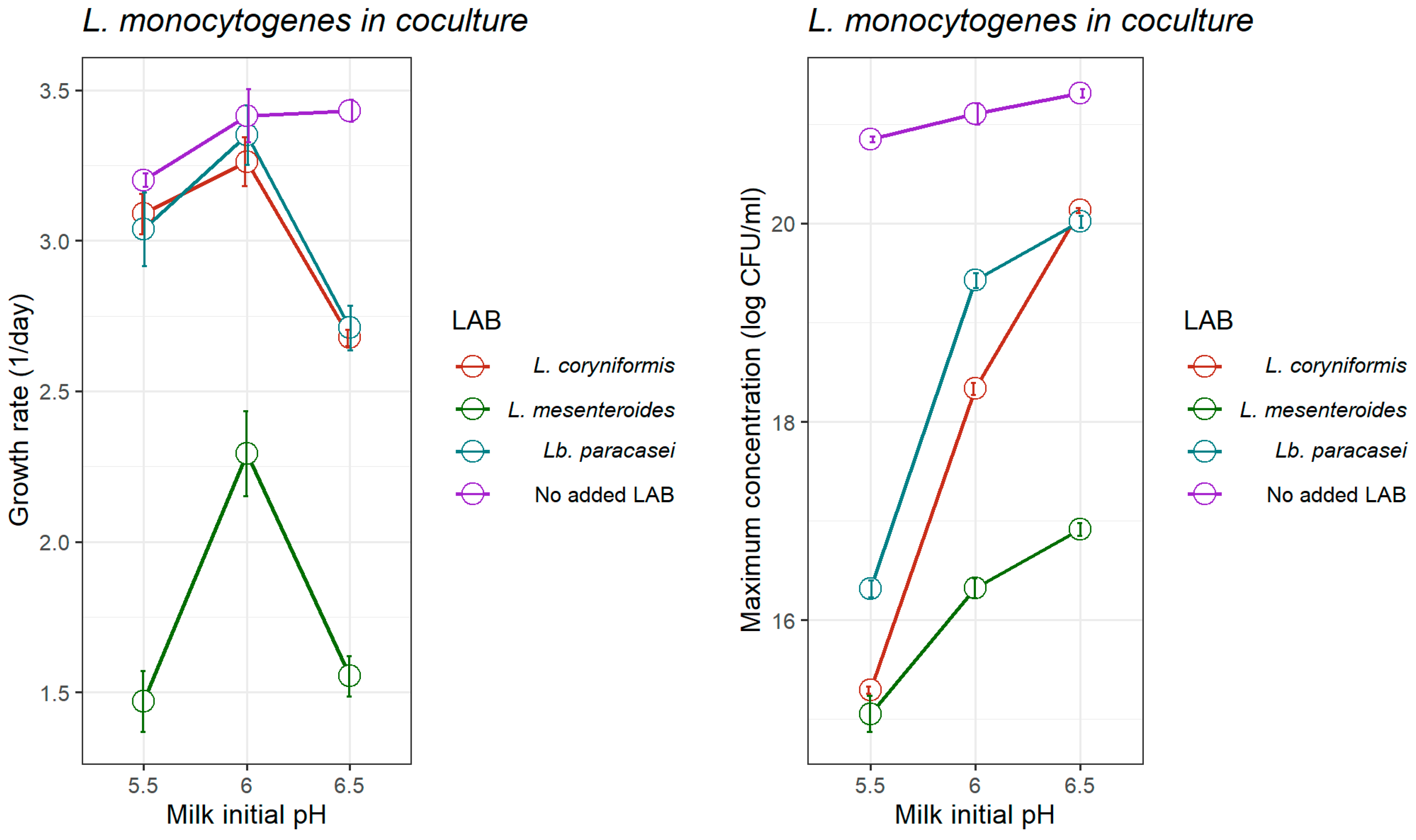
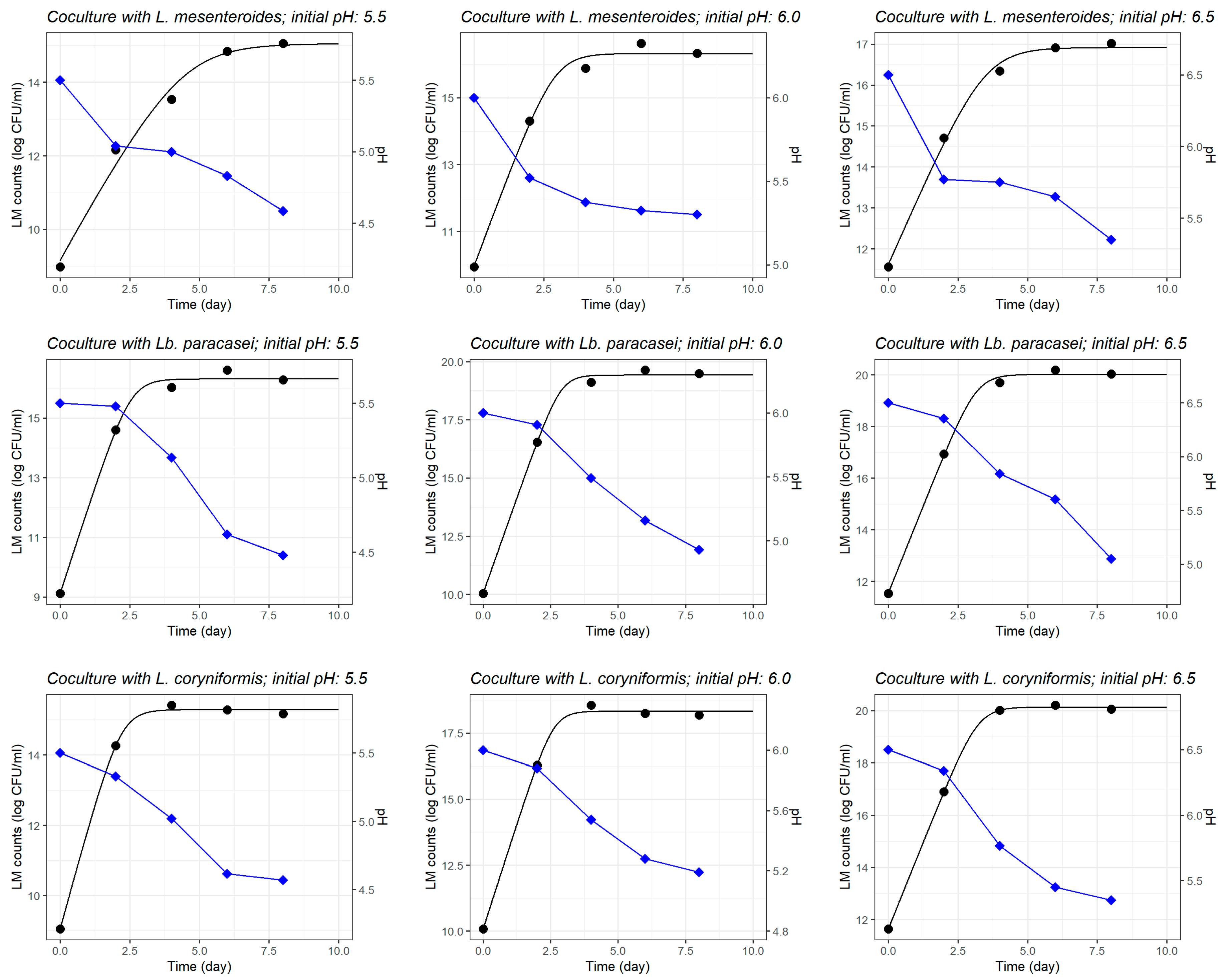
| Initial pH | L. monocytogenes | L. mesenteroides | Lb. paracasei | L. coryniformis | |||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | |
| 5.5 | Y0 | 8.897 (0.081) | <0.001 | 15.02 (0.063) | <0.001 | 13.77 (0.341) | <0.001 | 13.77 (0.169) | <0.001 |
| Ymax | 20.85 (0.060) | <0.001 | 20.54 (0.037) | <0.001 | 19.53 (0.219) | <0.001 | 18.92 (0.103) | <0.001 | |
| μopt | 3.201 (0.045) | <0.001 | 3.282 (0.109) | 0.001 | 1.910 (0.257) | 0.017 | 1.992 (0.143) | 0.005 | |
| Fit quality | |||||||||
| σ2 | 0.0037 | 0.0020 | 0.0592 | 0.0144 | |||||
| RMSE | 0.0544 | 0.0405 | 0.2177 | 0.1075 | |||||
| MAE | 0.0436 | 0.0260 | 0.1860 | 0.0816 | |||||
| 6.0 | Y0 | 8.483 (0.313) | 0.001 | 14.90 (0.144) | <0.001 | 13.44 (0.255) | <0.001 | 13.47 (0.313) | <0.001 |
| Ymax | 21.10 (0.212) | <0.001 | 20.45 (0.083) | <0.001 | 19.34 (0.152) | <0.001 | 19.47 (0.192) | <0.001 | |
| μopt | 3.416 (0.177) | 0.003 | 3.379 (0.322) | 0.009 | 2.194 (0.203) | 0.008 | 2.066 (0.239) | 0.0131 | |
| Fit quality | |||||||||
| σ2 | 0.0542 | 0.0104 | 0.0325 | 0.0493 | |||||
| RMSE | 0.2083 | 0.0912 | 0.1613 | 0.1985 | |||||
| MAE | 0.1852 | 0.0661 | 0.1221 | 0.1630 | |||||
| 6.5 | Y0 | 8.477 (0.124) | <0.001 | 14.00 (0.147) | <0.001 | 13.39 (0.445) | 0.001 | 13.11 (0.134) | <0.001 |
| Ymax | 21.31 (0.085) | <0.001 | 20.37 (0.085) | <0.001 | 19.59 (0.347) | <0.001 | 18.79 (0.080) | <0.001 | |
| μopt | 3.432 (0.073) | <0.001 | 3.000 (0.147) | 0.002 | 1.525 (0.241) | 0.024 | 2.143 (0.108) | 0.002 | |
| Fit quality | |||||||||
| σ2 | 0.0084 | 0.0108 | 0.1109 | 0.0090 | |||||
| RMSE | 0.0821 | 0.0928 | 0.2979 | 0.0849 | |||||
| MAE | 0.0731 | 0.0651 | 0.2794 | 0.0639 | |||||
| Initial pH | Jameson-Effect | Dynamic Model | |||
|---|---|---|---|---|---|
| Parameters | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | |
| 5.5 | LM0 | 8.783 (0.886) | <0.001 | 9.158 (0.357) | 0.002 |
| μmax LM/μopt LM | 2.334 (0.766) | 0.038 | 1.469 (0.205) | 0.019 | |
| LMmax | 14.51 (0.840) | <0.001 | 15.05 (0.367) | 0.001 | |
| LAB0 | 16.67 (0.945) | <0.001 | - | - | |
| μmax LAB | 2.047 (0.825) | 0.068 | - | - | |
| LABmax | 21.69 (0.720) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.3678 | 0.0721 | |||
| RMSE | 0.5754 | 0.2401 | |||
| MAE | 0.5059 | 0.1944 | |||
| 6.0 | LM0 | 9.808 (0.785) | <0.001 | 9.965 (0.335) | 0.001 |
| μmax LM/μopt LM | 3.302 (1.048) | 0.034 | 2.293 (0.284) | <0.001 | |
| LMmax | 16.22 (3.764) | 0.012 | 16.32 (0.204) | <0.001 | |
| LAB0 | 16.28 (0.811) | <0.001 | - | - | |
| μmax LAB | 2.423 (0.908) | 0.056 | - | - | |
| LABmax | 20.96 (2.344) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.2796 | 0.0548 | |||
| RMSE | 0.5016 | 0.2094 | |||
| MAE | 0.3918 | 0.1511 | |||
| 6.5 | LM0 | 11.40 (0.769) | <0.001 | 11.61 (0.182) | <0.001 |
| μmax LM/μopt LM | 2.653 (0.800) | 0.029 | 1.552 (0.132) | 0.007 | |
| LMmax | 16.59 (1.636) | <0.001 | 16.91 (0.132) | <0.001 | |
| LAB0 | 14.88 (0.791) | <0.001 | - | - | |
| μmax LAB | 3.118 (1.079) | 0.045 | - | - | |
| LABmax | 20.98 (1.943) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.2678 | 0.0168 | |||
| RMSE | 0.4909 | 0.1162 | |||
| MAE | 0.3843 | 0.0995 | |||
| Initial pH | Jameson-Effect | Dynamic Model | |||
|---|---|---|---|---|---|
| Parameters | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | |
| 5.5 | LM0 | 9.157 (0.259) | <0.001 | 9.120 (0.284) | <0.001 |
| μmax LM/μopt LM | 2.866 (0.209) | <0.001 | 3.038 (0.245) | 0.006 | |
| LMmax | 16.56 (0.406) | <0.001 | 16.31 (0.166) | <0.001 | |
| LAB0 | 13.85 (0.255) | <0.001 | - | - | |
| μmax LAB | 2.201 (0.167) | <0.001 | - | - | |
| LABmax | 19.52 (0.293) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.0303 | 0.0402 | |||
| RMSE | 0.1651 | 0.1793 | |||
| MAE | 0.1507 | 0.1244 | |||
| 6.0 | LM0 | 10.08 (0.149) | <0.001 | 10.05 (0.259) | <0.001 |
| μmax LM/μopt LM | 3.312 (0.109) | <0.001 | 3.351 (0.197) | 0.003 | |
| LMmax | 19.80 (0.577) | <0.001 | 19.42 (0.151) | <0.001 | |
| LAB0 | 13.76 (0.138) | <0.001 | - | - | |
| μmax LAB | 2.037 (0.071) | <0.001 | - | - | |
| LABmax | 19.65 (0.198) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.0103 | 0.0334 | |||
| RMSE | 0.0963 | 0.1635 | |||
| MAE | 0.0899 | 0.1185 | |||
| 6.5 | LM0 | 11.51 (0.132) | <0.001 | 11.55 (0.197) | <0.001 |
| μmax LM/μopt LM | 2.795 (0.092) | <0.001 | 2.711 (0.148) | 0.003 | |
| LMmax | 20.12 (0.120) | <0.001 | 20.02 (0.119) | <0.001 | |
| LAB0 | 13.47 (0.129) | <0.001 | - | - | |
| μmax LAB | 2.036 (0.075) | <0.001 | - | - | |
| LABmax | 20.04 (0.293) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.0080 | 0.0192 | |||
| RMSE | 0.0849 | 0.1239 | |||
| MAE | 0.0686 | 0.0931 | |||
| Initial pH | Jameson-Effect | Dynamic Model | |||
|---|---|---|---|---|---|
| Parameters | Mean (SE) | Pr > |t| | Mean (SE) | Pr > |t| | |
| 5.5 | LM0 | 9.049 (0.139) | <0.001 | 9.043 (0.130) | <0.001 |
| μmax LM/μopt LM | 2.908 (0.205) | <0.001 | 3.089 (0.133) | 0.002 | |
| LMmax | 17.70 (7.874) | 0.087 | 15.29 (0.076) | <0.001 | |
| LAB0 | 13.72 (0.139) | <0.001 | - | - | |
| μmax LAB | 2.493 (0.174) | <0.001 | - | - | |
| LABmax | 19.09 (0.078) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.0087 | 0.0085 | |||
| RMSE | 0.0886 | 0.0825 | |||
| MAE | 0.0669 | 0.0550 | |||
| 6.0 | LM0 | 10.05 (0.242) | <0.001 | 10.07 (0.209) | <0.001 |
| μmax LM/μopt LM | 3.241 (0.221) | <0.001 | 3.263 (0.163) | 0.002 | |
| LMmax | 18.31 (0.139) | <0.001 | 18.33 (0.122) | <0.001 | |
| LAB0 | 13.79 (0.240) | <0.001 | - | - | |
| μmax LAB | 1.797 (0.155) | <0.001 | - | - | |
| LABmax | 21.53 (9.912) | 0.096 | - | - | |
| Fit quality | |||||
| σ2 | 0.0265 | 0.0220 | |||
| RMSE | 0.1545 | 0.1327 | |||
| MAE | 0.1267 | 0.0979 | |||
| 6.5 | LM0 | 11.59 (0.212) | <0.001 | 11.64 (0.077) | <0.001 |
| μmax LM/μopt LM | 2.762 (0.143) | <0.001 | 2.677 (0.055) | <0.001 | |
| LMmax | 20.11 (0.135) | <0.001 | 20.13 (0.047) | <0.001 | |
| LAB0 | 13.25 (0.290) | <0.001 | - | - | |
| μmax LAB | 1.809 (0.114) | <0.001 | - | - | |
| LABmax | 20.23 (0.723) | <0.001 | - | - | |
| Fit quality | |||||
| σ2 | 0.0214 | 0.0030 | |||
| RMSE | 0.1389 | 0.0489 | |||
| MAE | 0.1213 | 0.0330 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loforte, Y.; Zanzan, M.; Cadavez, V.; Gonzales-Barron, U. Dynamic Modelling of Listeria monocytogenes Growth in a Milk Model Medium as Affected by pH and Selected Lactic Acid Bacteria Strains. Foods 2025, 14, 3999. https://doi.org/10.3390/foods14233999
Loforte Y, Zanzan M, Cadavez V, Gonzales-Barron U. Dynamic Modelling of Listeria monocytogenes Growth in a Milk Model Medium as Affected by pH and Selected Lactic Acid Bacteria Strains. Foods. 2025; 14(23):3999. https://doi.org/10.3390/foods14233999
Chicago/Turabian StyleLoforte, Yara, Mariem Zanzan, Vasco Cadavez, and Ursula Gonzales-Barron. 2025. "Dynamic Modelling of Listeria monocytogenes Growth in a Milk Model Medium as Affected by pH and Selected Lactic Acid Bacteria Strains" Foods 14, no. 23: 3999. https://doi.org/10.3390/foods14233999
APA StyleLoforte, Y., Zanzan, M., Cadavez, V., & Gonzales-Barron, U. (2025). Dynamic Modelling of Listeria monocytogenes Growth in a Milk Model Medium as Affected by pH and Selected Lactic Acid Bacteria Strains. Foods, 14(23), 3999. https://doi.org/10.3390/foods14233999







