Study on the Enrichment of the Main Active Components in Rhodococcus opacus PD630 Cell-Free Supernatant for the Degradation of Aflatoxin B1, the Degradation Products, and the Underlying Mechanisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Equipment
2.2. Preparation of R. opacus PD630 Seed Culture and Cell-Free Supernatant
2.3. HPLC Determination of AFB1
2.4. Enrichment of AFB1-Degrading Active Principles from RCFS by Ethanol Precipitation
2.5. Effect of Temperature on AFB1-Degrading Activity of RCFSC-EP
2.6. Effects of Proteinase K and EDTA-2Na on the AFB1-Degrading Activity of RCFSC-EP
2.7. Influence of Substrate Concentration on AFB1-Degrading Activity of RCFSC-EP
2.8. Non-Targeted Metabolomic Analysis of AFB1 Degradation Products by RCFS
2.8.1. Sample Preparation for Metabolomic Profiling of AFB1 Degradation by RCFS
2.8.2. Metabolomic Sample Pretreatment for RCFS
2.8.3. LC-MS Analysis of RCFS Non-Targeted Metabolomics
2.8.4. Non-Targeted Metabolomics Data Processing
2.8.5. Metabolite Identification
2.9. LC-MS/MS-Based Proteomic Profiling of RCFS
2.9.1. Protein Extraction
2.9.2. Proteolytic Digestion and Desalting
2.9.3. LC-MS/MS Instrument Conditions
2.10. Statistical Analysis
3. Results
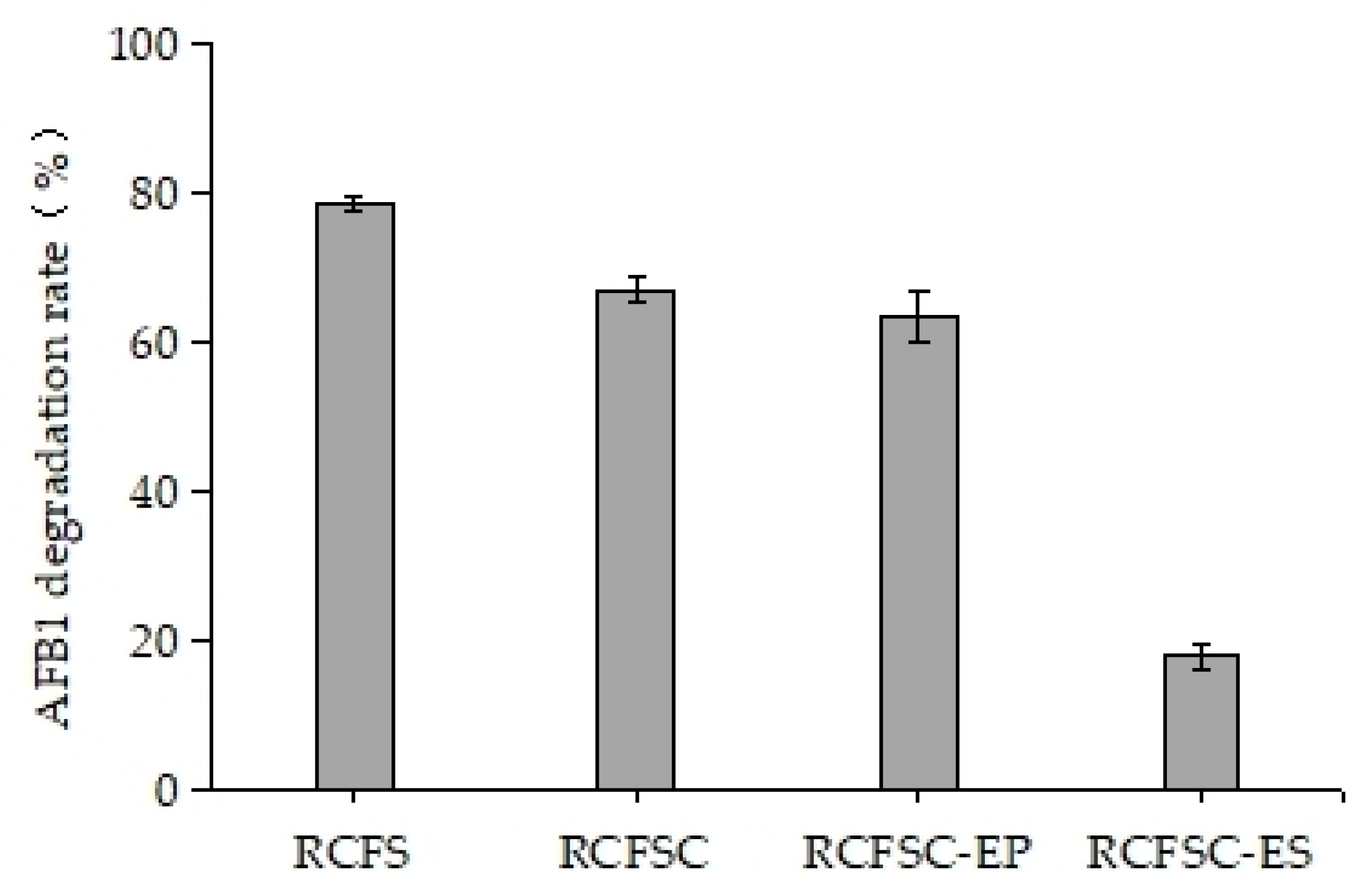
3.1. Ethanol PrecipitationEnrichment of AFB1-Degrading Active Constituents from RCFS
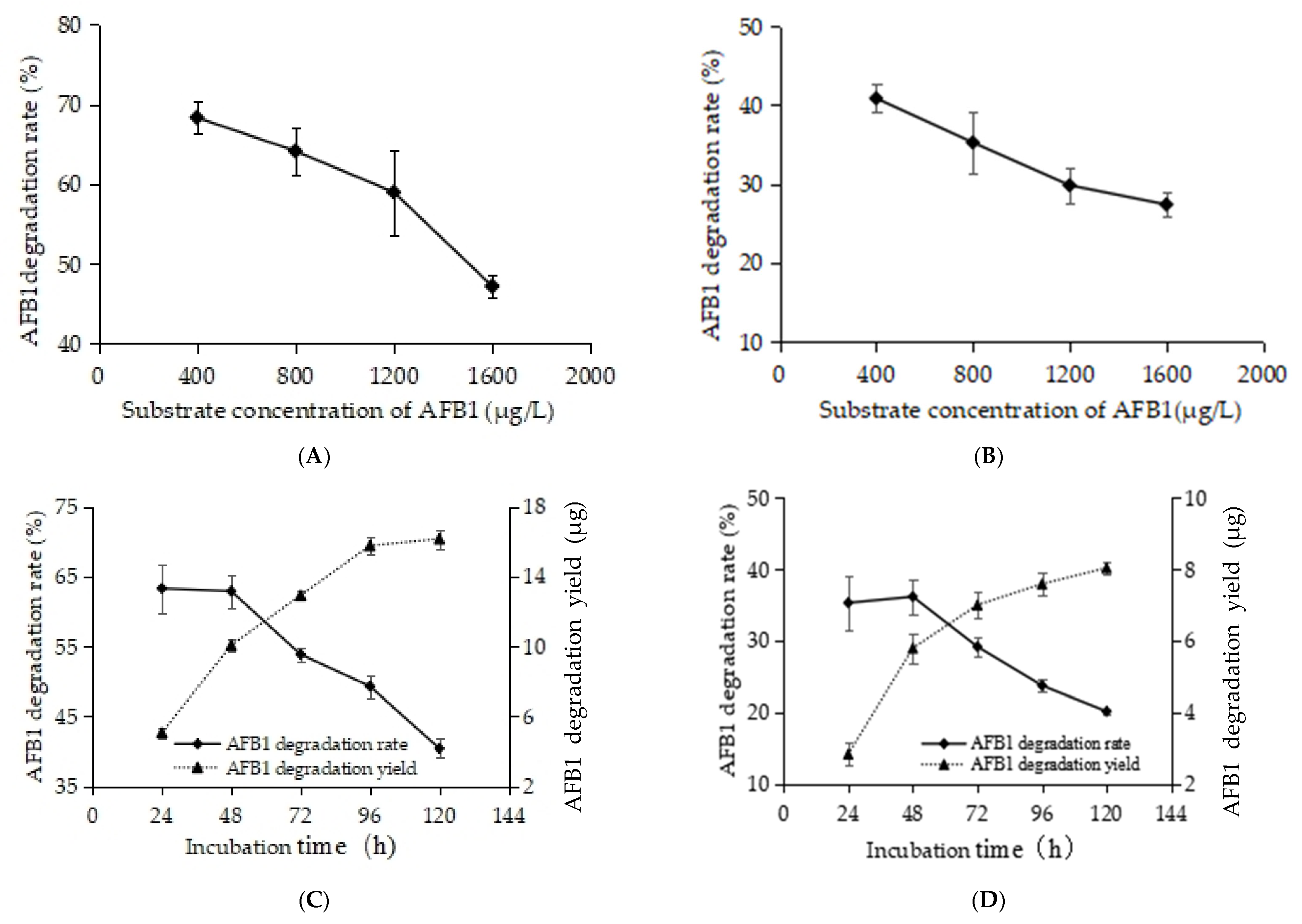
3.2. Effect of Temperature on AFB1-Degrading Activity of RCFSC-EP
3.3. Effects of Proteinase K and EDTA-2Na on AFB1-Degrading Activity of RCFSC-EP
3.4. Effect of Substrate on AFB1-Degrading Activity of RCFSC-EP
3.5. Characterization of AFB1 Degradation Products by RCFS
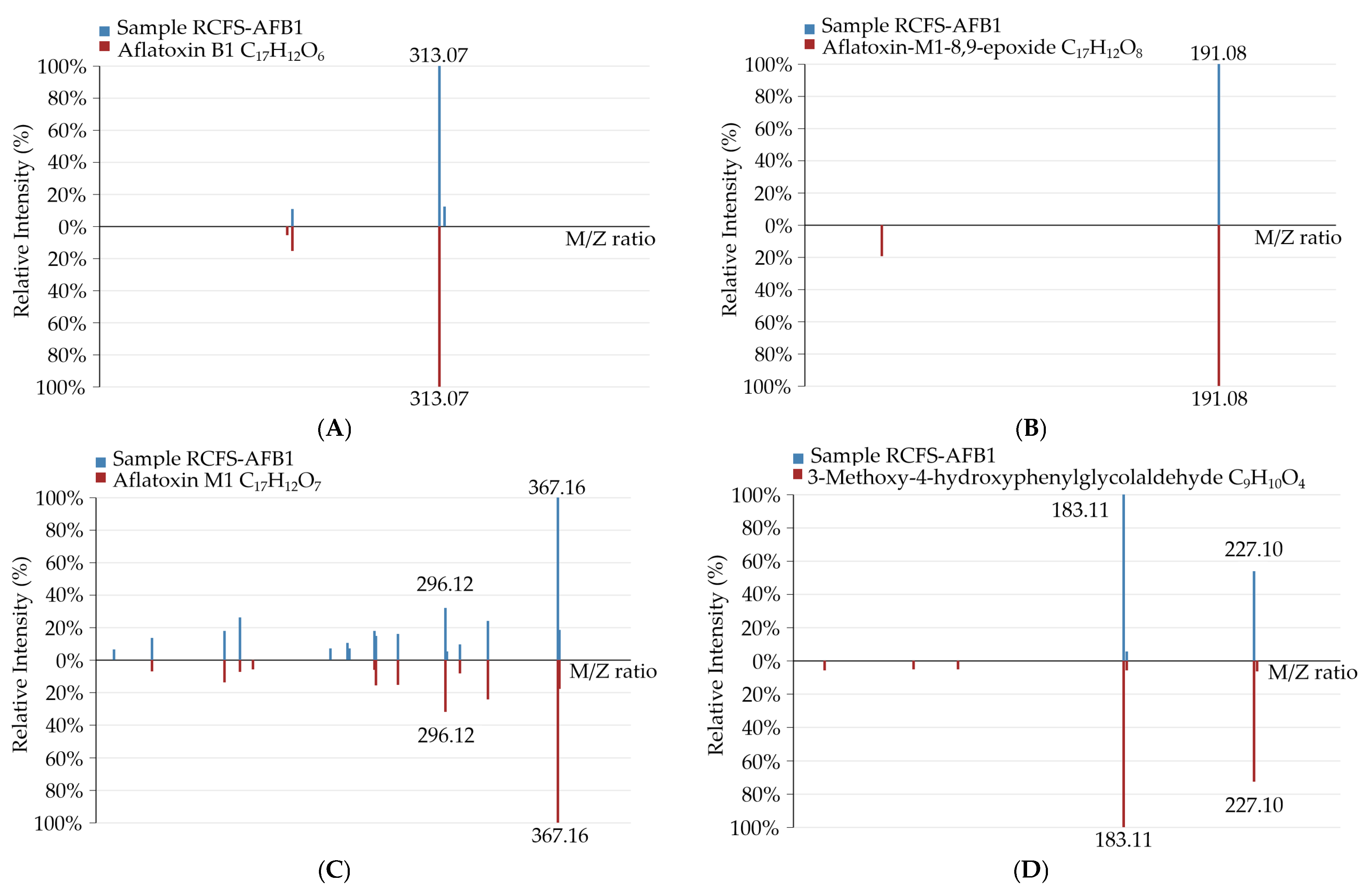
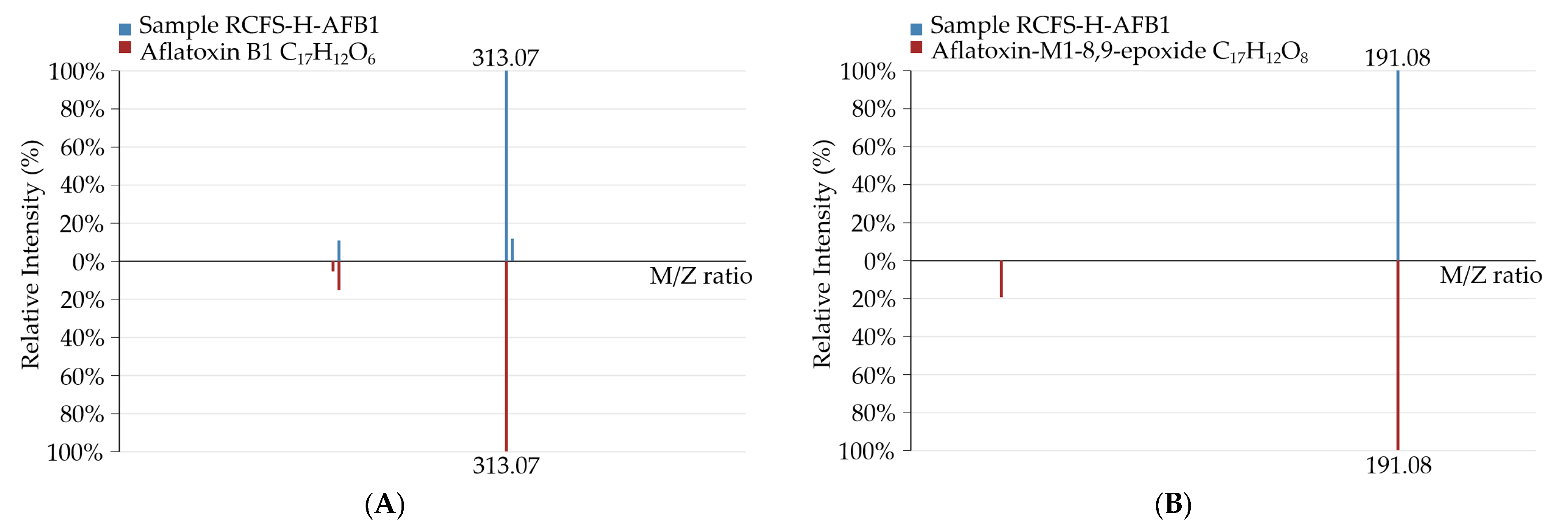
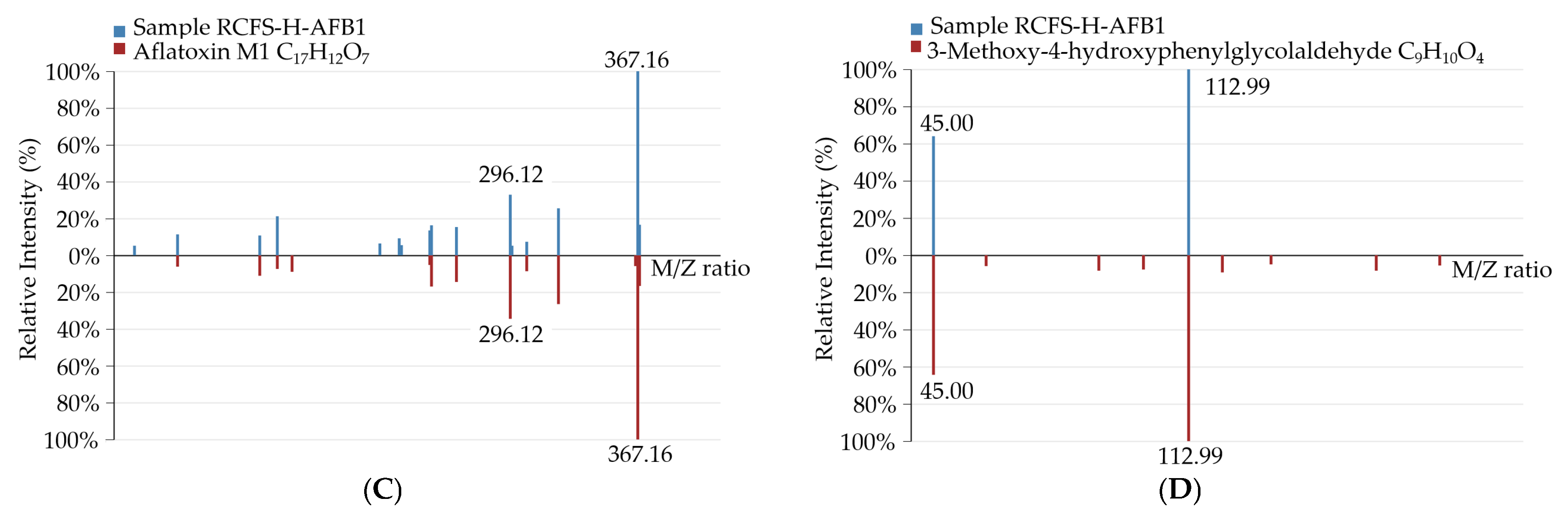
3.5.1. Identification of AFB1 Degradation Products Formed by RCFS
3.5.2. Identification and Profiling of AFB1 Degradation Products by RCFS
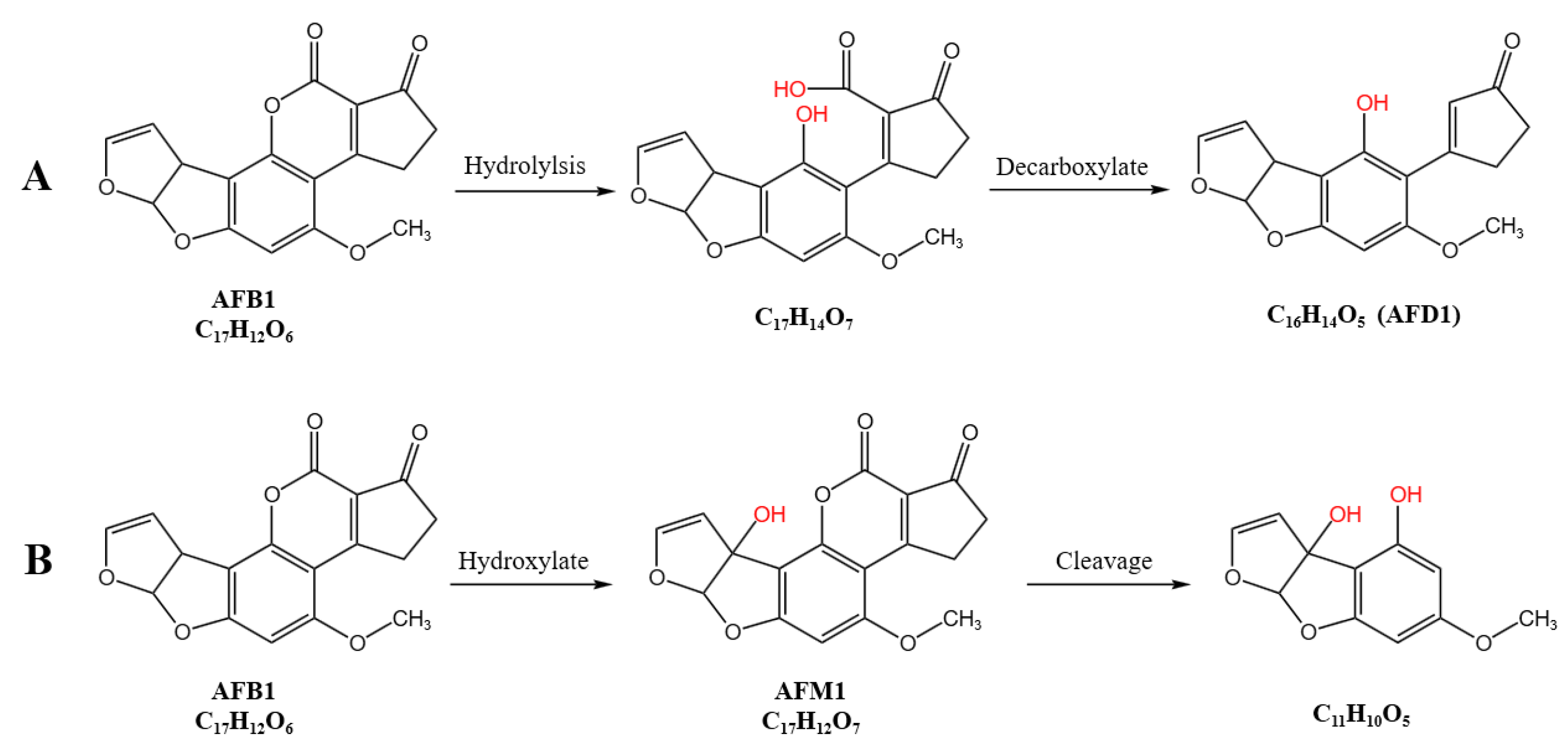
3.5.3. Proposed AFB1 Degradation Pathway by RCFS
3.6. Proteomic Profile of RCFS
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Khaneghah, A.M.; Fakhri, Y.; Gahruie, H.H.; Niakousari, M.; Sant’Ana, A.S. Mycotoxins in cereal-based products during 24 years (1983–2017): A global systematic review. Trends Food Sci. Technol. 2019, 91, 95–105. [Google Scholar] [CrossRef]
- EI-Sayed, R.A.; Jebur, A.; Kang, W.; EI-Esawi, M.; EI-Demerdash, F. An overview on the major mycotoxins in food products: Characteristics, toxicity, and analysis. J. Future Foods 2022, 2, 91–102. [Google Scholar] [CrossRef]
- Khaneghah, A.M.; Fakhri, Y.; Raeisi, S.; Armoon, B.; Sant’Ana, A.S. Prevalence and concentration of ochratoxin A, zearalenone, deoxynivalenol and total aflatoxin in cereal-based products: A systematic review and meta-analysis. Food Chem. Toxicol. 2018, 118, 830–848. [Google Scholar] [CrossRef]
- Khaneghah, A.M.; Fakhri, Y.; Silva, B.S.D.; Ana, A.S.S. Meta-analysis of occurrence of ochratoxin A (OTA), zearalenone (ZEN), Deoxynivalenol (DON) and total aflatoxin (TAF) in cereal based products. In Proceedings of the 26th International ICFMH Conference-Food Micro 2018, Berlin, Germany, 3–6 September 2018. Poster P5.14. [Google Scholar]
- Arce-López, B.; Coton, M.; Coton, E.; Hymery, N. Occurrence of the two major regulated mycotoxins, ochratoxin A and fumonisin B1, in cereal and cereal-based products in Europe and toxicological effects: A review. Environ. Toxicol. Pharmacol. 2024, 109, 104489. [Google Scholar] [CrossRef] [PubMed]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recher, T. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef]
- Emadi, A.; Eslami, M.; Yousefi, B.; Abdolshahi, A. In vitro strain specific reducing of aflatoxin B1 by probiotic bacteria: A systematic review and metaanalysis. Toxin Rev. 2022, 41, 995–1006. [Google Scholar] [CrossRef]
- Baan, R.; Grosse, Y.; Straif, K.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens-part f: Chemical agents and related occupations. Lancet Oncol. 2009, 10, 1143–1144. [Google Scholar] [CrossRef]
- Azziz-Baumgartner, E.; Lindblade, K.; Gieseker, K.; Rogers, H.S.; Kieszak, S.; Njapau, H.; Schleicher, R.; McCoy, L.F.; Misore, A.; DeCock, K.; et al. Case-control study of an acute aflatoxicosis outbreak, Kenya, 2004. Environ. Health Perspect. 2005, 113, 1779–1783. [Google Scholar] [CrossRef]
- Eaton, D.L.; Groopman, J.D. The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance; Academic Press: San Diego, CA, USA, 1993; p. 544. [Google Scholar]
- Raj, H.G.; Prasanna, H.R.; Magee, P.N.; Lotlikar, P.D. Effect of purified rat and hamster hepatic glutathione S-transferases on the microsome mediated binding of aflatoxin B1 to DNA. Cancer Lett. 1986, 33, 1–9. [Google Scholar] [CrossRef]
- Wochner, K.F.; Becker-Algeri, T.A.; Colla, E.; Badiale-Furlong, E.; Drunkler, D.A. The action of probiotic microorganisms on chemical contaminants in milk. Crit. Rev. Microbiol. 2018, 44, 112–123. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, J. A review of research progress on the microbial or enzymatic degradation and mechanism of aflatoxin, B1. J. Microbiol. Biotechnol. 2025, 35, e2504044. [Google Scholar] [CrossRef]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S.; Holzapfel, W.H.; Zyl, W. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef]
- Krifaton, C.S.; Kriszt, B.; Szoboszlay, S.; Mátyás, C.; József, K. Analysis of aflatoxin-B1-degrading microbes by use of a combined toxicity-profiling method. Mutat. Res. Toxicol. Env. Mutagen. 2011, 726, 1–7. [Google Scholar] [CrossRef]
- Risa, A.; Divinyi, D.M.; Erzsebet, B.; Krifaton, C. Aflatoxin B1detoxification by cell-free extracts of Rhodococcus strains. Acta Microbiol. Immunol. Hung. 2017, 64, 423–438. [Google Scholar] [CrossRef]
- Lapalikar, G.V.; Taylor, M.C.; Warden, A.C.; Scott, C.; Russell, R.J.; Oakeshott, J.G. F420H2-dependent degradation of aflatoxin and other furanocoumarins is widespread throughout the Actinomycetales. PLoS ONE 2012, 7, e30114. [Google Scholar] [CrossRef]
- Liu, H.; Tang, Y.; Si, W.; Yin, J.; Xu, Y.; Yang, J. Rhodococcus turbidus PD630 enables efficient biodegradation of aflatoxin B1. LWT-Food Sci. Technol. 2023, 186, 115225. [Google Scholar] [CrossRef]
- Li, Y.; He, Y.; Zhang, H.; Ma, X. Effects of ultrasonic-enzymatic-assisted ethanol precipitation method on the physicochemical characteristics, antioxidant and hypoglycemic activities of Tremella fuciformis polysaccharides. Ultrason. Sonochem. 2023, 101, 106682. [Google Scholar] [CrossRef]
- Shu, X.; Wang, Y.; Zhou, Q.; Li, M.; Hu, H.; Ma, Y.; Chen, X.; Ni, J.; Zhao, W.; Huang, S.; et al. Biological Degradation of Aflatoxin B1 by Cell-Free Extracts of Bacillus velezensis DY3108 with Broad PH Stability and Excellent Thermostability. Toxins 2018, 10, 330. [Google Scholar] [CrossRef]
- Sun, F.; Sun, Q.; Zhang, H.; Kong, B.; Xia, X. Purification and biochemical characteristics of the microbial extracellular protease from Lactobacillus curvatus isolated from Harbin dry sausages. Int. J. Biol. Macromol. 2019, 133, 987–997. [Google Scholar] [CrossRef]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 Degradation by Stenotrophomonas maltophilia and Other Microbes Selected Using Coumarin Medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef]
- Boyadzhieva, I.; Berberov, K.; Atanasova, N.; Krumov, N.; Kabaivanova, L. Extracellular Haloalkalophilic Pectinase Produced by Virgibacillus salarius Strain 434—A Useful Tool for Biotechnological Applications. Appl. Sci. 2024, 14, 9295. [Google Scholar] [CrossRef]
- He, J.; Zhang, H.; Liu, Y.; Ju, Y.; He, Y.; Jiang, Y.; Jiang, J. Interfacial Extraction to Trap and Characterize the Criegee Intermediates from Phospholipid Ozonolysis. Anal. Chem. 2023, 95, 5018–5023. [Google Scholar] [CrossRef]
- Zelena, E.; Dunn, W.B.; Broadhurst, D.; Francis-Mcintyre, S.; Carroll, K.M.; Begley, P.; O’Hagan, S.; Knowles, J.D.; Halsall, A.; Wilson, I.D.; et al. Development of a Robust and Repeatable UPLC-MS Method for the Long-Term Metabolomic Study of Human Serum. Anal. Chem. 2009, 81, 1357–1364. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Hirano, A.; Arakawa, T.; Shiraki, K. Mechanistic insights into protein precipitation by alcohol. Int. J. Biol. Macromol. 2012, 50, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Song, J.H.; Vasquez, R.; Lee, J.S.; Kim, I.H.; Kang, D.-K. Methods for Detection, Extraction, Purification, and Characterization of Exopolysaccharides of Lactic Acid Bacteria—A Systematic Review. Foods 2024, 13, 3687. [Google Scholar] [CrossRef] [PubMed]
- Sangare, L.; Zhao, Y.; Folly, Y.; Chang, J.; Li, J.; Selvaraj, J.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 Degradation by a Pseudomonas Strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef]
- Wang, C.; Li, Z.; Wang, H.; Qiu, H.; Zhang, M.; Li, S.; Luo, X.; Song, Y.; Zhou, H.; Ma, W.; et al. Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agric. 2017, 97, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Salihi, A.; Asoodeh, A.; Aliabadian, M. Production and biochemical characterization of an alkaline protease from Aspergillus oryzae CH93. Int. J. Biol. Macromol. 2016, 94, 827–835. [Google Scholar] [CrossRef]
- Becker-Algeri, T.A.; Castagnaro, D.; De Bortoli, K.; De Souza, C.; Drunkler, D.A.; Badiale-Furlong, E. Mycotoxins in bovine milk and dairy products: A review. J. Food Sci. 2016, 81, 544–552. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Luo, J.; Qi, B.; Chen, X.; Wan, Y. Horseradish peroxidase immobilized on multifunctional hybrid microspheres for aflatoxin B1 removal: Will enzymatic reaction be enhanced by adsorption? Ind. Eng. Chem. Res. 2019, 58, 11710–11719. [Google Scholar] [CrossRef]
- Alnasser, S.M. The role of glutathione S-transferases in human disease pathogenesis and their current inhibitors. Genes. Dis. 2025, 12, 101482. [Google Scholar] [CrossRef] [PubMed]
- Mazari, A.M.A.; Zhang, L.; Ye, Z.-W.; Zhang, J.; Tew, K.D.; Townsend, D.M. The Multifaceted Role of Glutathione S-Transferases in Health and Disease. Biomolecules 2023, 13, 688. [Google Scholar] [CrossRef]
- Dohnal, V.; Wu, Q.; Kuča, K. Metabolism of aflatoxins: Key enzymes and interindividual as well as interspecies differences. Arch. Toxicol. 2014, 88, 1635–1644. [Google Scholar] [CrossRef]
- Deng, J.; Peng, Z.; Xia, Z.; Mo, Y.; Guo, L.; Wei, J.; Sun, L.; Liu, M. Five glutathione S-transferase isozymes played crucial role in the detoxification of aflatoxin B1 in chicken live. J. Anim. Sci. Biotechnol. 2025, 16, 54. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, J.; Chen, H.; Zhang, J.; Li, D. Aldo-keto reductases 7A subfamily: A mini review. Chem. Biol. Interact. 2024, 391, 110896. [Google Scholar] [CrossRef]
- Bodreddigari, S.; Jones, L.K.; Egner, P.A.; Groopman, J.D.; Sutter, C.H.; Roebuck, B.D.; Guengerich, F.P.; Kensler, T.W.; Sutter, T.R. Protection against aflatoxin b1-induced cytotoxicity by expression of the cloned aflatoxin b1-aldehyde reductases rat AKR7A1 and human AKR7A3. Chem. Res. Toxicol. 2008, 21, 1134–1142. [Google Scholar] [CrossRef]
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef]
- Xie, C.; Li, F.; Jiang, T.; Yao, D. Armillariella tabescens aldo-keto reductase AtAKR297: Cloning, expression, and application in reduction of aflatoxin B1. Microbio. China 2025, 52, 2203–2215. [Google Scholar]
- Marimon, S.K.V.; Garcia, S.D.O.; Penteado, F.A.C.; Remedi, R.D.; Rodrigues, C.M.B.; Badiale-Furlong, E.; Garda-Buffon, J. Aflatoxin biotransformation by commercial peroxidase and its application in contaminated food. J. Chem. Techno. Biotechnol. 2019, 94, 1187–1194. [Google Scholar] [CrossRef]
- Qin, X.; Su, X.; Tu, T.; Zhang, J.; Wang, X.; Wang, Y.; Wang, Y.; Bai, Y.; Yao, B.; Luo, H.; et al. Enzymatic Degradation of Multiple Major Mycotoxins by Dye-Decolorizing Peroxidase from Bacillus subtilis. Toxins 2021, 13, 429. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Renaud, J.B.; Rosini, E.; Pollegioni, L.; Vignali, E.; Haidukowski, M.; Sumarah, M.W.; Logrieco, A.F.; Mulè, G. Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 2020, 250, 126296. [Google Scholar] [CrossRef]
- Forrest, G.L.; Gonzalez, B. Carbonyl reductase. Chem.Biol. Interact. 2000, 129, 21–40. [Google Scholar] [CrossRef]
- Oppermann, U. Carbonyl reductases: The complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 293–322. [Google Scholar] [CrossRef]



| Parameters | Value |
|---|---|
| Enzyme | Trypsin |
| Static modification | Carbamidomethyl/+57.021 Da (C) |
| Dynamic modification | Oxidation/+15.995 Da (M); Acetyl/+42.011 Da (N-Terminal) |
| Precursor charge range | 1–4 |
| Peptide length range | 7–30 |
| Precursor mz range | 300–1800 |
| Fragment ion mz range | 200–1800 |
| Max missed cleavages | 2 |
| Treatment Group | Molecular Formula | Molecular Weight | m/z Ratio | Identification Level |
|---|---|---|---|---|
| RCFS-AFB1 1 | C17H12O7 | 328.06 | 367.1554 | Level 1 |
| C17H12O8 | 344.05 | 327.0511 | Level 1 | |
| C9H10O4 | 182.06 | 227.0533 | Level 2 | |
| C17H14O7 | 330.074 | 313.0698 | Level 3 | |
| C16H14O5 | 286.08 | 285.0772 | Level 3 | |
| C11H10O5 | 222.05 | 223.0618 | Level 3 | |
| RCFS-H-AFB1 2 | C17H12O7 | 328.06 | 367.1581 | Level 1 |
| C17H12O8 | 344.05 | 327.0511 | Level 1 | |
| C9H10O4 | 182.06 | 181.0499 | Level 2 | |
| C11H10O5 | 222.05 | 223.0624 | Level 3 |
| Protein IDs | Genes | Protein Description | Protein Families |
|---|---|---|---|
| P40582 | GTT1 | Glutathione S-transferase 1 | GST superfamily |
| P80031 | GSTP1 | Glutathione S-transferase P | GST superfamily, Pi family |
| Q28035 | GSTA1 | Glutathione S-transferase A2 | GST superfamily, Alpha family |
| P09488 | GSTM1 | Glutathione S-transferase Mu 2 | GST superfamily, Mu family |
| Q9N0V4 | GSTM1 | Glutathione S-transferase 2 | GST superfamily, Mu family |
| Q9N1F5 | GSTO1 | Glutathione S-transferase omega-1 | GST superfamily, Omega family |
| Q3ZCJ2 | AKR1A1 | Aldo-keto reductase family 1 member A1 | Aldo/keto reductase family |
| O43488 | AKR7A2 | Aflatoxin B1 aldehyde reductase member 2 | Aldo/keto reductase family, Aldo/keto reductase 2 subfamily |
| Q8CG76 | Akr7a2 | Aflatoxin B1 aldehyde reductase member 2 | Aldo/keto reductase family, Aldo/keto reductase 2 subfamily |
| Q4PD66 | CCP2 | Putative heme-binding peroxidase | Peroxidase family, Cytochrome c peroxidase subfamily |
| Q0S5Y0 | katG | Catalase-peroxidase | Peroxidase family, Peroxidase/catalase subfamily |
| Q3SZD7 | CBR1 | Carbonyl reductase [NADPH] 1 | Short-chain dehydrogenases/reductases (SDR) family |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, A.; Zhang, X.; Yang, J. Study on the Enrichment of the Main Active Components in Rhodococcus opacus PD630 Cell-Free Supernatant for the Degradation of Aflatoxin B1, the Degradation Products, and the Underlying Mechanisms. Foods 2025, 14, 3772. https://doi.org/10.3390/foods14213772
Zhang A, Zhang X, Yang J. Study on the Enrichment of the Main Active Components in Rhodococcus opacus PD630 Cell-Free Supernatant for the Degradation of Aflatoxin B1, the Degradation Products, and the Underlying Mechanisms. Foods. 2025; 14(21):3772. https://doi.org/10.3390/foods14213772
Chicago/Turabian StyleZhang, Aiyuan, Xuewu Zhang, and Jiguo Yang. 2025. "Study on the Enrichment of the Main Active Components in Rhodococcus opacus PD630 Cell-Free Supernatant for the Degradation of Aflatoxin B1, the Degradation Products, and the Underlying Mechanisms" Foods 14, no. 21: 3772. https://doi.org/10.3390/foods14213772
APA StyleZhang, A., Zhang, X., & Yang, J. (2025). Study on the Enrichment of the Main Active Components in Rhodococcus opacus PD630 Cell-Free Supernatant for the Degradation of Aflatoxin B1, the Degradation Products, and the Underlying Mechanisms. Foods, 14(21), 3772. https://doi.org/10.3390/foods14213772







