From Waste to Wealth: Unlocking the Potential of Cellulase Characteristics for Food Processing Waste Management
Abstract
1. Background
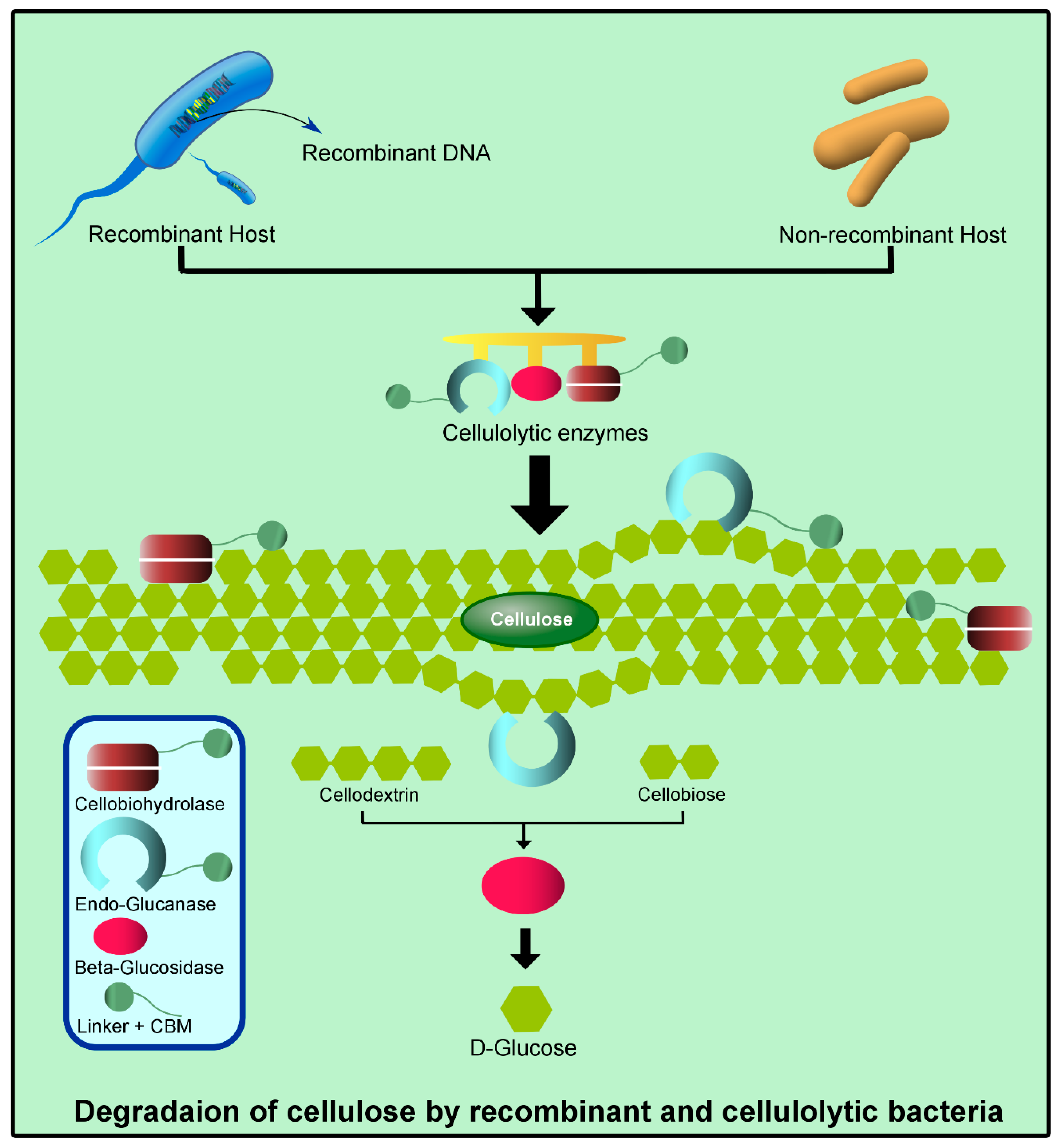
2. Basic Structural and Functional Features of Cellulase
3. Classification and Annotation of Cellulase
4. Evolution of Cellulase
5. Strategies for Cellulase Improvement
5.1. Rational Design Approach
5.2. Directed Evolution Method
5.3. Genetically Engineering the Strain
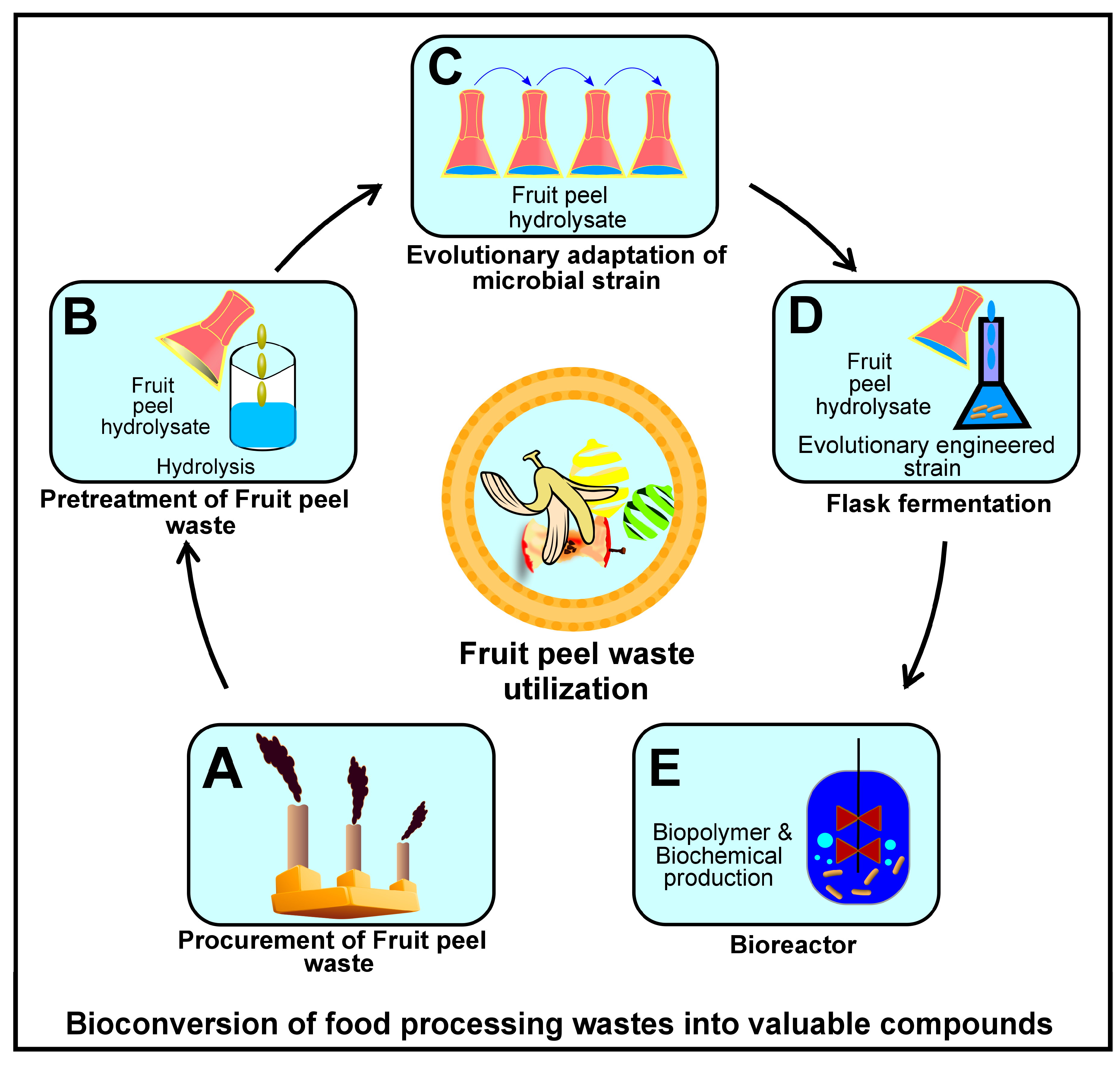
5.4. Improving Cellulase Production Through Microbial Fermentation
5.4.1. Solid State Fermentation (SSF)
5.4.2. Submerged Fermentation (SmF)
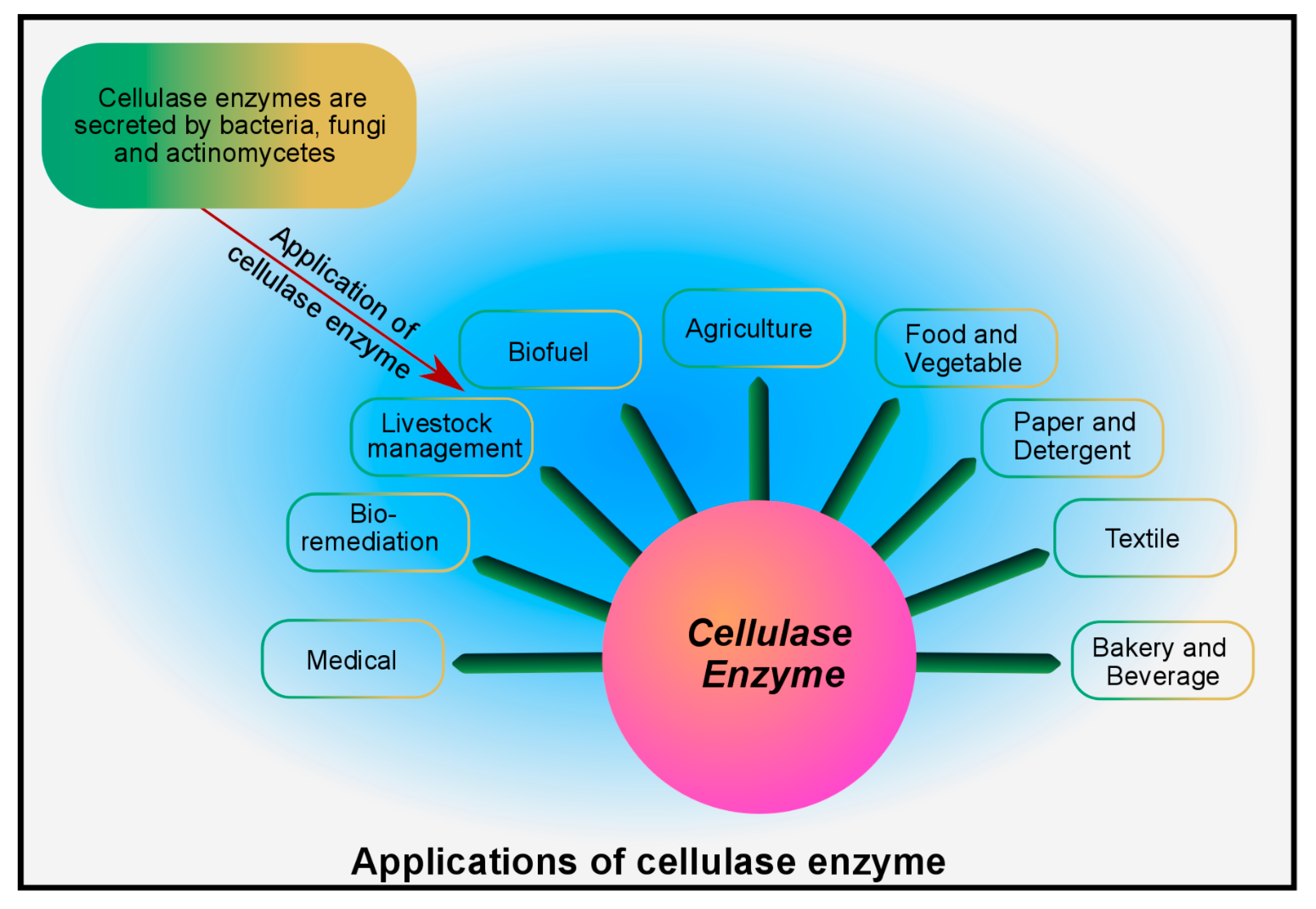
6. Industrial Applications of Microbial Cellulase
7. Role of Microbial Cellulase in Waste Management
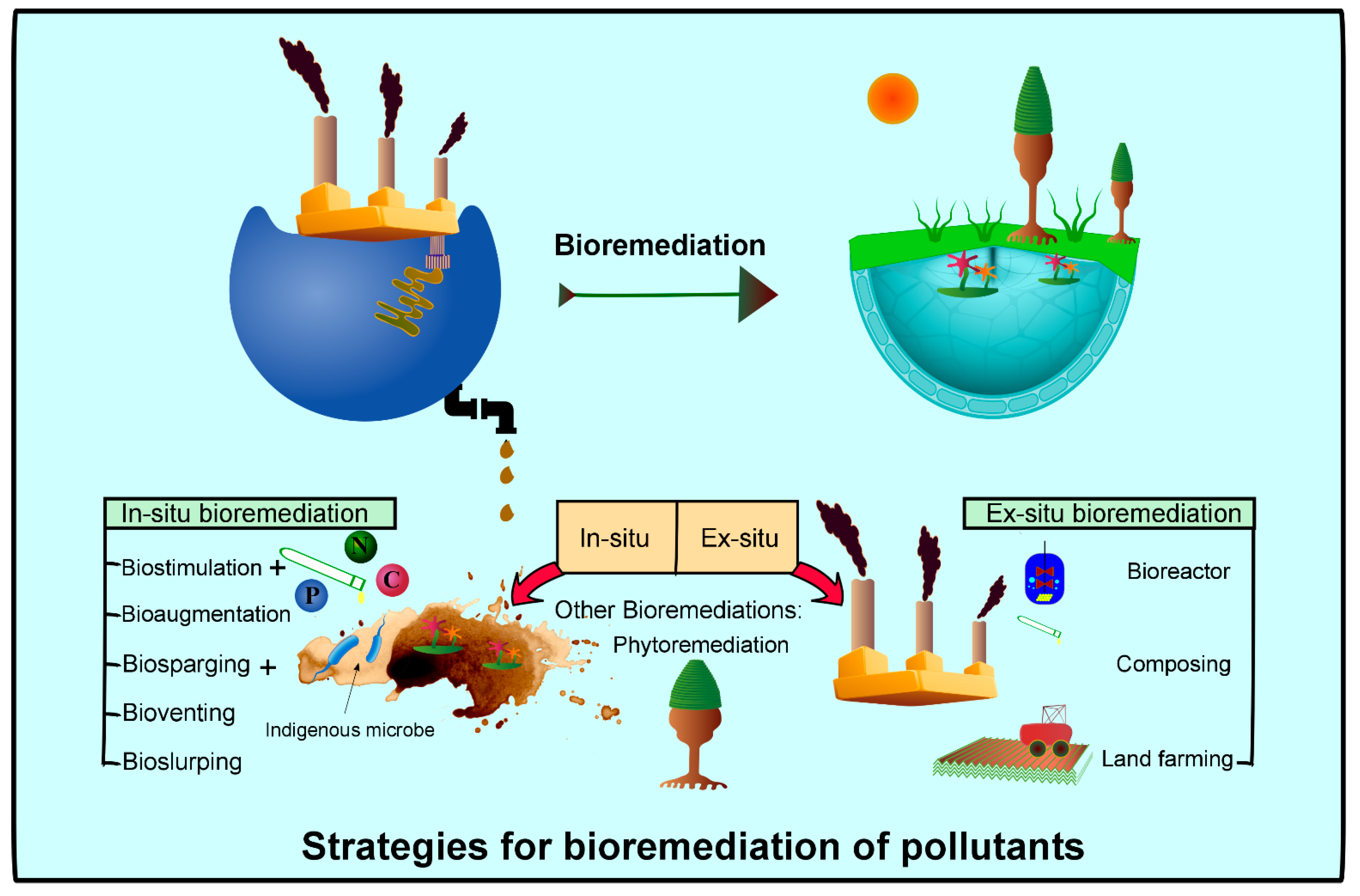
7.1. Bioconversion Through Ex Situ and In Situ Bioremediation Techniques
7.1.1. Composting
7.1.2. Anaerobic Digestion
7.1.3. Phytoremediation and Floating Treatment Wetland
7.2. Integrated Application of In Situ and Ex Situ Cellulase Bioremediation
7.3. Role of Cellulase Enzymes in Mitigating Global Warming and Environmental Impacts
8. Technoeconomic Analysis in Cellulase Biorefinery
9. Conclusions
10. Future Aspects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Conflicts of Interest
Abbreviations
| AD | Anaerobic digestion |
| AWB | Agricultural waste biomass |
| BES | Bio-electrochemical systems |
| CAZymes | Carbohydrate-active enzymes |
| CBDs | Carbohydrate-binding domains |
| Cd | Cadmium |
| CDs | Catalytic domains |
| CRC | Coffee residue compost |
| FPRs | Food processing residuals |
| GH | Glycoside hydrolase |
| LMEs | Lignin-modifying enzymes |
| MSW | Municipal solid waste |
| Sca | Scaffoldin |
| SDM | Site-directed mutagenesis |
| SmF | Solid submerged fermentation |
| SSF | Solid state fermentation |
| UV | Ultraviolet ray |
References
- Ribeiro, I.A.C.; Fontes, G.C.; Coelho, M.A.Z.; Ferreira-Leitão, V.S. Production of Sophorolipids by Candida Bombicola from Waste Frying Oil and its Potential Application in Oil Remediation. Process. Biochem. 2020, 92, 156–164. [Google Scholar] [CrossRef]
- Kaur, G.; Satyanarayana, T. Production of Xylitol from Agro-Residues using Candida Tropicalis and its Potential Industrial Applications. Bioresour. Technol. Rep. 2019, 7, 100236. [Google Scholar] [CrossRef]
- Mohsin, A.; Zhang, K.; Tariq, M.; Zaman, W.Q.; Khan, I.M.; Zhuang, Y.; Guo, M. Optimized Biosynthesis of Xanthan Via Effective Valorization of Orange Peels Using Response Surface Methodology: A Kinetic Model Approach. Carbohydr. Polym. 2018, 181, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.; Alidu, M. Recovery of Cellulose from Food and Agricultural Waste. In Cellulose-Biobased Solutions for Society; IntechOpen: London, UK, 2025. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Gupta, R.; Singh, A. Microbial Cellulases and Their Industrial Applications. Enzyme Res. 2011, 2011, 280696. [Google Scholar] [CrossRef]
- Tong, L.; Li, Y.; Lou, X.; Wang, B.; Jin, C.; Fang, W. Powerful Cell Wall Biomass Degradation Enzymatic System from Saprotrophic Aspergillus fumigatus. Cell Surf. 2024, 11, 100126. [Google Scholar] [CrossRef]
- Jayasekara, S.; Ratnayake, R. Microbial Cellulases: An Overview and Applications; Martín, M.E.E., Ed.; IntechOpen: Rijeka, Croatia, 2019; p. 5. ISBN 978-1-83968-057-1. [Google Scholar]
- Ejaz, U.; Sohail, M.; Ghanemi, A. Cellulases: From Bioactivity to a Variety of Industrial Applications. Biomimetic 2021, 6, 44. [Google Scholar] [CrossRef]
- United Nations Environment Programme (UNEP); United Nations Office for Sustainable Development (UNOSD). Global Waste Management Outlook 2021; United Nations: New York, NY, USA, 2021. [Google Scholar]
- Uddin, M.S.; Abedin, M.Z. Segregation of Plastic Waste from Solid Wastes: Bangladesh Perspective. Int. J. Eng. Mater. Manufact. 2021, 6, 324–331. [Google Scholar] [CrossRef]
- Nazir, M.; Iram, A.; Cekmecelioglu, D.; Demirci, A. Approaches for Producing Fungal Cellulases Through Submerged Fermentation. Front. Biosci. 2024, 16, 5. [Google Scholar] [CrossRef]
- Lakhundi, S.; Siddiqui, R.; Khan, N.A. Cellulose Degradation: A Therapeutic Strategy in the Improved Treatment of Acanthamoeba Infections. Parasit. Vectors 2015, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, H.; Ran, Q.; Liu, J.; Zhou, S.; Qiao, Q.; Song, H.; Peng, F.; Jiang, Z. Effect of Carbohydrate Binding Modules Alterations on Catalytic Activity of Glycoside Hydrolase Family 6 Exoglucanase from Chaetomium thermophilum to Cellulose. Int. J. Biol. Macromol. 2021, 191, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Doi, R.H.; Kosugi, A. Cellulosomes: Plant-Cell-Wall-Degrading Enzyme Complexes. Nat. Rev. Microbiol. 2004, 2, 541–551. [Google Scholar] [CrossRef]
- Ohmiya, K.; Sakka, K.; Karita, S.; Kimura, T. Structure of Cellulases and Their Applications. Biotechnol. Genet. Eng. Rev. 1997, 14, 365–414. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Tavares, G.A.; Béguin, P.; Alzari, P.M. The Crystal Structure of a Type I Cohesin Domain at 1.7 Å Resolution. J. Mol. Biol. 1997, 273, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, N.R.; Henrissat, B.; Kilburn, D.G.; Miller, R.C.; Warren, R.A. Domains in Microbial Beta-1, 4-Glycanases: Sequence Conservation, Function, and Enzyme Families. Microbiol. Rev. 1991, 55, 303–315. [Google Scholar] [CrossRef]
- Harris, P.V.; Xu, F.; Kreel, N.E.; Kang, C.; Fukuyama, S. New Enzyme Insights Drive Advances in Commercial Ethanol Production. Curr. Opin. Chem. Biol. 2014, 19, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Book, A.J.; Lewin, G.R.; McDonald, B.R.; Takasuka, T.E.; Wendt-Pienkowski, E.; Doering, D.T.; Suh, S.; Raffa, K.F.; Fox, B.G.; Currie, C.R. Evolution of High Cellulolytic Activity in Symbiotic Streptomyces through Selection of Expanded Gene Content and Coordinated Gene Expression. PLoS Biol. 2016, 14, e1002475. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. The Hologenome Concept of Evolution after 10 Years. Microbiome 2018, 6, 78. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, T.; Homer, R.J.; Kim, Y.-K.; Chen, N.Y.; Cohn, L.; Hamid, Q.; Elias, J.A. Acidic Mammalian Chitinase in Asthmatic Th2 Inflammation and IL-13 Pathway Activation. Science 2004, 304, 1678–1682. [Google Scholar] [CrossRef]
- Davison, A.; Blaxter, M. Ancient Origin of Glycosyl Hydrolase Family 9 Cellulase Genes. Mol. Biol. Evol. 2005, 22, 1273–1284. [Google Scholar] [CrossRef]
- Libertini, E.; Li, Y.; McQueen-Mason, S.J. Phylogenetic Analysis of the Plant Endo-Beta-1,4-Glucanase Gene Family. J. Mol. Evol. 2004, 58, 506–515. [Google Scholar] [CrossRef]
- Kukor, J.J.; Martin, M.M. Acquisition of Digestive Enzymes by Siricid Woodwasps from their Fungal Symbiont. Science 1983, 220, 1161–1163. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Czjzek, M.; Michel, G. Transfer of Carbohydrate-Active Enzymes from Marine Bacteria to Japanese Gut Microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef]
- Sukharnikov, L.O.; Alahuhta, M.; Brunecky, R.; Zhulin, I.B. Sequence, Structure, and Evolution of Cellulases in Glycoside Hydrolase Family 48. J. Biol. Chem. 2012, 287, 41068–41077. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Sieriebriennikov, B.; Susoy, V.; Dong, C.; Sommer, R.J. Horizontally Acquired Cellulases Assist the Expansion of Dietary Range in Pristionchus Nematodes. Mol. Biol. Evol. 2022, 2022, 39. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Jones, J.T.; Aikawa, T.; Ogura, N. A Family of Glycosyl Hydrolase Family 45 Cellulases from the Pine Wood Nematode Bursaphelenchus xylophilus. FEBS Lett. 2004, 572, 201–205. [Google Scholar] [CrossRef]
- Wang, F.; Li, D.; Wang, Z.; Wang, B.; Liu, X. Transcriptomic Analysis of the Rice White Tip Nematode, Aphelenchoides besseyi (Nematoda: Aphelenchoididae). PLoS ONE 2014, 9, E91591. [Google Scholar] [CrossRef] [PubMed]
- Boto, L. Horizontal Gene Transfer in the Acquisition of Novel Traits by Metazoans. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132450. [Google Scholar] [CrossRef]
- Brune, A. Symbiotic Digestion of Lignocellulose in Termite Guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Berlemont, R.; Martiny, A.C. Phylogenetic Distribution of Potential Cellulases in Bacteria. Appl. Environ. Microbiol. 2013, 79, 1545–1554. [Google Scholar] [CrossRef]
- Li, S.; Cao, L.; Yang, X.; Wu, X.; Liu, Y. Simultaneously Optimizing Multiple Properties of Β-Glucosidase Bgl6 using Combined (Semi-)Rational Design Strategies and Investigation of the Underlying Mechanisms. Bioresour. Technol. 2023, 374, 128792. [Google Scholar] [CrossRef]
- Lin, H.; Wang, Q.; Shen, Q.; Zhao, Y. Engineering Aspergillus oryzae A-4 Through the Chromosomal Insertion of Foreign Cellulase Expression Cassette to Improve Conversion of Cellulosic Biomass into Lipids. PLoS ONE 2023, 9, E108442. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yano, S.; Inoue, H.; Sawayama, S. Strain Improvement of Acremonium cellulolyticus for Cellulase Production by Mutation. J. Biosci. Bioeng. 2009, 107, 256–261. [Google Scholar] [CrossRef]
- Durand, H.; Clanet, M.; Tiraby, G. Genetic Improvement of Trichoderma reesei for Large Scale Cellulase Production. Enzyme Microb. Technol. 1988, 10, 341–346. [Google Scholar] [CrossRef]
- Escovar-Kousen, J.M.; Wilson, D.; Irwin, D. Integration of Computer Modeling and Initial Studies of Site-Directed Mutagenesis to Improve Cellulase Activity on Cel9A from Thermobifida fusca. Appl. Biochem. Biotechnol. 2004, 113, 287–298. [Google Scholar] [CrossRef]
- Baker, J.O.; Mccarley, J.R.; Lovett, R.; Adney, W.S.; Sakon, J.; Himmel, M.E. Catalytically Enhanced Endocellulase Cel5A from Acidothermus cellulolyticus. Appl. Biochem. Biotechnol. 2005, 121, 129–148. Available online: https://pubmed.ncbi.nlm.nih.gov/15917594 (accessed on 10 October 2025). [CrossRef]
- Mu, Y.; Ju, X.; Fu, J.; Meng, F.; Li, L. Surface Charge Engineering of Β-Glucosidase using Rational Design Improves Catalytic Capacity and Ionic Liquid Tolerance. J. Mol. Liq. 2022, 367, 120577. [Google Scholar] [CrossRef]
- Dotsenko, A.S.; Dotsenko, G.S.; Zorov, I.N.; Sinitsyn, A.P. Rational Design and Structure Insights for Thermostability Improvement of Penicillium verruculosum Cel7A Cellobiohydrolase. Biochimie 2020, 176, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhai, L.; Geng, A. Enhanced Cellulase and Reducing Sugar Production by a New Mutant Strain Trichoderma harzianum EUA20. J. Biosci. Bioeng. 2020, 129, 242–249. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current Perspective on Production and Applications of Microbial Cellulases: A Review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Harkki, A.; Mäntylä, A.; Penttilä, M.; Muttilainen, S. Genetic Engineering of Trichoderma to Produce Strains with Novel Cellulase Profiles. Enzyme Microb. Technol. 1991, 13, 227–233. [Google Scholar] [CrossRef]
- Kashima, Y.; Udaka, S. High-Level Production of Hyperthermophilic Cellulase in the Bacillus brevis Expression and Secretion System. Biosci. Biotechnol. Biochem. 2004, 68, 235–237. [Google Scholar] [CrossRef]
- Hong, J.; Tamaki, H. Cloning and Functional Expression of Thermostable Beta-Glucosidase Gene from Thermoascus aurantiacus. Appl. Microbiol. Biotechnol. 2007, 73, 1331–1339. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Wang, S.; Xing, M.; Liu, G. Achieving Efficient Protein Expression in Trichoderma Reesei by Using Strong Constitutive Promoters. Microb. Cell Fact. 2012, 11, 84. [Google Scholar] [CrossRef]
- Singh, J.; Kaur, P. Optimization of Process Parameters for Cellulase Production from Bacillus sp. JS14 in Solid Substrate Fermentation using Response Surface Methodology. Braz. Arch. Biol. Technol. 2012, 55, 505–512. [Google Scholar] [CrossRef]
- Liao, H.; Xu, C.; Tan, S.; Zhang, R.; Shen, Q.; Xu, Y. Production and Characterization of Acidophilic Xylanolytic Enzymes from Penicillium oxalicum GZ-2. Bioresour. Technol. 2012, 23, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Chysirichote, T.; Phaiboonsilpa, N.; Laosiripojana, N. High Production of Cellulase and Xylanase in Solid-State Fermentation by Trichoderma Reesei Using Spent Copra and Wheat Bran in Rotary Bioreactor. Ind. Eng. Chem. Res. 2013, 62, 3087–3097. [Google Scholar] [CrossRef]
- Cerda, A.; Mejías, L.; Gea, T.; Sánchez, A. Cellulase and Xylanase Production at Pilot Scale by Solid-State Fermentation from Coffee Husk Using Specialized Consortia: The Consistency of the Process and the Microbial Communities Involved. Bioresour. Technol. 2017, 243, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Saroj, P.; Narasimhulu, M.P.K. Assessment and Evaluation of Cellulase Production Using Ragi (Eleusine coracana) Husk as a Substrate from Thermo-Acidophilic Aspergillus fumigatus JCM 10253. Bioprocess Biosyst. Eng. 2021, 44, 113–126. [Google Scholar] [CrossRef]
- Saini, R.; Saini, J.K.; Mathur, A.; Tuli, D.; Singhania, R.R. Enhanced Cellulase Production by Penicillium oxalicum for Bio-Ethanol Application. Bioresour. Technol. 2015, 188, 240–246. [Google Scholar] [CrossRef]
- Prajapati, A.S.; Panchal, K.J.; Pawar, V.A.; Noronha, M.J.; Subramanian, R.B. Review on Cellulase and Xylanase Engineering for Biofuel Production. Ind. Biotechnol. 2018, 14, 38–44. [Google Scholar] [CrossRef]
- Vu, V.H.; Pham, T.A.; Kim, K. Fungal Strain Improvement for Cellulase Production Using Repeated and Sequential Mutagenesis. Mycobiology 2009, 37, 267. [Google Scholar] [CrossRef]
- Ranganathan, S.; Mahesh, S.; Suresh, S.; Nagarajan, A.Z.; Sen, T.; Yennamalli, R.M. Experimental and Computational Studies of Cellulases as Bioethanol Enzymes. Bioengineered 2022, 13, 14028–14046. [Google Scholar] [CrossRef]
- Arumugam Mahadevan, S.; Gon Wi, S.; Lee, D.-S.; Bae, H.-J. Site-Directed Mutagenesis and CBM Engineering of Cel5A (Thermotoga maritima). FEMS Microbiol. Lett. 2008, 287, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.R.; Liao, J.C. Engineering of an Escherichia coli Strain for the Production of 3-Methyl-1-Butanol. Appl. Environ. Microbiol. 2008, 74, 5769–5775. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Saito, H.; Nishiyama, R.; Yoshida, N.; Matsubayashi, T.; Teshima, Y.; Yamada, C.; Hiramatsu, S.; Yamada, K.; Kagawa, Y. Isolation of a Cellulase Hyperproducing Mutant Strain of Trichoderma reesei. Bioresour. Technol. Reports 2021, 15, 100733. [Google Scholar] [CrossRef]
- Adsul, M.G.; Dixit, P.; Saini, J.K.; Gupta, R.P.; Ramakumar, S.S.V.; Mathur, A.S. Morphologically Favorable Mutant of Trichoderma reesei for Low Viscosity Cellulase Production. Biotechnol. Bioeng. 2022, 119, 2167–2181. [Google Scholar] [CrossRef]
- Chand, P.; Aruna, A.; Maqsood, A.M.; Rao, L.V. Novel Mutation Method for Increased Cellulase Production. J. Appl. Microbiol. 2005, 98, 318–323. [Google Scholar] [CrossRef]
- Singh, A.; Patel, A.K.; Adsul, M.; Mathur, A.; Singhania, R.R. Genetic Modification: A Tool for Enhancing Cellulase Secretion. Biofuel Res. J. 2017, 4, 600–610. [Google Scholar] [CrossRef]
- Chica, R.A.; Doucet, N.; Pelletier, J.N. Semi-Rational Approaches to Engineering Enzyme Activity: Combining the Benefits of Directed Evolution and Rational Design. Curr. Opin. Biotechnol. 2005, 16, 378–384. [Google Scholar] [CrossRef]
- Haakana, H.; Miettinen-Oinonen, A.; Joutsjoki, V.; Mäntylä, A.; Suominen, P.; Vehmaanperä, J. Cloning of Cellulase Genes from Melanocarpus Albomyces and Their Efficient Expression in Trichoderma reesei. Enzyme Microb. Technol. 2004, 34, 159–167. [Google Scholar] [CrossRef]
- da Silva Delabona, P.; Rodrigues, G.N.; Zubieta, M.P.; Ramoni, J.; Codima, C.A.; Lima, D.J.; Farinas, C.S.; da Cruz Pradella, J.G.; Seiboth, B. The Relation between Xyr1 Overexpression in Trichoderma harzianum and Sugarcane Bagasse Saccharification Performance. J. Biotechnol. 2017, 246, 24–32. [Google Scholar] [CrossRef]
- Gilmore, S.P.; Lillington, S.P.; Haitjema, C.H.; de Groot, R.; O’Malley, M.A. Designing Chimeric Enzymes Inspired by Fungal Cellulosomes. Synth. Syst. Biotechnol. 2020, 5, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kawaguchi, T.; Sumitani, J.; Takada, G.; Izumori, K.; Arai, M. Cloning and Sequencing of an Exoglucanase Gene from Streptomyces sp. M23, and Its Expression in Streptomyces lividans TK-24. J. Biosci. Bioeng. 2005, 99, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, W.-W.; Yang, M.-M.; Chen, Y.-L. Cloning of the Thermostable Cellulase Gene from Newly Isolated Bacillus subtilis and Its Expression in Escherichia coli. Mol. Biotechnol. 2008, 40, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Seiboth, B.; Karimi, R.A.; Phatale, P.A.; Linke, R.; Hartl, L.; Sauer, D.G.; Smith, K.M.; Baker, S.E.; Freitag, M.; Kubicek, C.P. The Putative Protein Methyltransferase LAE1 Controls Cellulase Gene Expression in Trichoderma reesei. Mol. Microbiol. 2012, 84, 1150–1164. [Google Scholar] [CrossRef]
- Yao, G.; Li, Z.; Gao, L.; Wu, R.; Kan, Q.; Liu, G.; Qu, Y. Redesigning the Regulatory Pathway to Enhance Cellulase Production in Penicillium oxalicum. Biotechnol. Biofuels 2015, 8, 71. [Google Scholar] [CrossRef]
- Häkkinen, M.; Valkonen, M.J.; Westerholm-Parvinen, A.; Aro, N.; Arvas, M.; Vitikainen, M.; Penttilä, M.; Saloheimo, M.; Pakula, T.M. Screening of Candidate Regulators for Cellulase and Hemicellulase Production in Trichoderma reesei and Identification of a Factor Essential for Cellulase Production. Biotechnol. Biofuels 2014, 7, 14. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, K.; Ren, Y.; Zhang, J. The Optimization of Culture Conditions for the Cellulase Production of a Thermostable Cellulose-Degrading Bacterial Strain and Its Application in Environmental Sewage Treatment. Water 2025, 17, 2225. [Google Scholar] [CrossRef]
- Cavka, A.; Alriksson, B.; Rose, S.H. Production of cellulosic ethanol and enzyme from waste fiber sludge using SSF, recycling of hydrolytic enzymes and yeast, and recombinant cellulase-producing Aspergillus niger. J. Ind. Microbiol. Biotechnol. 2014, 41, 1191–1200. [Google Scholar] [CrossRef]
- Moran-Aguilar, M.G.; Costa-Trigo, I.; Calderón-Santoyo, M.; Domínguez, J.M.; Aguilar-Uscanga, M.G. Production of Cellulases and Xylanases in Solid-State Fermentation by Different Strains of Aspergillus niger Using Sugarcane Bagasse and Brewery Spent Grain. Biochem. Eng. J. 2021, 172, 108060. [Google Scholar] [CrossRef]
- Prasetyo, J.; Naruse, K.; Kato, T. Bioconversion of paper sludge to biofuel by simultaneous saccharification and fermentation using a cellulase of paper sludge origin and thermotolerant Saccharomyces cerevisiae TJ14. Biotechnol. Biofuels 2011, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Gao, M.; Li, Y.; Wang, Q.; Sun, X. Production of Cellulase by Aspergillus niger through Fermentation of Spent Mushroom Substance: Glucose Inhibition and Elimination Approaches. Process Biochem. 2022, 122, 26–35. [Google Scholar] [CrossRef]
- Kumar Ramamoorthy, N.; Sambavi, T.R.; Renganathan, S. A Study on Cellulase Production from a Mixture of Lignocellulosic Wastes. Process Biochem. 2019, 83, 148–158. [Google Scholar] [CrossRef]
- Kurt, A.S.; Cekmecelioglu, D. Bacterial cellulase production using grape pomace hydrolysate by shake-flask submerged fermentation. Biomass Convers. Biorefinery 2023, 13, 6981–6988. [Google Scholar] [CrossRef]
- Sirohi, R.; Singh, A.; Tarafdar, A.; Shahi, N.C.; Kushwaha, A. Cellulase Production from Pre-Treated Pea Hulls Using Trichoderma reesei Under Submerged Fermentation. Waste Biomass Valoriz. 2019, 10, 2651–2659. [Google Scholar] [CrossRef]
- Thegarathah, P.; Jewaratnam, J.; Simarani, K. Aspergillus niger as An Efficient Biological Agent for Separator Sludge Remediation: Two-Level Factorial Design for Optimal Fermentation. Peer J. 2024, 12, E17151. [Google Scholar] [CrossRef]
- Libardi, N.; Soccol, C.R.; de Carvalho, J.C.; de Souza Vandenberghe, L.P. Simultaneous Cellulase Production Using Domestic Wastewater and Bioprocess Effluent Treatment—A Biorefinery Approach. Bioresour. Technol. 2019, 276, 42–50. [Google Scholar] [CrossRef]
- Cheah, C.; Cheow, Y.L.; Ting, A.S.Y. Co-Cultivation, Metal Stress and Molasses: Strategies to Improving Exopolymeric Yield and Metal Removal Efficacy. Sustain. Environ. Res. 2022, 32, 9. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.A.; Blanch, H.W. The Challenge of Enzyme Cost in the Production of Lignocellulosic Biofuels. Biotechnol. Bioeng. 2012, 109, 1083–1087. [Google Scholar] [CrossRef]
- Kumar, B.; Bhardwaj, N.; Alam, A.; Agrawal, K.; Prasad, H.; Verma, P. Production, Purification and Characterization of an Acid/Alkali and Thermo Tolerant Cellulase from Schizophyllum commune NAIMCC-F-03379 and Its Application in Hydrolysis of Lignocellulosic Wastes. AMB Express 2018, 8, 173. [Google Scholar] [CrossRef]
- Shah, F.; Ranawat, B.; Dubey, S.; Mishra, S. Optimization of Fermentation Conditions for Higher Cellulase Production Using Marine Bacillus licheniformis KY962963: An Epiphyte of Chlorococcum sp. Biocatal. Agric. Biotechnol. 2021, 35, 102047. [Google Scholar] [CrossRef]
- Sukumaran, R.K.; Singhania, R.R.; Pandey, A. Microbial Cellulases Production, Applications and Challenges. J. Sci. Ind. Res. 2005, 64, 832–844. [Google Scholar]
- Singh, A.; Bajar, S.; Devi, A.; Pant, D. An Overview on the Recent Developments in Fungal Cellulase Production and Their Industrial Applications. Bioresour. Technol. Rep. 2021, 14, 100652. [Google Scholar] [CrossRef]
- Bhatia, R.; Singh, P.; Kumar, A.; Verma, S. Economic Analysis of Cellulase Production and Process Bottlenecks in Lignocellulosic Biorefineries. Low-Carbon Technol. J. 2023, 8, 112–125. [Google Scholar]
- Patel, A.; Sharma, K.; Lee, D.; Gupta, R. Thermodynamic and Kinetic Limitations of Cellulase-Mediated Hydrolysis of Biomass. Front. Bioeng. Biotechnol. 2024, 12, 141–152. [Google Scholar]
- An, J.; Lee, H.; Park, S.; Kim, Y. Enzyme-Assisted Processes for Biomass Valorization: A Review of Environmental Benefits. Sustainability 2023, 15, 10512. [Google Scholar] [CrossRef]
- Rani, R.; Gupta, A.; Verma, P.; Singh, R. Cellulases in Sustainable Industry: Applications, Challenges and Future Directions. Foods 2024, 13, 3846. [Google Scholar] [CrossRef]
- Ogundele, O.M.; Rapheal, O.M.; Abiodun, A.M. Effects of Municipal Waste Disposal Methods on Community Health in Ibadan-Nigeria. Polytechnica 2018, 1, 61–72. [Google Scholar] [CrossRef]
- Barlaz, M.A.; Staley, B.F.; de los Reyes, F.L. Anaerobic Biodegradation of Solid Waste. In Environmental Microbiology; Wiley: Hoboken, NJ, USA, 2009; pp. 281–299. [Google Scholar]
- Patel, A.K.; Singhania, R.R.; Albarico, F.P.J.B.; Pandey, A.; Chen, C.-W.; Dong, C.-D. Organic Wastes Bioremediation and Its Changing Prospects. Sci. Total Environ. 2022, 824, 153889. [Google Scholar] [CrossRef]
- Solórzano-Acosta, R.; Cruz-Luis, J.; Chuchon-Remon, R.; Romero-Chávez, L.E.; Lozano, A.; Gaona-Jimenez, N.; Vallejos-Torres, G. The Conversion of Forests to Agricultural Croplands Significantly Depletes Soil Organic Carbon Reserves, Total Nitrogen, and Available Potassium, Reaching Critical Thresholds in the Peruvian Amazon. Front. Soil Sci. 2025, 5, 1662180. [Google Scholar] [CrossRef]
- Sahu, A.; Manna, M.C.; Bhattacharjya, S.; Rahman, M.M.; Mandal, A.; Thakur, J.K.; Sahu, K.; Bhargav, V.K.; Singh, U.B.; Sahu, K.P.; et al. Dynamics of Maturity and Stability Indices during Decomposition of Biodegradable City Waste Using Rapo-Compost Technology. Appl. Soil Ecol. 2020, 155, 103670. [Google Scholar] [CrossRef]
- Awasthi, M.K.; Wang, Q.; Ren, X.; Zhao, J.; Huang, H.; Awasthi, S.K.; Lahori, A.H.; Li, R.; Zhou, L.; Zhang, Z. Role of Biochar Amendment in Mitigation of Nitrogen Loss and Greenhouse Gas Emission during Sewage Sludge Composting. Bioresour. Technol. 2016, 219, 270–280. [Google Scholar] [CrossRef]
- Fathallh Eida, M.; Nagaoka, T.; Wasaki, J.; Kouno, K. Isolation and Characterization of Cellulose-Decomposing Bacteria Inhabiting Sawdust and Coffee Residue Composts. Microbes Environ. 2012, 27, 226–233. [Google Scholar] [CrossRef]
- Rastogi, G.; Bhalla, A.; Adhikari, A.; Bischoff, K.M.; Hughes, S.R.; Christopher, L.P.; Sani, R.K. Characterization of Thermostable Cellulases Produced by Bacillus and Geobacillus Strains. Bioresour. Technol. 2010, 101, 8798–8806. [Google Scholar] [CrossRef]
- Sizova, M.V.; Izquierdo, J.A.; Panikov, N.S.; Lynd, L.R. Cellulose- and Xylan-Degrading Thermophilic Anaerobic Bacteria from Biocompost. Appl. Environ. Microbiol. 2011, 77, 2282–2291. [Google Scholar] [CrossRef]
- Kato, S.; Haruta, S.; Cui, Z.J.; Ishii, M.; Yokota, A.; Igarashi, Y. Clostridium Straminisolvens sp. Nov., a Moderately Thermophilic, Aerotolerant and Cellulolytic Bacterium Isolated from a Cellulose-Degrading Bacterial Community. Int. J. Syst. Evol. Microbiol. 2004, 54, 2043–2047. [Google Scholar] [CrossRef]
- Antoniou, A.; Tsolakidou, M.-D.; Stringlis, I.A.; Pantelides, I.S. Rhizosphere Microbiome Recruited from a Suppressive Compost Improves Plant Fitness and Increases Protection against Vascular Wilt Pathogens of Tomato. Front. Plant Sci. 2017, 8, 2022. [Google Scholar] [CrossRef]
- Guidi Nissim, W.; Cincinelli, A.; Martellini, T.; Alvisi, L.; Palm, E.; Mancuso, S.; Azzarello, E. Phytoremediation of Sewage Sludge Contaminated by Trace Elements and Organic Compounds. Environ. Res. 2018, 164, 356–366. [Google Scholar] [CrossRef]
- Mejáre, M.; Bülow, L. Metal-Binding Proteins and Peptides in Bioremediation and Phytoremediation of Heavy Metals. Trends Biotechnol. 2001, 19, 67–73. [Google Scholar] [CrossRef]
- Poszytek, K.; Ciężkowska, M.; Skłodowska, A.; Drewniak, Ł. Microbial Consortium with High Cellulolytic Activity (MCHCA) for Enhanced Biogas Production. Front. Microbiol. 2016, 7, 324. [Google Scholar] [CrossRef]
- Rahimnejad, M.; Adhami, A.; Darvari, S.; Zirepour, A.; Oh, S.-E. Microbial Fuel Cell as New Technology for Bioelectricity Generation: A Review. Alex. Eng. J. 2015, 54, 745–756. [Google Scholar] [CrossRef]
- Ellacuriaga, M.; Cascallana, J.G.; González, R.; Gómez, X. High-Solid Anaerobic Digestion: Reviewing Strategies for Increasing Reactor Performance. Environments 2021, 8, 80. [Google Scholar] [CrossRef]
- Ziemiński, K.; Kowalska-Wentel, M. Effect of Enzymatic Pretreatment on Anaerobic Co-Digestion of Sugar Beet Pulp Silage and Vinasse. Bioresour. Technol. 2015, 180, 274–280. [Google Scholar] [CrossRef]
- Karray, R.; Hamza, M.; Sayadi, S. Evaluation of Ultrasonic, Acid, Thermo-Alkaline and Enzymatic Pre-Treatments on Anaerobic Digestion of Ulva Rigida for Biogas Production. Bioresour. Technol. 2015, 187, 205–213. [Google Scholar] [CrossRef]
- Zhao, X.; Luo, K.; Zhang, Y.; Zheng, Z.; Cai, Y.; Wen, B.; Cui, Z.; Wang, X. Improving the Methane Yield of Maize Straw: Focus on the Effects of Pretreatment with Fungi and Their Secreted Enzymes Combined with Sodium Hydroxide. Bioresour. Technol. 2018, 250, 204–213. [Google Scholar] [CrossRef]
- Martin-Ryals, A.; Schideman, L.; Li, P.; Wilkinson, H.; Wagner, R. Improving Anaerobic Digestion of a Cellulosic Waste via Routine Bioaugmentation with Cellulolytic Microorganisms. Bioresour. Technol. 2015, 189, 62–70. [Google Scholar] [CrossRef]
- Kupski, L.; Telles, A.C.; Gonçalves, L.M.; Nora, N.S.; Furlong, E.B. Recovery of Functional Compounds from Lignocellulosic Material: An Innovative Enzymatic Approach. Food Biosci. 2018, 22, 26–31. [Google Scholar] [CrossRef]
- Weide, T.; Baquero, C.D.; Schomaker, M.; Brügging, E.; Wetter, C. Effects of Enzyme Addition on Biogas and Methane Yields in the Batch Anaerobic Digestion of Agricultural Waste (Silage, Straw, and Animal Manure). Biomass Bioenerg. 2020, 132, 105442. [Google Scholar] [CrossRef]
- Regueiro, L.; Veiga, P.; Figueroa, M.; Alonso-Gutierrez, J.; Stams, A.J.M.; Lema, J.M.; Carballa, M. Relationship between Microbial Activity and Microbial Community Structure in Six Full-Scale Anaerobic Digesters. Microbiol. Res. 2012, 167, 581–589. [Google Scholar] [CrossRef]
- Lone, M.I.; He, Z.; Stoffella, P.J.; Yang, X. Phytoremediation of Heavy Metal Polluted Soils and Water: Progresses and Perspectives. J. Zhejiang Univ. Sci. B 2008, 9, 210–220. [Google Scholar] [CrossRef]
- Khatoon, Z.; Khatoon, Z.; Orozco-Mosqueda, M.D.C.; Santoyo, G. Microbial Contributions to Heavy Metal Phytoremediation in Agricultural Soils: A Review. Microorganisms 2024, 12, 1945. [Google Scholar] [CrossRef]
- Jia, T.; Guo, T.; Chai, B. Bacterial Community Characteristics and Enzyme Activities in Imperata cylindrica Litter as Phytoremediation Progresses in a Copper Tailings Dam. Peer J. 2020, 8, e9612. [Google Scholar] [CrossRef]
- Xu, Q.-F.; Liang, C.-F.; Chen, J.-H.; Li, Y.-C.; Qin, H.; Fuhrmann, J.J. Rapid Bamboo Invasion (Expansion) and Its Effects on Biodiversity and Soil Processes. Glob. Ecol. Conserv. 2020, 21, e00787. [Google Scholar] [CrossRef]
- Wu, Z.; Kong, Z.; Lu, S.; Huang, C.; Huang, S.; He, Y.; Wu, L. Isolation, Characterization and the Effect of Indigenous Heavy Metal-Resistant Plant Growth-Promoting Bacteria on Sorghum Grown in Acid Mine Drainage Polluted Soils. J. Gen. Appl. Microbiol. 2019, 65, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Uchida, E.; Ouchi, T.; Suzuki, Y.; Yoshida, T.; Habe, H.; Yamaguchi, I.; Omori, T.; Nojiri, H. Secretion of Bacterial Xenobiotic-Degrading Enzymes from Transgenic Plants by an Apoplastic Expressional System: An Applicability for Phytoremediation. Environ. Sci. Technol. 2005, 39, 7671–7677. [Google Scholar] [CrossRef]
- Shahid, M.J.; AL-surhanee, A.A.; Kouadri, F.; Ali, S.; Nawaz, N.; Afzal, M.; Rizwan, M.; Ali, B.; Soliman, M.H. Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review. Sustainability 2020, 12, 5559. [Google Scholar] [CrossRef]
- Sillu, D.; Agnihotri, S. Cellulase Immobilization onto Magnetic Halloysite Nanotubes: Enhanced Enzyme Activity and Stability with High Cellulose Saccharification. ACS Sustain. Chem. Eng. 2020, 8, 900–913. [Google Scholar] [CrossRef]
- Yin, H.; Su, Z.; Shao, H.; Cai, J.; Wang, X.; Yin, H. Immobilization of Cellulase on Modified Mesoporous Silica Shows Improved Thermal Stability and Reusability. Afr. J. Microbiol. Res. 2013, 7, 3248–3253. [Google Scholar] [CrossRef]
- Lombardi, V.V.; Trande, M.M.; Back, M.M.; Patwardhan, S.V.; Benedetti, A.A. Facile Cellulase Immobilisation on Bioinspired Silica. Catalysts 2022, 12, 626. [Google Scholar] [CrossRef]
- Zhou, M.; Ju, X.; Zhou, Z.; Yan, L.; Chen, J.; Yao, X.; Xu, X.; Li, L.-Z. Development of an Immobilized Cellulase System Based on Metal–Organic Frameworks for Improving Ionic Liquid Tolerance and In Situ Saccharification of Bagasse. ACS Sustain. Chem. Eng. 2019, 7, 19185–19193. [Google Scholar] [CrossRef]
- Ungurean, M.; Paul, C.; Peter, F. Cellulase Immobilized by Sol-Gel Entrapment for Efficient Hydrolysis of Cellulose. Bioprocess Biosyst. Eng. 2013, 36, 1327–1338. [Google Scholar] [CrossRef]
- Bansal, M.; Sharma, R.; Kaur, G. Life Cycle Assessment of Cellulase-Based Bioethanol Production: A Step toward Low-Carbon Fuel Transition. Renew. Sustain. Energy Rev. 2023, 189, 113989. [Google Scholar] [CrossRef]
- Benavides, P.T.; Balchandani, S.; Gracida-Alvarez, U.R. Environmental analysis of biotechnologies for biofuels, bioplastics, and bioproducts: A greenhouse gas (GHG) emissions review. Biotechnol. Environ. 2024, 1, 10. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2023: Synthesis Report; Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2023; Available online: https://www.ipcc.ch/report/ar6/syr/ (accessed on 10 October 2025).
- Zhao, H.; Li, X.; Chen, W. Cellulase-Assisted Composting and Its Impact on Carbon Cycling and Soil Quality under Variable Seasonal Conditions. Bioresour. Technol. Rep. 2022, 20, 101287. [Google Scholar] [CrossRef]
- Kumar, S.; Jain, R.; Mehta, N. Mitigating Eutrophication through Cellulase-Based Wastewater Treatment: A Sustainable Bioprocess Approach. Environ. Res. 2024, 247, 118433. [Google Scholar] [CrossRef]
- Roman, H.J.; Burgess, J.E.; Pletschke, B.I. Enzyme Treatment to Decrease Solids and Improve Digestion of Primary Sewage Sludge. Afr. J. Biotechnol. 2006, 5, 963–967. [Google Scholar]
- Parmar, N.; Singh, A.; Ward, O.P. Enzyme Treatment to Reduce Solids and Improve Settling of Sewage Sludge. J. Ind. Microbiol. Biotechnol. 2001, 26, 383–386. [Google Scholar] [CrossRef]
- Siqueira, J.G.W.; Rodrigues, C.; de Souza Vandenberghe, L.P.; Woiciechowski, A.L.; Soccol, C.R. Current Advances in On-Site Cellulase Production and Application on Lignocellulosic Biomass Conversion to Biofuels: A Review. Biomass Bioenerg. 2020, 132, 105419. [Google Scholar] [CrossRef]
- Khan, R.J.; Guan, J.; Lau, C.Y.; Zhuang, H.; Rehman, S.; Leu, S. Monolignol Potential and Insights into Direct Depolymerization of Fruit and Nutshell Remains for High Value Sustainable Aromatics. Chem. Sus. Chem. 2024, 17, e202301306. [Google Scholar] [CrossRef] [PubMed]
- Taiwo, A.E.; Tom-James, A.; Musonge, P. Economic Assessment of Cellulase Production in Batch and Semi-Batch Solid-State Fermentation Processes. Int. J. Low-Carbon Technol. 2023, 18, 204–211. [Google Scholar] [CrossRef]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Chupka, G.; Strathmann, T.J.; Beckham, G.T. Lignin Valorization through Integrated Biological Funneling and Chemical Catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, V.; Singh, B. Cellulosic and Hemicellulosic Fractions of Sugarcane Bagasse: Potential, Challenges and Future Perspective. Int. J. Biol. Macromol. 2021, 169, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Leu, S.-Y.; Zhu, J.Y.; Gleisner, R.; Sessions, J.; Marrs, G. Robust Enzymatic Saccharification of a Douglas-Fir Forest Harvest Residue by SPORL. Biomass Bioenerg. 2013, 59, 393–401. [Google Scholar] [CrossRef]
- Shankar, A.; Saini, S.; Sharma, K.K. Fungal-Integrated Second-Generation Lignocellulosic Biorefinery: Utilization of Agricultural Biomass for Co-Production of Lignocellulolytic Enzymes, Mushroom, Fungal Polysaccharides, and Bioethanol. Biomass Convers. Biorefin. 2024, 14, 1117–1131. [Google Scholar] [CrossRef]
- Palazzolo, M.A.; Postemsky, P.D.; Kurina-Sanz, M. From Agro-Waste to Tool: Biotechnological Characterization and Application of Ganoderma lucidum E47 Laccase in Dye Decolorization. 3 Biotech 2019, 9, 213. [Google Scholar] [CrossRef]
- Zanuso, E.; Ruiz, H.A.; Domingues, L.; Teixeira, J.A. Magnetic Nanoparticles as Support for Cellulase Immobilization Strategy for Enzymatic Hydrolysis Using Hydrothermally Pretreated Corn Cob Biomass. BioEnergy Res. 2022, 15, 1946–1957. [Google Scholar] [CrossRef]
- Ferreira, R.D.G.; Azzoni, A.R.; Freitas, S. Techno-Economic Analysis of the Industrial Production of a Low-Cost Enzyme Using E. coli: The Case of Recombinant β-Glucosidase. Biotechnol. Biofuels 2018, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Brondi, M.G.; Elias, A.M.; Furlan, F.F.; Farinas, C.S. Performance Targets Defined by Retro-Techno-Economic Analysis for the Use of Soybean Protein as Saccharification Additive in an Integrated Biorefinery. Sci. Rep. 2020, 10, 7367. [Google Scholar] [CrossRef]
- Barta, Z.; Kovacs, K.; Reczey, K.; Zacchi, G. Process Design and Economics of On-Site Cellulase Production on Various Carbon Sources in a Softwood-Based Ethanol Plant. Enzyme Res. 2010, 2010, 1–8. [Google Scholar] [CrossRef]
- Dong, C.; Wang, Y.; Zhang, H.; Leu, S.-Y. Feasibility of High-Concentration Cellulosic Bioethanol Production from Undetoxified Whole Monterey Pine Slurry. Bioresour. Technol. 2018, 250, 102–109. [Google Scholar] [CrossRef] [PubMed]


| Organism | Origin | Acquired Enzyme | Significance | Reference |
|---|---|---|---|---|
| Macrotermes spp. | Termitomyces spp. | Exoglucanases | Both nodules and comb materials are eaten by termites. The acquired digestive enzymes play a role in the biology of fungus-feeding invertebrates. | [25] |
| Siricid wood wasps | Wood-rot fungi | Cellulase, Xylanase | Wood wasp larvae devoid of Cx-cellulases and the xylanases. The larvae acquire these enzymes while ingesting tissue of Amylostereum chailletii (fungal symbiont). | [26] |
| Resident gut bacterium | Marine bacterium | Agarase, porphyranase | Seaweeds with associated bacteria may have been the route through which these CAZymes are acquired in human gut bacteria (Bacteroides thetaiotaomicron). | [27] |
| Fungi and insect | Firmicutes, Actinobacteria | GH48-type enzymes | The enzymatic activity of GH48 proteins coded by horizontally transferred genes had been verified via experiments. | [28] |
| Pristionchus pacificus | Eukaryotic host | Cellulase | Pristionchus pacificus cellulases are embedded in a cluster of cellulases from amoeba and algae and have been reported to bring diverse resources into the ecosystem. | [29] |
| Bursaphelenchus spp. | Fungal origin | GHF45 cellulase | Nematode stylet secretes Bx-ENG-1, 2, and 3 into plant tissues. HGT made a significant contribution to the evolution of plant parasitism in nematodes. | [30] |
| Aphelenchoides besseyi | Fungal origin | GH45 cellulases | Fungi-consuming nematodes achieved the endo-1,3-β-glucanase genes from bacteria and obtained cellulase genes from fungi via HGT. | [31] |
| Urochordate Ciona intestinalis | Bacteria | Cellulose synthase gene | Ci-CesA is a fusion that consists of a cellulase domain and cellulose synthase domain, and both have no animal homologs. There is proof of likely lateral transfer of the desired gene into the urochordate lineage. | [32] |
| Lower termite & Cockroach Cryptocercus | Flagellate derived from parabasalid and oxymonadid lineages | GHs in flagellate | Before the evolution of eusociality, the vertical transmission of symbionts and metabolic interdependence between the host and flagellates existed. Digest cellulose via symbiotic relationship. | [33] |
| Sea squirts, termites, abalone | Primitive metazoan ancestor | GHF9 gene | All contain GHF9 genes with introns in identical positions, indicating that they inherited it vertically from ancient metazoan ancestors. | [34] |
| Microbial Strain | Improvement Strategies | Improved Characteristics | Reference |
|---|---|---|---|
| Gloeophyllum trabeum | Mutagenesis | Site-directed mutagenesis on loop 6 to improve the activity of cellulase (GtCel5) | [34] |
| Coniophora puteana | Mutagenesis | Site-directed mutagenesis was applied on β-glucosidase to enhance the enzyme activity of mutants CpBgl-A240S and CpBgl-Q20C by 58.5% and 65.7% | [35] |
| Aspergillus oryzae A4 | Mutagenesis | Mutagenized via Recombinant DNA technology in which four cellulase genes, such as cel A, cel B, cel C, and cel D, are inserted, it further leads to increased secretion of cellulase and enhanced lipid production. | [36] |
| Acidothermus cellulolyticus | Mutagenesis | Parental strain C-1 was treated with two mutagens (UV-irradiation and NTG), and the FPase activity (17.8 U/mL) of the mutant strain CF-2612 was also increased. At the same time, its cellulase productivity by using batch culture reached 240.3 FPU/l/h. | [37] |
| Trichoderma reesei RUT C30 | Mutagenesis | The six-step mutation caused mutant strain CL 847 formation and generated a two-fold increase in cellulase production compared to Rut C30. | [38] |
| Thermobifida fusca | Mutagenesis | Mutation of the conserved residue F476 to Y476 from Cel9A results in a 40% improvement in catalytic activity. | [39] |
| Acidothermus cellulolyticus | Mutagenesis | Substitution of Tyr245 to Gly (Y245G) in endocellulase Cel5A enzyme alleviates the product inhibition and results in a 40% increase in the release of soluble sugar. | [40] |
| β-glucosidase mutants BGL-1, BGL-14 | Rational design | Surface charge is altered under applied zeta-potential gradient to improve catalytic efficiency and rate of hydrolysis by 42% and 14% in the β-glucosidase (BGL)-14 mutant. | [41] |
| Penicillium verruculosum | Rational design | The proline substitution enhances the thermal stability of cellobiohydrolase (Cel7A), and the 3.5-fold increase in the half-life of the resulting protein (G415P) was observed at 60 °C. | [42] |
| Trichoderma harzianum | Directed evolution | Trichoderma harzianum EU2–77 mutation using UV, NTG, and ethyl methyl sulfonate improved the activity of FPase (14.79 IU/mL) | [43] |
| Trichoderma reesei QM9414 | Directed evolution | A T-DNA-tagged mutant library created by the AMT method is used to increase cellulase production in three mutants, TE-6, TA-32, and TB-87, showing 31%, 38%, and 51% increased cellulase activity compared to the parental strain. | [44] |
| Trichoderma reesei | Genetic engineering | pAMH110 vector carrying cellobiohydrolase I gene’s promoter and terminator sequences are used to enhance endoglucanase productivity by a factor of 2–4 | [45] |
| Brevibacillus brevis | Genetic engineering | Pyrococcus horikoshii’s cellulase was cloned and expressed in B. brevis, resulting in a 20-fold increase in cellulase production. | [46] |
| Pichia pastoris | Genetic engineering | T. aurantiacus’s β-glucosidase cloned and expressed in Pichia pastoris to enhance its cellobiose utilization | [47] |
| Trichoderma reesei | Genetic engineering | The Pyruvate decarboxylase (pdc) and enolase (eno) promoters of T. reesei were used to express xylanase II and its productivity was 1.52 g/L with the eno promoter and 1.61 g/L with the pdc promoter. | [48] |
| Serratia rubidaea | Fermentation | During submerged fermentation, the microbial hydrolysis of alkaline pretreated pulpy biomass leads to significant FPase (0.5 U/mL) and xylanase (11.98 U/mL) activities (pH 8, 55 °C). | [49] |
| Penicillium oxalicum GZ-2 | Fermentation | Wheat straw as an inducer enhanced β-xylosidase (89 mU/mL) and xylanase (115.2 U/mL) activities during submerged fermentation. | [50] |
| Trichoderma reesei | Fermentation | During solid-state fermentation, Trichoderma reesei has been reported with enhanced CMCase (8.66 U/g) and FPase (5.68 U/g) activity by using copra and wheat bran waste in a 30 L rotary fermentation. | [51] |
| Consortia | Fermentation | SSF of microbial consortia (Sphingobacterium composti, Barnettozyma californica, Pseudoxanthonomas taiwanensis, and Cyberlindnera jardinii) was performed in a 50 L bioreactor for improved cellulase production on coffee husk. | [52] |
| Aspergillus fumigatus | Fermentation | In SSF, the Aspergillus fumigatus and Eleusine coracana husk were used under optimized parameters of substrate concentration (1–2%), temperature (60 °C), and pH (between 2 and 4) to achieve optimal CMCase (95.2 U/mL) and β-glucosidase (0.174 U/mL) activity. | [53] |
| Penicillium oxalicum | Fermentation | In submerged fermentation, Penicillium oxalicum generated a 1.7-fold increase in cellulase production, with an optimal cellulase activity of 1.2 FPU/mL after optimizing processing conditions. | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, M.H.; Ashraf, K.; Abdullah Alqudaimi, R.E.; Martuscelli, M.; Leu, S.-Y.; Rehman, S.-u.; Aslam, M.S.; Li, Z.; Khaliq, A.; Zhuang, Y.; et al. From Waste to Wealth: Unlocking the Potential of Cellulase Characteristics for Food Processing Waste Management. Foods 2025, 14, 3639. https://doi.org/10.3390/foods14213639
Hussain MH, Ashraf K, Abdullah Alqudaimi RE, Martuscelli M, Leu S-Y, Rehman S-u, Aslam MS, Li Z, Khaliq A, Zhuang Y, et al. From Waste to Wealth: Unlocking the Potential of Cellulase Characteristics for Food Processing Waste Management. Foods. 2025; 14(21):3639. https://doi.org/10.3390/foods14213639
Chicago/Turabian StyleHussain, Muhammad Hammad, Kamran Ashraf, Redhwan Ebrahim Abdullah Alqudaimi, Maria Martuscelli, Shao-Yuan Leu, Salim-ur Rehman, Muhammad Shahbaz Aslam, Zhanao Li, Adnan Khaliq, Yingping Zhuang, and et al. 2025. "From Waste to Wealth: Unlocking the Potential of Cellulase Characteristics for Food Processing Waste Management" Foods 14, no. 21: 3639. https://doi.org/10.3390/foods14213639
APA StyleHussain, M. H., Ashraf, K., Abdullah Alqudaimi, R. E., Martuscelli, M., Leu, S.-Y., Rehman, S.-u., Aslam, M. S., Li, Z., Khaliq, A., Zhuang, Y., Guo, M., & Mohsin, A. (2025). From Waste to Wealth: Unlocking the Potential of Cellulase Characteristics for Food Processing Waste Management. Foods, 14(21), 3639. https://doi.org/10.3390/foods14213639








