Physiologically Relevant Simulation of Carbohydrate Digestion: From Glycemic Index Estimation to Intestinal Cellular Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
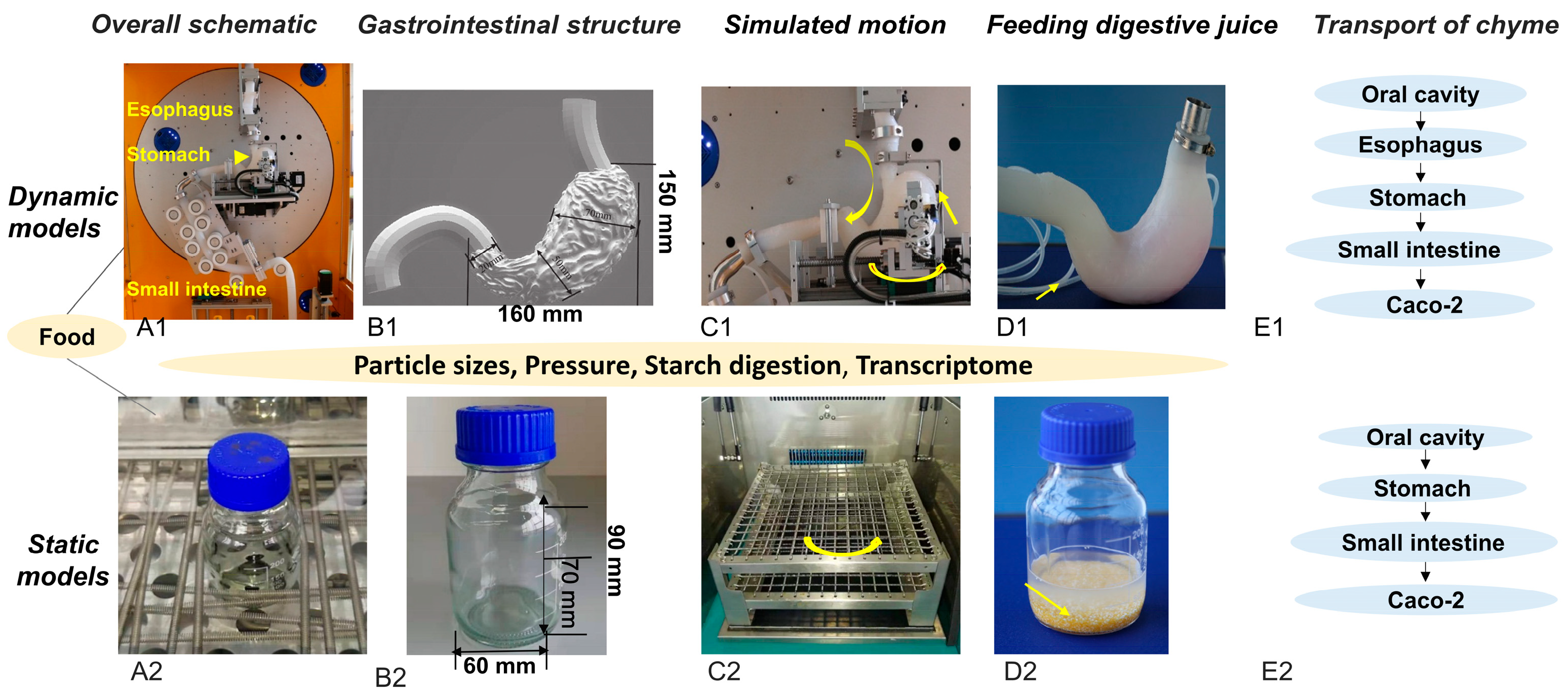
2.2. Overview of the Static and Dynamic In Vitro Models
2.3. In Vitro Digestion
2.3.1. Static Digestion Using a Water Bath Shaker
2.3.2. Dynamic Digestion Using the DIVHS System
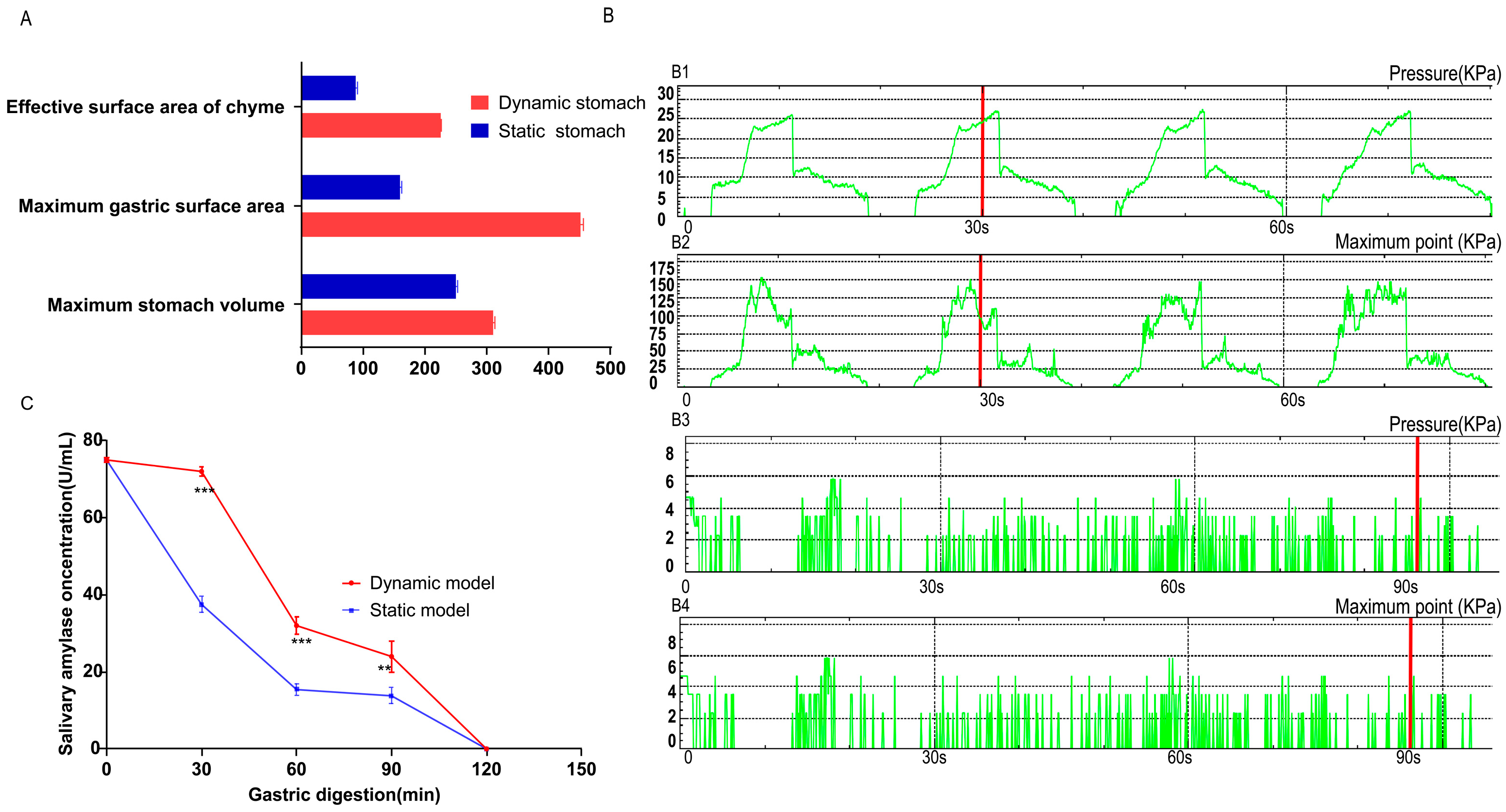
2.4. Pressure Measurement During Digestion
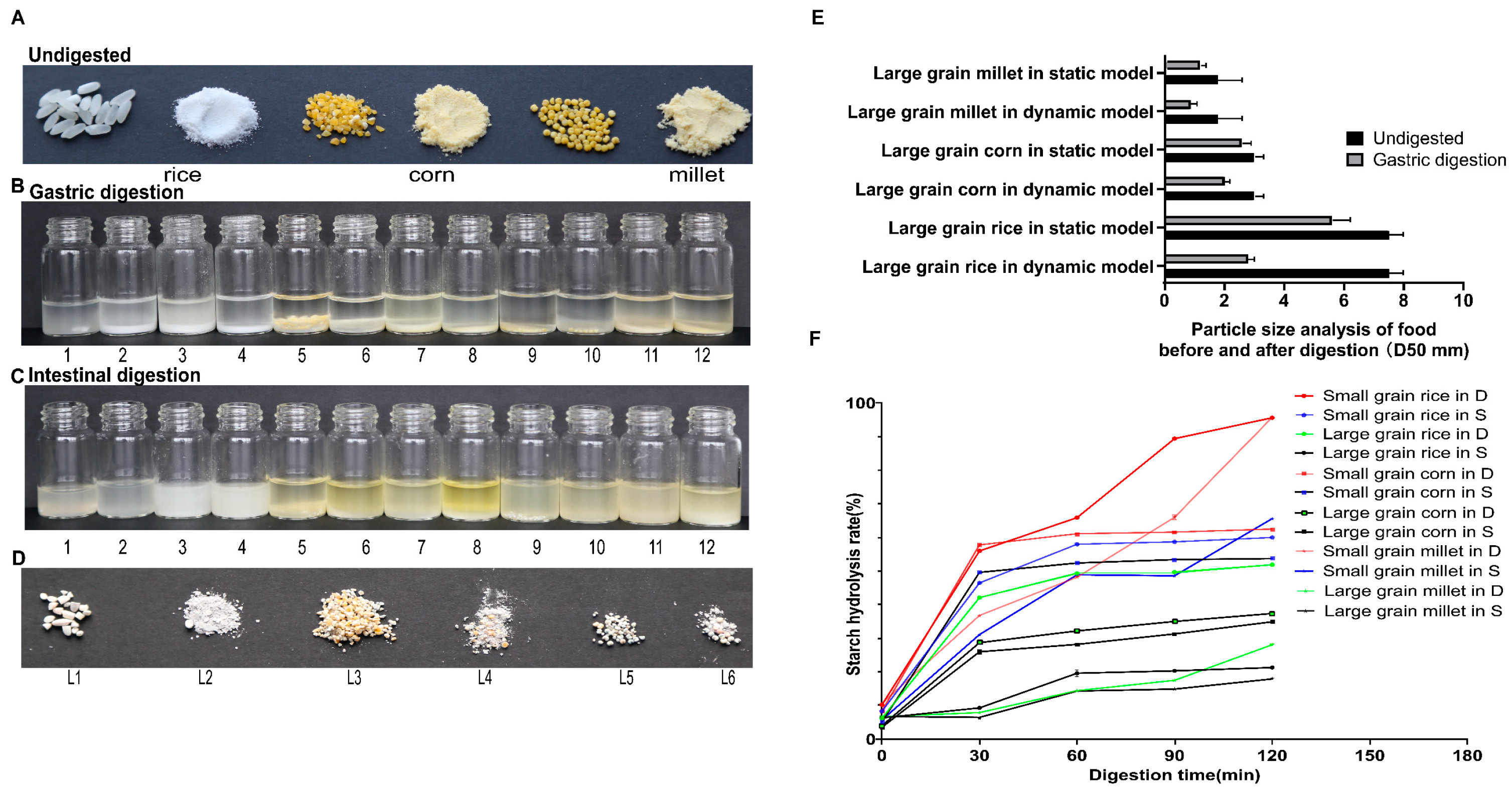
2.5. Particle Size and Starch Digestion Analysis
2.6. Cell Culture and Caco-2 Stimulation with Digested Products
2.7. RNA Quality Control, Sequencing, and Quantitative Real-Time PCR
2.8. In Vitro Food Glycemic Index Prediction (eGI)
2.9. Data Analysis
3. Results
3.1. Comparison of Digestion Outcomes for 3 Cereals in Static and Dynamic Models
3.2. Mechanistic Exploration of Digestion Differences Between Static and Dynamic Models
3.3. In Vitro Glycemic Index Prediction of 14 Foods
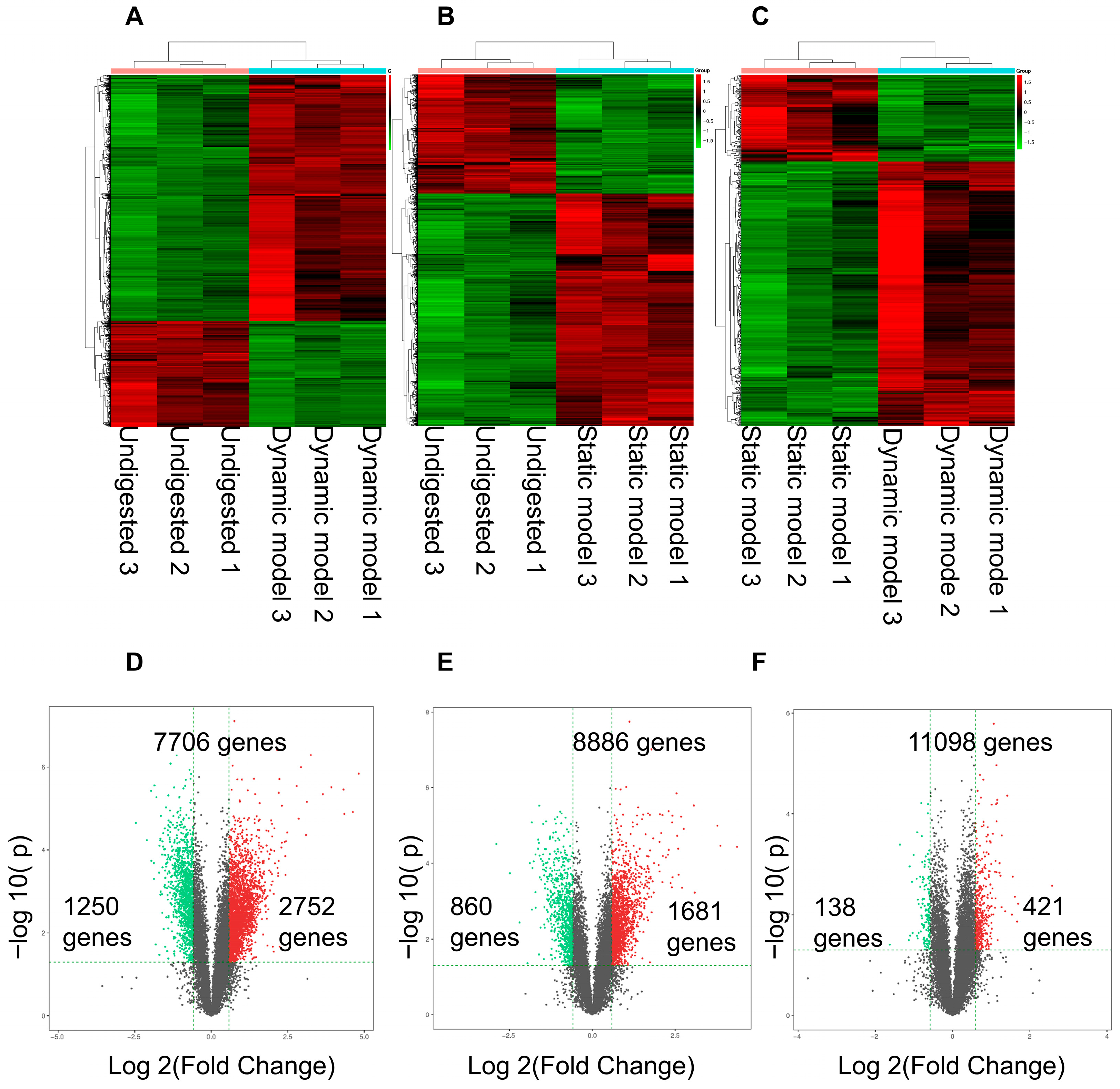
3.4. Differentially Expressed Genes Stimulated by Digestion Products in Caco-2 Cells
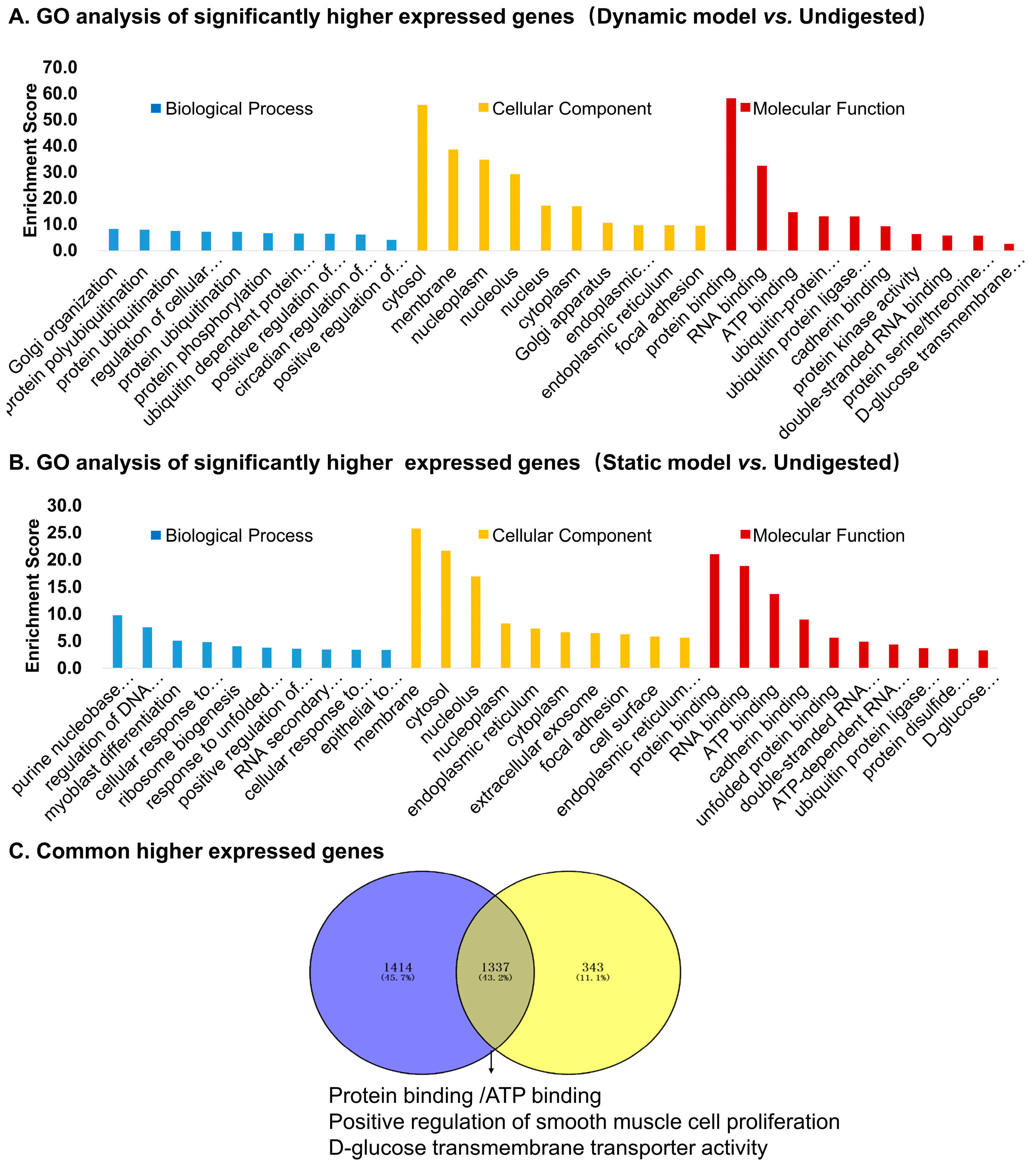
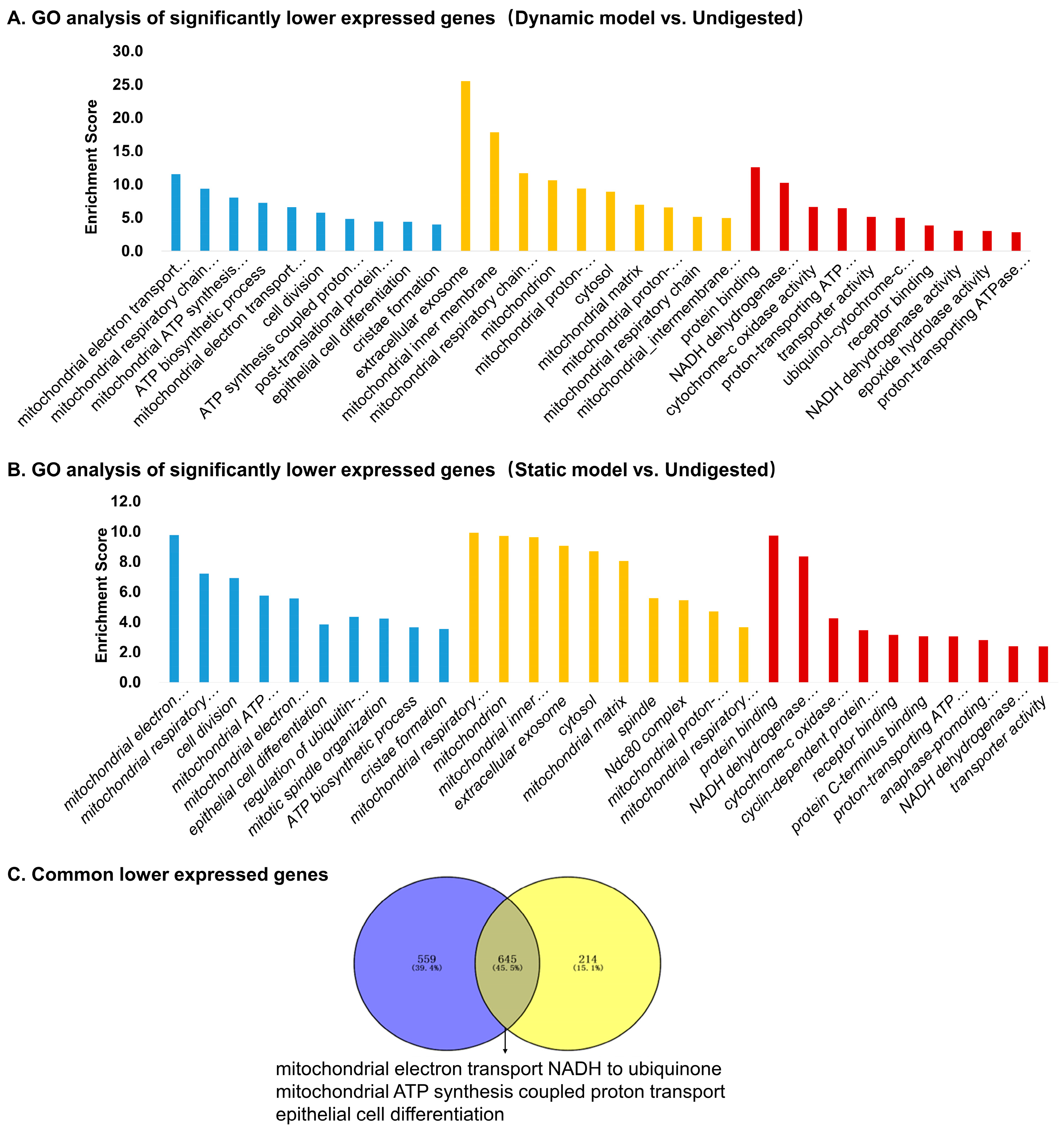
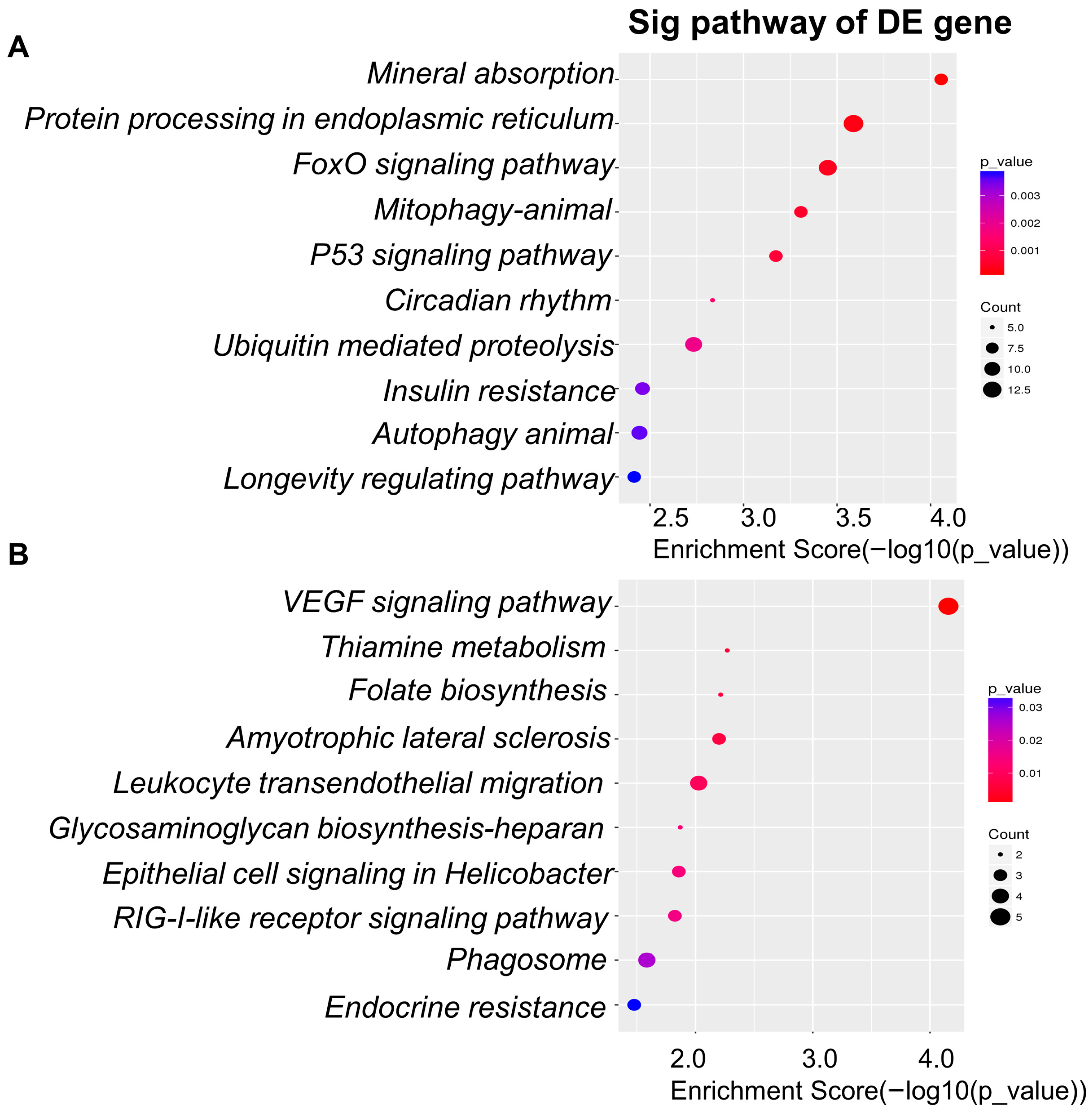
3.5. Differentially Expressed Genes: Functional Enrichment and Key Gene Identification
3.5.1. Upregulated Genes
3.5.2. Downregulated Genes
3.6. Differentially Expressed Genes with Two Digested Products
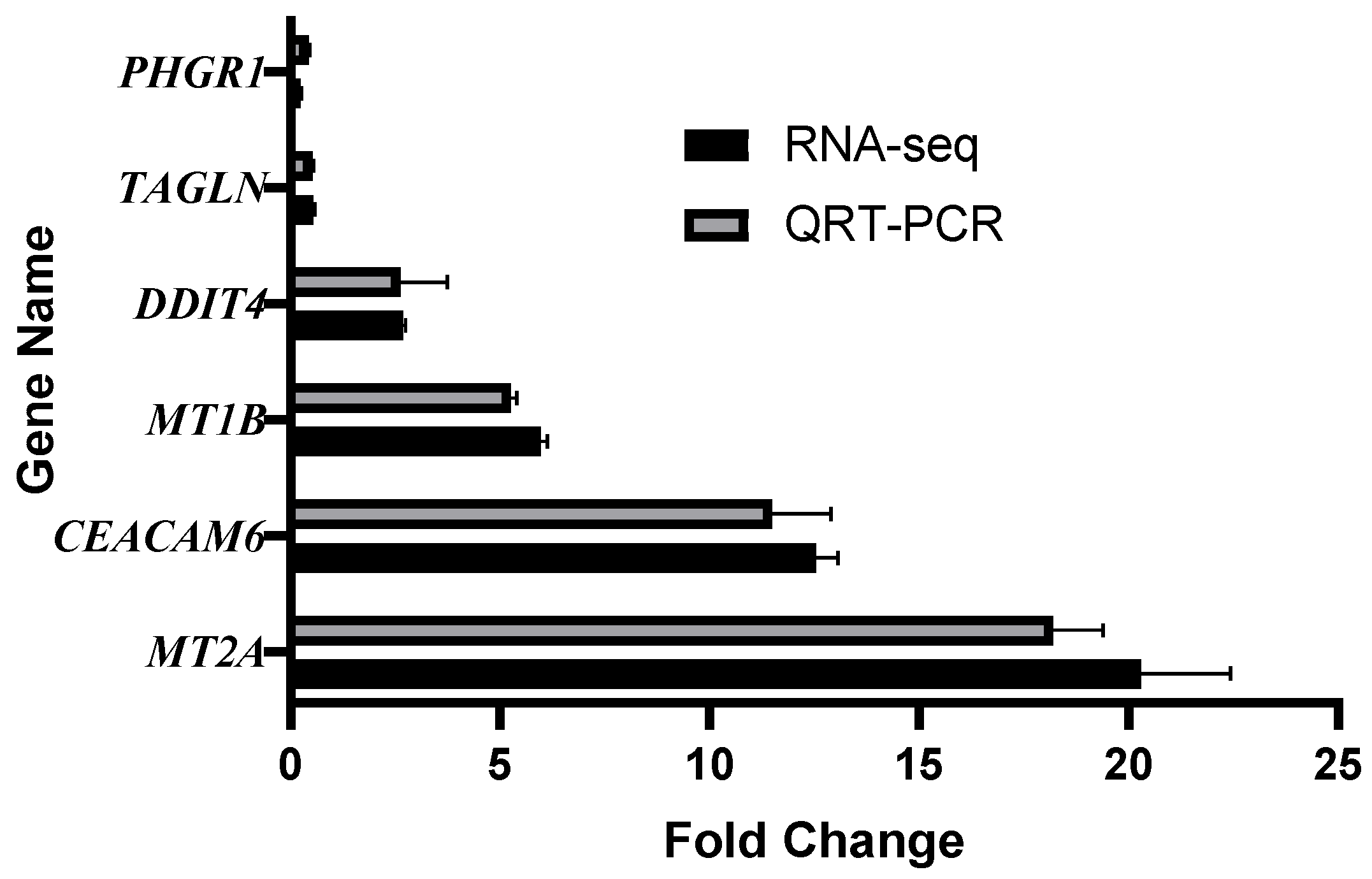
3.7. Validation of RNA-Sequencing by qRT-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seidelmann, S.B.; Claggett, B.; Cheng, S.; Henglin, M.; Shah, A.; Steffen, L.M.; Folsom, A.R.; Rimm, E.B.; Willett, W.C.; Solomon, S.D. Dietary carbohydrate intake and mortality: A prospective cohort study and meta-analysis. Lancet Public Health 2018, 3, e419–e428. [Google Scholar] [CrossRef]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- Eyinla, T.E.; Sanusi, R.A.; Maziya-Dixon, B. Evaluation of in vitro and in vivo Glycemic Index of common staples made from varieties of White Yam (Dioscorea rotundata). Front. Nutr. 2022, 9, 983212. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, J.M. The food matrix: Implications in processing, nutrition and health. Crit. Rev. Food Sci. Nutr. 2019, 59, 3612–3629. [Google Scholar] [CrossRef]
- Mackie, A.; Mulet-Cabero, A.I.; Torcello-Gómez, A. Simulating human digestion: Developing our knowledge to create healthier and more sustainable foods. Food Funct. 2020, 11, 9397–9431. [Google Scholar] [CrossRef]
- Kong, F.; Singh, R.P. A human gastric simulator (HGS) to study food digestion in human stomach. J. Food Sci. 2010, 75, E627–E635. [Google Scholar] [CrossRef]
- Dall’Asta, M.; Del Rio, D.; Tappy, L.; Potì, F.; Agostoni, C.; Brighenti, F. Critical and emerging topics in dietary carbohydrates and health. Int. J. Food Sci. Nutr. 2020, 71, 286–295. [Google Scholar] [CrossRef]
- Guan, Y.; Toommuangpak, W.; Zhao, G.; Thaiudom, S. The Microstructure, Rheological Characteristics, and Digestibility Properties of Binary or Ternary Mixture Systems of Gelatinized Potato Starch/Milk Protein/Soybean Oil during the In Vitro Digestion Process. Foods 2023, 12, 2451. [Google Scholar] [CrossRef]
- Shahidi, F.; Danielski, R. Review on the Role of Polyphenols in Preventing and Treating Type 2 Diabetes: Evidence from In Vitro and In Vivo Studies. Nutrients 2024, 16, 3159. [Google Scholar] [CrossRef]
- Wang, J.; Wu, P.; Liu, M.; Liao, Z.; Wang, Y.; Dong, Z.; Chen, X.D. An advanced near real dynamic in vitro human stomach system to study gastric digestion and emptying of beef stew and cooked rice. Food Funct. 2019, 10, 2914–2925. [Google Scholar] [CrossRef]
- Zhang, P.; Iqbal, S.; Deng, R.; Duan, X.; Han, H.; Dong Chen, X.; Wu, P. Impact of elderly gastrointestinal alterations on gastric emptying and enzymatic hydrolysis of skim milk: An in vitro study using a dynamic stomach system. Food Chem. 2023, 402, 134365. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wu, P.; Wang, J.; Dupont, D.; Menard, O.; Jeantet, R.; Chen, X.D. Achieving realistic gastric emptying curve in an advanced dynamic in vitro human digestion system: Experiences with cheese—A difficult to empty material. Food Funct. 2021, 12, 3965–3977. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Goñi, I.; Garcia-Alonso, A.; Saura-Calixto, F. A starch hydrolysis procedure to estimate glycemic index. Nutr. Res. 1997, 17, 427–437. [Google Scholar] [CrossRef]
- Aziz, A. The glycemic index: Methodological aspects related to the interpretation of health effects and to regulatory labeling. J. AOAC Int. 2009, 92, 879–887. [Google Scholar] [CrossRef]
- Li, Y.; Kong, F. Simulating human gastrointestinal motility in dynamic in vitro models. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3804–3833. [Google Scholar] [CrossRef]
- Zhong, C.; Langrish, T. A comparison of different physical stomach models and an analysis of shear stresses and strains in these system. Food Res. Int. 2020, 135, 109296. [Google Scholar] [CrossRef]
- Cassilly, D.; Kantor, S.; Knight, L.C.; Maurer, A.H.; Fisher, R.S.; Semler, J.; Parkman, H.P. Gastric emptying of a non-digestible solid: Assessment with simultaneous SmartPill pH and pressure capsule, antroduodenal manometry, gastric emptying scintigraphy. Neurogastroenterol. Motil. 2008, 20, 311–319. [Google Scholar] [CrossRef]
- Laulicht, B.; Tripathi, A.; Schlageter, V.; Kucera, P.; Mathiowitz, E. Understanding gastric forces calculated from high-resolution pill tracking. Proc. Natl. Acad. Sci. USA 2010, 107, 8201–8206. [Google Scholar] [CrossRef]
- van Kempen, T.A.; Regmi, P.R.; Matte, J.J.; Zijlstra, R.T. In vitro starch digestion kinetics, corrected for estimated gastric emptying, predict portal glucose appearance in pigs. J. Nutr. 2010, 140, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Bornhorst, G.M.; Paul Singh, R. Gastric digestion in vivo and in vitro: How the structural aspects of food influence the digestion process. Annu. Rev. Food Sci. Technol. 2014, 5, 111–132. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Liu, H.; Li, X.; Wang, H.; Zhang, K.; Li, S.; Bao, X.; Zou, W.; Yu, W. Predicting the Glycemic Index of Biscuits Using Static In Vitro Digestion Protocols. Foods 2023, 12, 404. [Google Scholar] [CrossRef] [PubMed]
- Grenko, C.M.; Taylor, H.J.; Bonnycastle, L.L.; Xue, D.; Lee, B.N.; Weiss, Z.; Yan, T.; Swift, A.J.; Mansell, E.C.; Lee, A.; et al. Single-cell transcriptomic profiling of human pancreatic islets reveals genes responsive to glucose exposure over 24 h. Diabetologia 2024, 67, 2246–2259. [Google Scholar] [CrossRef]
- Miao, B.; Mohiuddin, M.S.; Barua, R.; Wahiduzzaman, M.; Fang, Z.; Hu, W.; Tirumalasetty, M.B.; Sun, X.; Choubey, M.; Miao, Q.R. Glucose transporter 1 is essential to maintain brain endothelial cell homeostasis under hyperglycemia condition. Am. J. Physiol. Cell Physiol. 2025, 329, C341–C354. [Google Scholar] [CrossRef]
- Huang, B.; Bao, J.; Cao, Y.R.; Gao, H.F.; Jin, Y. Cytochrome P450 1A1 (CYP1A1) Catalyzes Lipid Peroxidation of Oleic Acid-Induced HepG2 Cells. Biochemistry 2018, 83, 595–602. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Shin, S.; Chun, Y.J. Biological roles of cytochrome P450 1A1, 1A2, and 1B1 enzymes. Arch. Pharmacal Res. 2021, 44, 63–83. [Google Scholar] [CrossRef]
- Ma, X.; Jin, H.; Chu, X.; Dai, W.; Tang, W.; Zhu, J.; Wang, F.; Yang, X.; Li, W.; Liu, G.; et al. The Host CYP1A1-Microbiota Metabolic Axis Promotes Gut Barrier Disruption in Methicillin-Resistant Staphylococcus aureus-Induced Abdominal Sepsis. Front. Microbiol. 2022, 13, 802409. [Google Scholar] [CrossRef]
- Yang, R.; Roshani, D.; Gao, B.; Li, P.; Shang, N. Metallothionein: A Comprehensive Review of Its Classification, Structure, Biological Functions, and Applications. Antioxidants 2024, 13, 825. [Google Scholar] [CrossRef]
- Rios, C.; Santander, I.; Méndez-Armenta, M.; Nava-Ruiz, C.; Orozco-Suárez, S.; Islas, M.; Barón-Flores, V.; Diaz-Ruiz, A. Metallothionein-I + II Reduces Oxidative Damage and Apoptosis after Traumatic Spinal Cord Injury in Rats. Oxidative Med. Cell. Longev. 2018, 2018, 3265918. [Google Scholar] [CrossRef]
- Wu, G.; Wang, D.; Xiong, F.; Wang, Q.; Liu, W.; Chen, J.; Chen, Y. The emerging roles of CEACAM6 in human cancer (Review). Int. J. Oncol. 2024, 64, 27. [Google Scholar] [CrossRef]
- Ru, G.Q.; Han, Y.; Wang, W.; Chen, Y.; Wang, H.J.; Xu, W.J.; Ma, J.; Ye, M.; Chen, X.; He, X.L.; et al. CEACAM6 is a prognostic biomarker and potential therapeutic target for gastric carcinoma. Oncotarget 2017, 8, 83673–83683. [Google Scholar] [CrossRef]
- Jones, S.K.; McCarthy, D.M.; Vied, C.; Stanwood, G.D.; Schatschneider, C.; Bhide, P.G. Transgenerational transmission of aspartame-induced anxiety and changes in glutamate-GABA signaling and gene expression in the amygdala. Proc. Natl. Acad. Sci. USA 2022, 119, e2213120119. [Google Scholar] [CrossRef]
- Larson, E.A.; Accardi, M.V.; Wang, Y.; D’Antoni, M.; Karimi, B.; Siddiqui, T.J.; Bowie, D. Nitric Oxide Signaling Strengthens Inhibitory Synapses of Cerebellar Molecular Layer Interneurons through a GABARAP-Dependent Mechanism. J. Neurosci. 2020, 40, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Qiang, S.; Cheng, C.; Dong, Y.; Tang, C.; Zheng, J.; Liu, Y. DDIT4 participates in high glucose-induced fibroblast-like synoviocytes overactivation and cartilage injury by regulating glycolysis. Regen. Ther. 2025, 29, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, Z.; Liu, C.; Zhao, M.; Wang, X.; Zhai, J.; Wang, C.; Song, G. Circulating levels of DDIT4 and mTOR, and contributions of BMI, inflammation and insulin sensitivity in hyperlipidemia. Exp. Ther. Med. 2022, 24, 666. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, X.; Ouyang, Y.; Fei, X.; He, C.; Yang, X.; Ren, Y.; Zhou, Y.; Chen, S.; Hu, Y.; et al. The Protective Role of DDIT4 in Helicobacter pylori-induced Gastric Metaplasia Through Metabolic Regulation of Ferroptosis. Cell. Mol. Gastroenterol. Hepatol. 2025, 19, 101448. [Google Scholar] [CrossRef]
- Du, F.; Sun, L.; Chu, Y.; Li, T.; Lei, C.; Wang, X.; Jiang, M.; Min, Y.; Lu, Y.; Zhao, X.; et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun. 2018, 38, 45. [Google Scholar] [CrossRef]
- Pinto, J.A.; Rolfo, C.; Raez, L.E.; Prado, A.; Araujo, J.M.; Bravo, L.; Fajardo, W.; Morante, Z.D.; Aguilar, A.; Neciosup, S.P.; et al. In silico evaluation of DNA Damage Inducible Transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci. Rep. 2017, 7, 1526. [Google Scholar] [CrossRef]
- Satokari, R. High Intake of Sugar and the Balance between Pro- and Anti-Inflammatory Gut Bacteria. Nutrients 2020, 12, 1348. [Google Scholar] [CrossRef]
- Carvalho, M.R.; Yan, L.P.; Li, B.; Zhang, C.H.; He, Y.L.; Reis, R.L.; Oliveira, J.M. Gastrointestinal organs and organoids-on-a-chip: Advances and translation into the clinics. Biofabrication 2023, 15, 042004. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, F.; Jin, Y.; Ma, Y. Applications of human organoids in the personalized treatment for digestive diseases. Signal Transduct. Target. Ther. 2022, 7, 336. [Google Scholar] [CrossRef]
| Gene Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| CEACAM6 | ACGTCACCCAGAATGACACA | GACCATTTACCCACCACAGG |
| PHGR1 | CCCTGCTCTGCACTCTCAG | CGCAGTGACCTGGAGGAT |
| MT2A | CCTCGAGGATGGATCCCAAC | GGGTACCCGGCGCAGCAG |
| DDIT4 | CTGGACAGCAGCAACAGTG | ACACCCCATCCAGGTAAGC |
| MT1B | GCCTTGGCTGACTTGGTGAT | GACCCAACCGTCACGGATAA |
| TAGLN | GAGCAAGCTGGTGAACAGCC | GACCATGGAGGGTGGGTTCT |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
| Num. | Food | Carbohydrates (g/100 g) | Protein (g/100 g) | Fat (g/100 g) | Fiber (g/100 g) | Human GI *** | eGI [15] * | Deviation % **** | eGI (This Study) ** | Deviation % **** |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lotus root powder (pure) | 93.0 | 0.2 | 0.0 | 0.1 | 39 | 46 | −17.9 | 36 | 7.7 |
| 2 | Grain millet | 73.5 | 9.0 | 3.1 | 1.6 | 71 | 78 | −9.9 | 69 | 2.8 |
| 3 | Rice | 71.8 | 12.7 | 0.9 | 0.6 | 82 | 86 | −4.9 | 75 | 8.5 |
| 4 | Chinese yam | 69.4 | 9.1 | 1.0 | 1.4 | 51 | 56 | −9.8 | 45 | 11.8 |
| 5 | Lotus seed powder | 64.2 | 17.2 | 2 | 3.0 | 51 | 56 | −9.8 | 41 | 19.6 |
| 6 | Dumpling wrapper | 57.0 | 9.3 | 1.4 | 2.2 | 70 | 83 | −18.6 | 68 | 2.9 |
| 7 | Biscuits | 55.0 | 12.0 | 10.0 | 10.0 | 52 | 49 | 5.8 | 38 | 26.9 |
| 8 | Milk powder | 51.7 | 20.1 | 21.2 | 0.0 | 40 | 42 | −5.0 | 40 | 0.0 |
| 9 | Dumplings | 26.6 | 8.8 | 12.3 | 3.2 | 50 | 72 | −44.0 | 55 | −10.0 |
| 10 | Black soybean | 23.4 | 36 | 15.9 | 10.2 | 42 | 51 | −21.4 | 27 | 35.7 |
| 11 | Potato | 16.5 | 2 | 0.2 | 0.7 | 65 | 74 | −13.8 | 63 | 3.1 |
| 12 | Apple | 12.3 | 0.2 | 0.2 | 1.2 | 36 | 64 | −77.8 | 39 | −8.3 |
| 13 | Pear | 10.2 | 0.4 | 0.2 | 3.1 | 36 | 51 | −41.7 | 30 | 16.7 |
| 14 | Milk (pure) | 3.4 | 3 | 3.2 | 0 | 27 | 52 | −92.6 | 28 | −3.7 |
| Rank | Gene | Full Name | Dynamic Model vs. Undigested | Static Model vs. Undigested | ||
|---|---|---|---|---|---|---|
| FC | q Value | FC | q Value | |||
| 1 | CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 | 28.24 | 1.49 × 10−3 | 20.57 | 4.88 × 10−3 |
| 2 | MT2A | Metallothionein 2A | 20.36 | 2.73 × 10−3 | 14.56 | 4.85 × 10−3 |
| 3 | MT1H | Metallothionein 1H | 20.01 | 1.71 × 10−3 | 6.53 | 1.04 × 10−2 |
| 4 | CEACAM6 | CEA Cell Adhesion Molecule 6 | 12.53 | 1.76 × 10−3 | 13.68 | 2.88 × 10−3 |
| 5 | NGFR | Nerve Growth Factor Receptor | 7.62 | 1.49 × 10−3 | 6.41 | 2.76 × 10−3 |
| 6 | ABCG2 | ATP Binding Cassette Subfamily G Member 2 | 7.24 | 2.75 × 10−3 | 6.03 | 5.09 × 10−3 |
| Rank | Gene | Full Name | Dynamic Model vs. Undigested | Static Model vs. Undigested | ||
|---|---|---|---|---|---|---|
| FC | q Value | FC | q Value | |||
| 1 | TAC4 | Tachykinin Precursor 4 | 0.18 | 2.75 × 10−3 | 0.13 | 4.66 × 10−3 |
| 2 | PHGR1 | Proline, Histidine And Glycine Rich 1 | 0.23 | 3.44 × 10−3 | 0.27 | 6.19 × 10−3 |
| 3 | ACSS1 | Acyl-CoA Synthetase Short Chain Family Member 1 | 0.28 | 1.71 × 10−3 | 0.28 | 2.76 × 10−3 |
| 4 | MMP9 | Matrix Metallopeptidase 9 | 0.31 | 5.96 × 10−3 | 0.33 | 8.47 × 10−3 |
| 5 | DEGS2 | Delta 4-Desaturase, Sphingolipid 2 | 0.31 | 3.76 × 10−3 | 0.37 | 1.07 × 10−2 |
| 6 | NPR2 | Natriuretic Peptide Receptor 2 | 0.32 | 3.44 × 10−3 | 0.32 | 5.09 × 10−3 |
| Rank | Gene | Full Name | FC | q-Value |
|---|---|---|---|---|
| 1 | MT1B | Metallothionein 1B | 5.96 | 2.69 × 10−3 |
| 2 | GABARAP | GABA Type A Receptor Associated | 3.18 | 1.36 × 10−2 |
| 3 | MT1H | Metallothionein 1H | 3.07 | 4.35 × 10−3 |
| 4 | MT1X | Metallothionein 1X | 2.95 | 1.77 × 10−3 |
| 5 | DDIT4 | DNA Damage Inducible Transcript 4 | 2.67 | 4.33 × 10−5 |
| 6 | SLC38A2 | Solute Carrier Family 38 Member 2 | 2.57 | 8.45 × 10−3 |
| 7 | ATP6V1B1 | ATPase H+ Transporting V1 Subunit B1 | 0.57 | 6.13 × 10−5 |
| 8 | PMPCA | Peptidase Mitochondrial Processing Subunit Alpha | 0.64 | 6.38 × 10−5 |
| 9 | RASL10B | RAS Like Family 10 Member B | 0.64 | 9.71 × 10−5 |
| 10 | TAGLN | Transgelin | 0.53 | 2.33 × 10−4 |
| 11 | RARRES4 | Phospholipase A and Acyltransferase 4 | 0.58 | 2.76 × 10−4 |
| 12 | CRCT1 | Cysteine Rich C-Terminal 1 | 0.66 | 5.31 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.; Sun, Y.; Wu, P.; Dong, Z.; Qin, Y.; Wang, L.; Xiao, J.; Hou, C.; Ying, X.; Gao, J.; et al. Physiologically Relevant Simulation of Carbohydrate Digestion: From Glycemic Index Estimation to Intestinal Cellular Responses. Foods 2025, 14, 3864. https://doi.org/10.3390/foods14223864
Meng J, Sun Y, Wu P, Dong Z, Qin Y, Wang L, Xiao J, Hou C, Ying X, Gao J, et al. Physiologically Relevant Simulation of Carbohydrate Digestion: From Glycemic Index Estimation to Intestinal Cellular Responses. Foods. 2025; 14(22):3864. https://doi.org/10.3390/foods14223864
Chicago/Turabian StyleMeng, Jinfeng, Ying Sun, Peng Wu, Zhizhong Dong, Yuhan Qin, Liming Wang, Jie Xiao, Can Hou, Xin Ying, Jiaxing Gao, and et al. 2025. "Physiologically Relevant Simulation of Carbohydrate Digestion: From Glycemic Index Estimation to Intestinal Cellular Responses" Foods 14, no. 22: 3864. https://doi.org/10.3390/foods14223864
APA StyleMeng, J., Sun, Y., Wu, P., Dong, Z., Qin, Y., Wang, L., Xiao, J., Hou, C., Ying, X., Gao, J., Huan, M., Chen, R., Wang, Y., Wang, Y., Wang, J., Chen, X., & An, T. (2025). Physiologically Relevant Simulation of Carbohydrate Digestion: From Glycemic Index Estimation to Intestinal Cellular Responses. Foods, 14(22), 3864. https://doi.org/10.3390/foods14223864






