Regulatory Mechanisms of Free Umami Amino Acid Accumulation in Fresh Waxy Kernels: Insights from Transcriptome and Metabolomics Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Test Materials
2.2. Determination of Free Amino Acid Contents
2.2.1. Sample Extraction
2.2.2. Liquid Chromatography Conditions
2.2.3. Mass Spectrum Reference Conditions
2.2.4. Qualitative Determination and Limit of Quantification (LOQ)
2.3. Detection of Non-Targeted Metabolites [32]
2.3.1. Reagents and Instruments
2.3.2. Dry Sample Extraction
2.3.3. HPLC Conditions
2.3.4. MS Conditions (AB)
2.3.5. Data Analysis
2.3.6. PCA and Differential Metabolites Selected
2.4. RNA Extraction, Library Preparation and Sequencing
2.5. Quality Control (QC) and Read Mapping
TPMi = FPKMi/∑FPKMj × 106
2.6. Co-Expression Correlation Analysis
2.7. bZIPs Binding Motifs Analysis
2.8. Quantitative PCR
3. Results
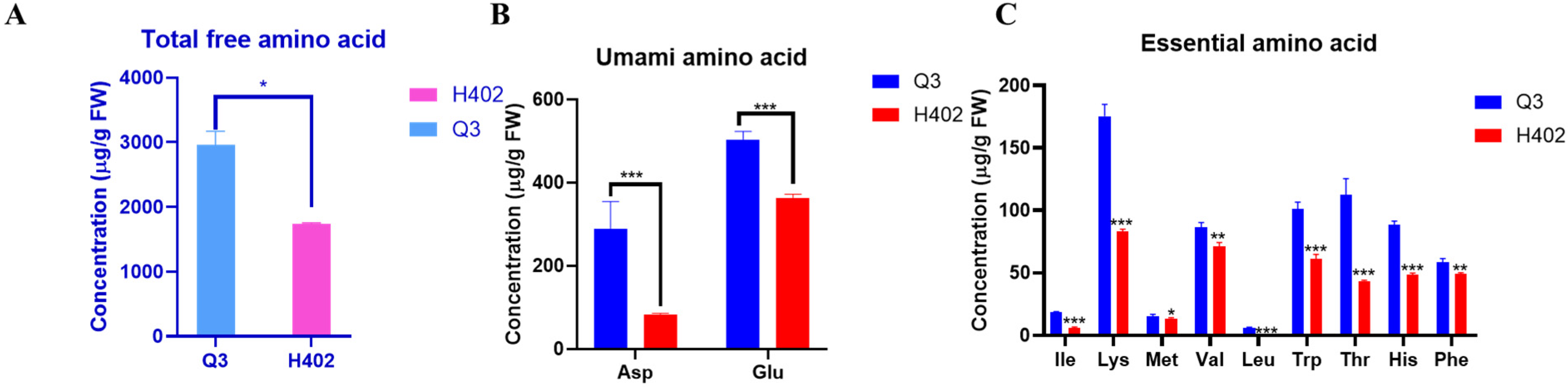
3.1. Free Amino Acid Content in Kernels of Q3 and H402 Varieties
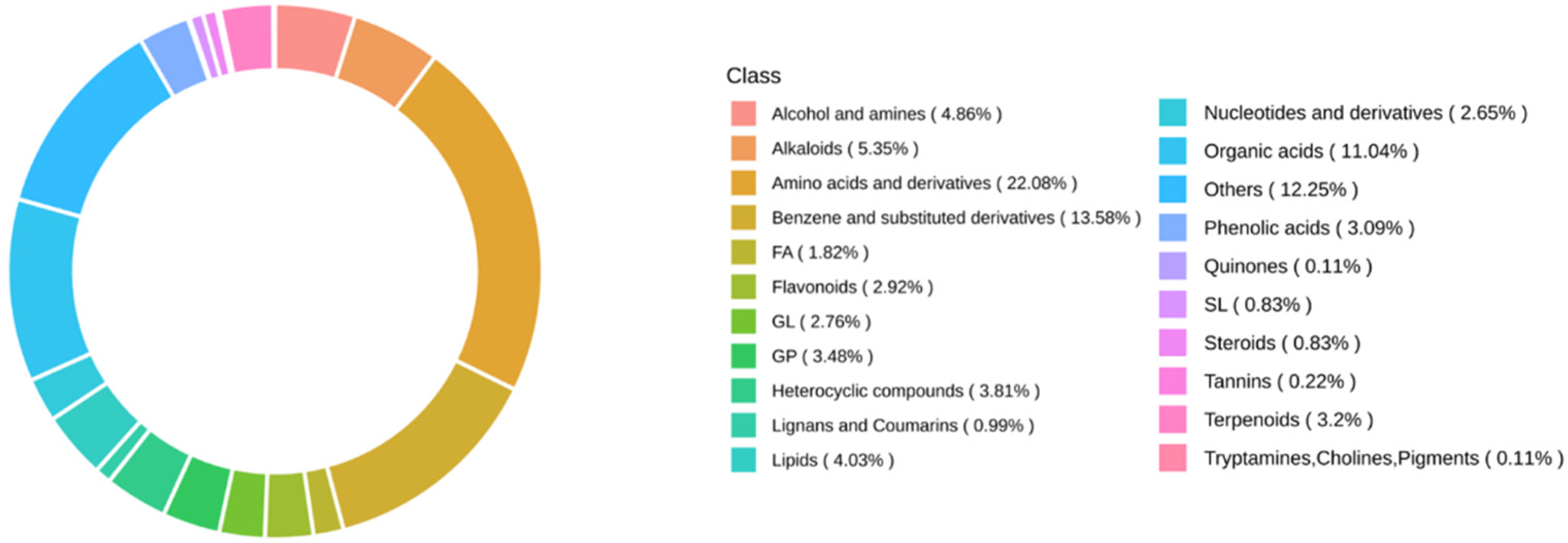
3.2. Non-Targeted Metabolomic Analysis of Amino Acids in Isolated Sweet and Waxy Kernels from Hybrid Varieties
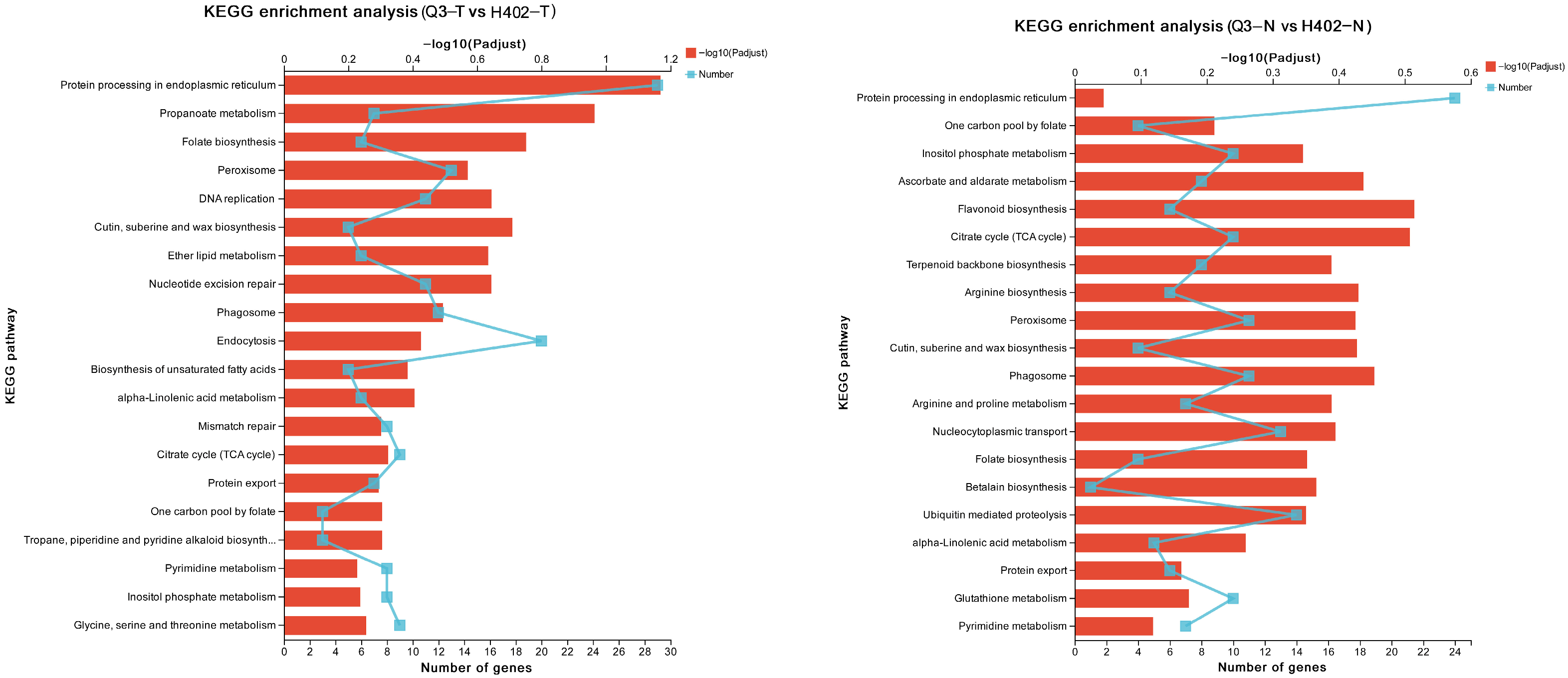
3.3. Differential Gene Expression in Sweet and Waxy Kernels
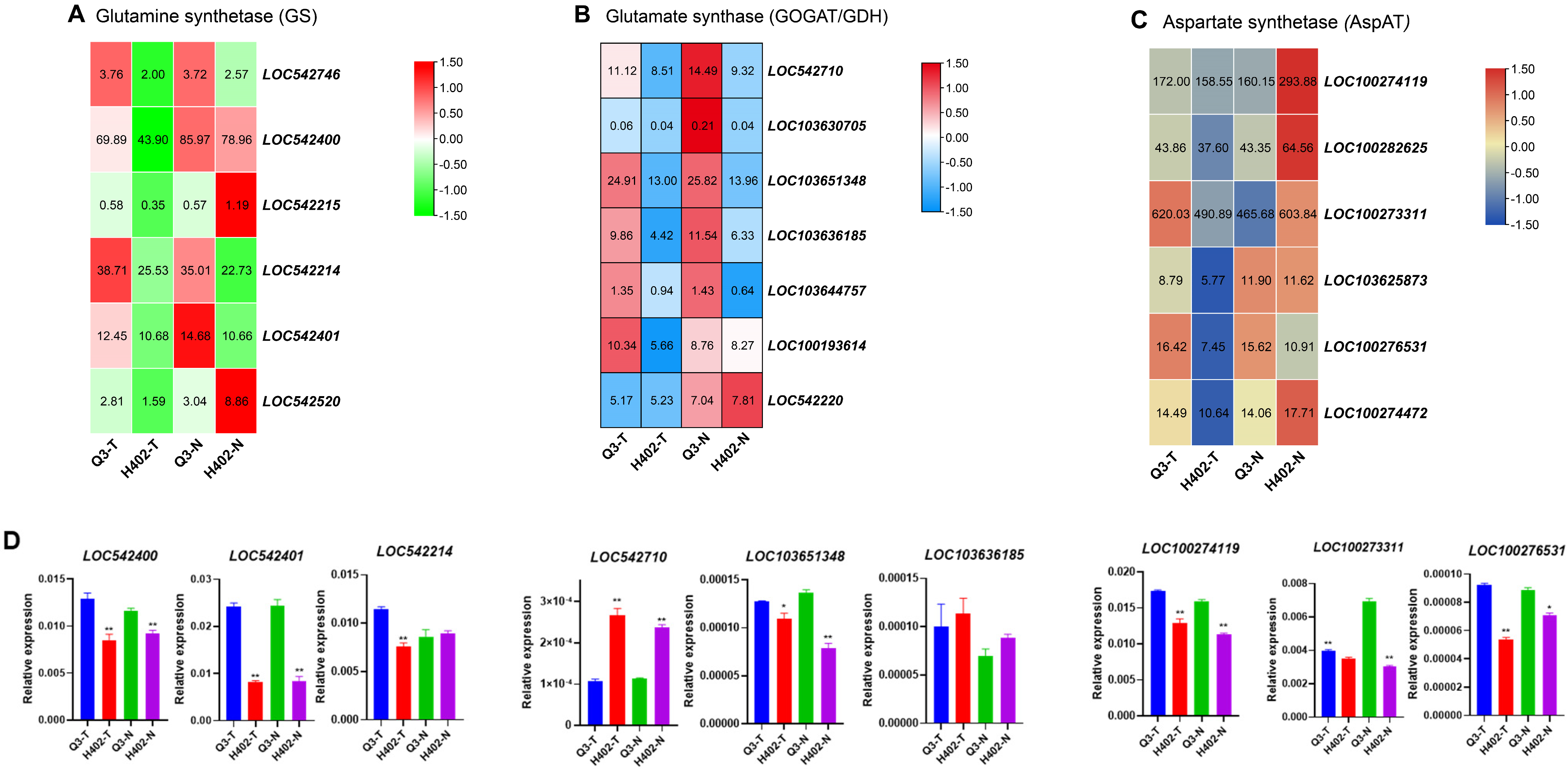
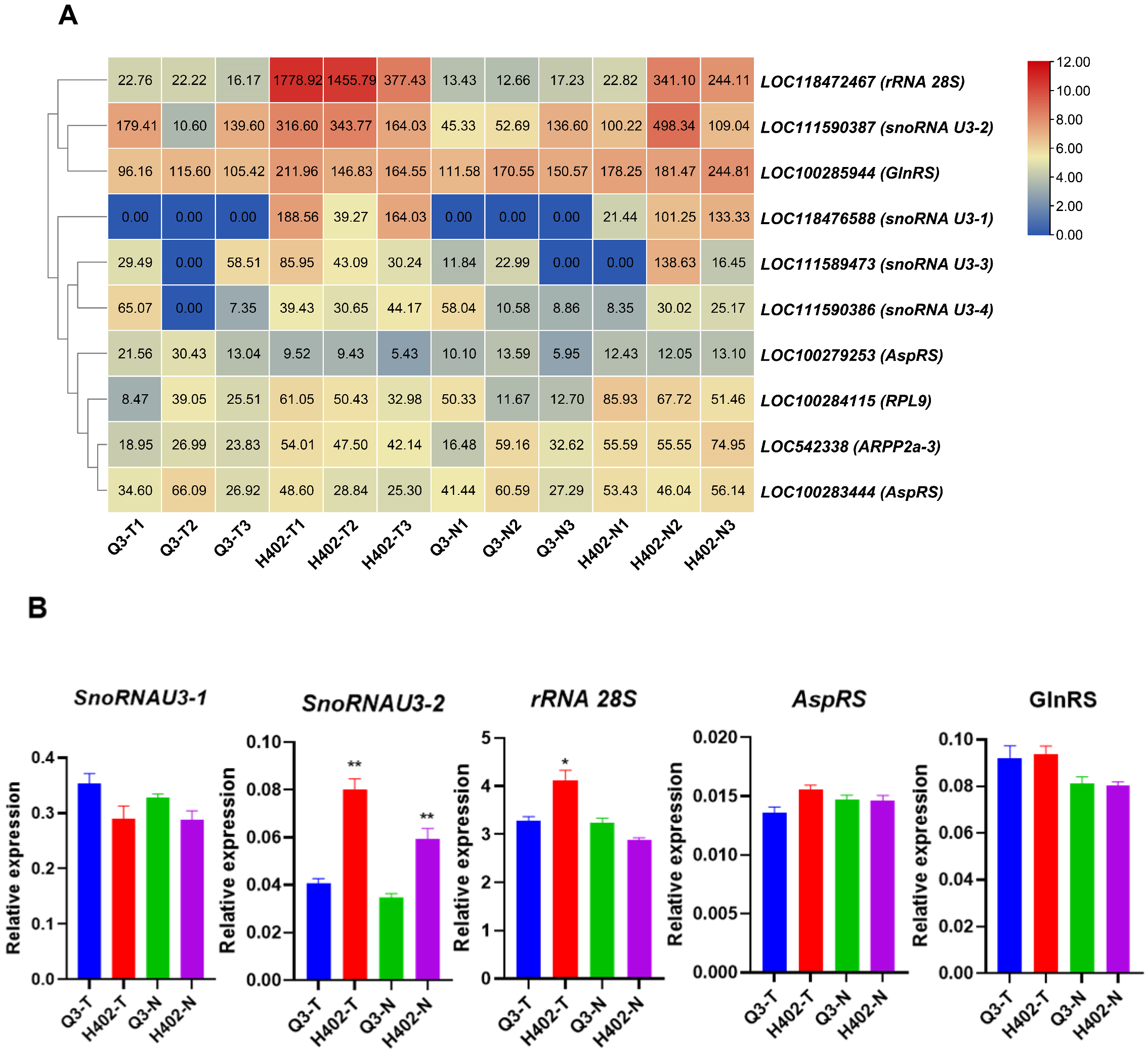
3.4. Expression Profiles of Umami Amino Acid Synthesis Genes in Sweet and Waxy Kernels
3.5. Protein Translation Related Genes in Sweet and Waxy Kernels
3.6. Transcription Factors Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabrera-Soto, L.; Pixley, K.V.; Rosales-Nolasco, A.; Galicia-Flores, L.A.; Palacios-Rojas, N. Carotenoid and Tocochromanol Profiles during Kernel Development Make Consumption of Biofortified “Fresh” Maize an Option to Improve Micronutrient Nutrition. J. Agric. Food Chem. 2018, 66, 9391–9398. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hong, T.; Shen, G.; Gu, Y.; Guo, Y.; Han, J. Amino Acid Profiles and Nutritional Evaluation of Fresh Sweet-Waxy Corn from Three Different Regions of China. Nutrients 2022, 14, 3887. [Google Scholar] [CrossRef]
- Ghirri, A.; Bignetti, E. Occurrence and role of umami molecules in foods. Int. J. Food Sci. Nutr. 2012, 63, 871–881. [Google Scholar] [CrossRef]
- Cairoli, P.; Pieraccini, S.; Sironi, M.; Morelli, C.F.; Speranza, G.; Manitto, P. Studies on umami taste. Synthesis of new guano sine 5′-phosphate derivatives and their synergistic effect with monosodium glutamate. J. Agric. Food Chem. 2008, 56, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, Y.; Zhang, Y.; Huang, J.; Liu, J.; Lin, Z.; Yu, Q.; Wu, A.; Wang, B. Widely Targeted Metabolomics Analysis Reveals Key Quality-Related Metabolites in Kernels of Sweet Corn. Int. J. Genom. 2021, 2021, 2654546. [Google Scholar] [CrossRef]
- Hou, Y.; He, W.; Hu, S.; Wu, G. Composition of polyamines and amino acids in plant-source foods for human consumption. Amino Acids 2019, 51, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Song, L.; Zhang, X.; Li, X.; Yan, N.; Xia, R.; Zhu, H.; Weng, J.; Hao, Z.; Zhang, D.; et al. Introgression of opaque2 into Waxy Maize Causes Extensive Biochemical and Proteomic Changes in Endosperm. PLoS ONE 2016, 11, e0158971. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Pan, X.; Wang, R.; Li, D.; Ren, W.; Hao, Z.; Shi, X.; Guo, J.; Ramarojaona, E.; et al. ZmCCD8 regulates sugar and amino acid accumulation in maize kernels via strigolactone signalling. Plant Biotechnol. J. 2025, 23, 492–508. [Google Scholar] [CrossRef]
- Cliquet, J.B.; Deléens, E.; Mariotti, A. C and N Mobilization from Stalk and Leaves during Kernel Filling by C and N Tracing in Zea mays L. Plant Physiol. 1990, 94, 1547–1553. [Google Scholar] [CrossRef]
- Bass, H.W.; Goode, J.H.; Greene, T.W.; Boston, R.S. Control of ribosome-inactivating protein (RIP) RNA levels during maize seed development. Plant Sci. 1994, 101, 17–30. [Google Scholar] [CrossRef]
- He, J.; Wang, J.; Zhang, Z. Toward unveiling transcriptome dynamics and regulatory modules at the maternal/filial interface of developing maize kernel. Plant J. 2024, 118, 2124–2140. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Impli cations for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef]
- Schultz, C.J.; Coruzzi, G.M. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995, 7, 61–75. [Google Scholar] [CrossRef]
- Kaachra, A.; Vats, S.K.; Kumar, S. Heterologous Expression of Key C and N Metabolic Enzymes Improves Re-assimila tion of Photorespired CO(2) and NH(3), and Growth. Plant Physiol. 2018, 177, 1396–1409. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Intine, R.V. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Expr. 2002, 10, 41–57. [Google Scholar] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Mleczko, A.M.; Bąkowska-Żywicka, K. When small RNAs become smaller: Emerging functions of snoRNAs and their derivatives. Acta Biochim. Pol. 2016, 63, 601–607. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, S.; Zhang, H.; Lee, J.S.; Ni, C.; Guo, J.; Chen, E.; Wang, S.; Acharya, A.; Chang, T.C.; et al. A non-canonical role for a small nucleolar RNA in ribosome biogenesis and senescence. Cell 2024, 187, 4770–4789.e4723. [Google Scholar] [CrossRef]
- Chen, B.; Li, L.; Huang, Y.; Ma, J.; Ji, F.; Chen, Y.; Wu, L.; Peng, H. N(6)-methyladenosine in 28S rRNA promotes oncogenic mRNA translation and tyrosine catabolism. Cell Rep. 2025, 44, 115139. [Google Scholar] [CrossRef]
- Fan, W.; Liu, H.; Stachelek, G.C.; Begum, A.; Davis, C.E.; Dorado, T.E.; Ernst, G.; Reinhold, W.C.; Ozbek, B.; Zheng, Q.; et al. Ribosomal RNA transcription governs splicing through ribosomal protein RPL22. bioRxiv 2024. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, J.; Zhang, Z.; Wu, Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020, 18, 1897–1907. [Google Scholar] [CrossRef]
- Gontarek, B.C.; Neelakandan, A.K.; Wu, H.; Becraft, P.W. NKD Transcription Factors Are Central Regulators of Maize Endosperm Development. Plant Cell 2016, 28, 2916–2936. [Google Scholar] [CrossRef]
- Huang, S.; Frizzi, A.; Florida, C.A.; Kruger, D.E.; Luethy, M.H. High lysine and high tryptophan transgenic maize resulting from the reduction of both 19- and 22-kD alpha-zeins. Plant Mol. Biol. 2006, 61, 525–535. [Google Scholar] [CrossRef]
- Zhan, J.; Li, G.; Ryu, C.H.; Ma, C.; Zhang, S.; Lloyd, A.; Hunter, B.G.; Larkins, B.A.; Drews, G.N.; Wang, X.; et al. Opaque-2 Regulates a Complex Gene Network Associated with Cell Differentiation and Storage Functions of Maize Endosperm. Plant Cell 2018, 30, 2425–2446. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Galli, M.; Spears, C.J.; Zhan, J.; Liu, P.; Yadegari, R.; Dannenhoffer, J.M.; Gallavotti, A.; Becraft, P.W. NAKED ENDOSPERM1, NAKED ENDOSPERM2, and OPAQUE2 interact to regulate gene networks in maize endosperm development. Plant Cell 2023, 36, 19–39. [Google Scholar] [CrossRef]
- Yang, T.; Guo, L.; Ji, C.; Wang, H.; Wang, J.; Zheng, X.; Xiao, Q.; Wu, Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 2021, 33, 104–128. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yue, Y.; Chen, H.; Qi, W.; Song, R. The ZmbZIP22 Transcription Factor Regulates 27-kD γ-Zein Gene Transcription during Maize Endosperm Development. Plant Cell 2018, 30, 2402–2424. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Yu, H.; He, J.; Peng, D.; Zhu, P.; Pan, S.; Wu, X.; Wang, J.; Ji, C.; Chao, Z.; et al. The transcription factors ZmNAC128 and ZmNAC130 coordinate with Opaque2 to promote endosperm filling in maize. Plant Cell 2023, 35, 4066–4090. [Google Scholar] [CrossRef]
- Ning, L.; Wang, Y.; Shi, X.; Zhou, L.; Ge, M.; Liang, S.; Wu, Y.; Zhang, T.; Zhao, H. Nitrogen-dependent binding of the transcription factor PBF1 contributes to the balance of protein and carbohydrate storage in maize endosperm. Plant Cell 2023, 35, 409–434. [Google Scholar] [CrossRef]
- Tian, R.; Yang, Z.; Yang, R.; Wang, S.; Shen, Q.; Wang, G.; Wang, H.; Zhou, Q.; Tang, J.; Fu, Z. Regulation of maize kernel development via divergent activation of α-Zein genes by transcription factors O11, O2, and PBF1. J. Genet. Genom. 2025. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
- Maoz, I.; Lewinsohn, E.; Gonda, I. Amino acids metabolism as a source for aroma volatiles biosynthesis. Curr. Opin. Plant Biol. 2022, 67, 102221. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Duan, J.; Liu, X.; Zhang, M.; Bao, X. Preparation of sunflower seed-derived umami protein hydrolysates and their synergistic effect with monosodium glutamate and disodium inosine-5′-monophosphate. J. Food Sci. 2023, 88, 3332–3340. [Google Scholar] [CrossRef]
- Pasini, E.; Corsetti, G.; Aquilani, R.; Romano, C.; Picca, A.; Calvani, R.; Dioguardi, F.S. Protein-Amino Acid Metabolism Disarrangements: The Hidden Enemy of Chronic Age-Related Conditions. Nutrients 2018, 10, 391. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Dietary protein intake and human health. Food Funct. 2016, 7, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Zhang, B.; Duan, W.; Zhang, J.; Wang, M. Nutrients and bioactive components from vinegar: A fermented and functional food. J. Funct. Foods 2020, 64, 103681. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, L.; He, Z. Understanding the regulation of cereal grain filling: The way forward. J. Integr. Plant Biol. 2023, 65, 526–547. [Google Scholar] [CrossRef]
- Doll, N.M.; Depège-Fargeix, N.; Rogowsky, P.M.; Widiez, T. Signaling in Early Maize Kernel Development. Mol. Plant 2017, 10, 375–388. [Google Scholar] [CrossRef]
- Larkins, B.A. Chapter 12—Proteins of the Kernel. In Corn, 3rd ed.; Serna-Saldivar, S.O., Ed.; AACC International Press: Oxford, UK, 2019; pp. 319–336. [Google Scholar]
- Monné, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial Carriers for Aspartate, Glutamate and Other Amino Acids: A Review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef]
- Zhang, W.; Ni, K.; Long, L.; Ruan, J. Nitrogen transport and assimilation in tea plant (Camellia sinensis): A review. Front. Plant Sci. 2023, 14, 1249202. [Google Scholar] [CrossRef]
- Wang, J.Y.; Rao, Z.M.; Xu, J.A.-O.; Zhang, W.G. Enhancing β-alanine production from glucose in genetically modified Corynebacterium glutamicum by metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2021, 105, 9153–9166. [Google Scholar] [CrossRef]
- Mair, A.; Pedrotti, L.; Wurzinger, B.; Anrather, D.; Simeunovic, A.; Weiste, C.; Valerio, C.; Dietrich, K.; Kirchler, T.; Nägele, T.; et al. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLife 2015, 4, e05828. [Google Scholar] [CrossRef][Green Version]
- Garg, A.; Kirchler, T.; Fillinger, S.; Wanke, F.; Stadelhofer, B.; Stahl, M.; Chaban, C. Targeted manipulation of bZIP53 DNA-binding properties influences Arabidopsis metabolism and growth. J. Exp. Bot. 2019, 70, 5659–5671. [Google Scholar] [CrossRef]
- Lorenzo, O. bZIP edgetic mutations: At the frontier of plant metabolism, development and stress trade-off. J. Exp. Bot. 2019, 70, 5517–5520. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Hanssen, M.; Wiese, A.; Hendriks, M.M.; Smeekens, S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008, 53, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.A.-O.; Zhang, J.; Su, L.; Jia, M.; Tian, Y.; Pan, H.A.-O.; Zhang, X.A.-O.X. ChCpc1, a bZIP transcription factor, coordinates amino acid synthesis and autophagy and modulates conidiation and virulence in Cochliobolus heterostrophus. mBio 2025, 13, e0084525. [Google Scholar] [CrossRef] [PubMed]




| Time (min) | A (%) | B (%) |
|---|---|---|
| 0 | 95 | 5 |
| 5 | 35 | 65 |
| 6 | 1 | 99 |
| 7.5 | 1 | 99 |
| 7.6 | 95 | 5 |
| 10 | 95 | 5 |
| Number | Parent Ion | Daughter Ion | Dwell Time (ms) | Amino Acids | DP | CE |
|---|---|---|---|---|---|---|
| 1 | 132.1 | 86.4 | 50 | Ile-1 | 40 | 14 |
| 2 | 132.1 | 103.8 | 50 | Ile-2 | 40 | 11 |
| 3 | 133.9 | 74.1 | 50 | Asp-1 | 30 | 19 |
| 4 | 133.9 | 88.0 | 50 | Asp-2 | 40 | 13 |
| 5 | 147.1 | 84.3 | 50 | Lys-1 | 30 | 22 |
| 6 | 147.1 | 130.2 | 50 | Lys-2 | 17 | 14 |
| 7 | 106.2 | 60.2 | 50 | Ser-1 | 23 | 16 |
| 8 | 106.2 | 88.2 | 50 | Ser-2 | 6 | 13 |
| 9 | 150.1 | 132.9 | 50 | Met-1 | 33 | 13 |
| 10 | 150.1 | 104.3 | 50 | Met-2 | 34 | 15 |
| 11 | 147.1 | 130.1 | 50 | Gln-1 | 29 | 14 |
| 12 | 147.1 | 84.1 | 50 | Gln-2 | 24 | 23 |
| 13 | 118.1 | 72.1 | 50 | Val-1 | 23 | 15 |
| 14 | 118.1 | 89.7 | 50 | Val-2 | 23 | 9 |
| 15 | 116.2 | 70.1 | 50 | Pro-1 | 34 | 21 |
| 16 | 116.2 | 59.9 | 50 | Pro-2 | 34 | 15 |
| 17 | 90.8 | 65.0 | 50 | Ala-1 | 234 | 26 |
| 18 | 90.8 | 72.9 | 50 | Ala-2 | 94 | 12 |
| 19 | 148.2 | 84.2 | 50 | Glu-1 | 15 | 20 |
| 20 | 148.2 | 102.1 | 50 | Glu-2 | 20 | 14 |
| 21 | 132.1 | 85.9 | 50 | Leu-1 | 19 | 14 |
| 22 | 132.1 | 104.1 | 50 | Leu-2 | 53 | 10 |
| 23 | 76.0 | 30.0 | 50 | Gly-1 | 40 | 14 |
| 24 | 76.0 | 46.0 | 50 | Gly-2 | 40 | 17 |
| 25 | 175.2 | 70.1 | 50 | Arg-1 | 32 | 26 |
| 26 | 175.2 | 116.2 | 50 | Arg-2 | 50 | 19 |
| 27 | 205.1 | 188.3 | 50 | Trp-1 | 16 | 15 |
| 28 | 205.1 | 86.7 | 50 | Trp-2 | 20 | 14 |
| 29 | 182.1 | 165.1 | 50 | Tyr-1 | 35 | 14 |
| 30 | 182.1 | 136.2 | 50 | Tyr-2 | 20 | 19 |
| 31 | 120.1 | 102.1 | 50 | Thr-1 | 35 | 12 |
| 32 | 120.1 | 74.0 | 50 | Thr-2 | 32 | 15 |
| 33 | 156.0 | 110.1 | 50 | His-1 | 41 | 20 |
| 34 | 156.0 | 95.2 | 50 | His-2 | 25 | 21 |
| 35 | 166.1 | 120.1 | 50 | Phe-1 | 44 | 18 |
| 36 | 166.1 | 148.9 | 50 | Phe-2 | 41 | 13 |
| 37 | 133.1 | 87.2 | 50 | Asn-1 | 38 | 14 |
| 38 | 133.1 | 116.0 | 50 | Asn-2 | 49 | 14 |
| 39 | 241.1 | 152.0 | 50 | Cys-1 | 33 | 17 |
| 40 | 241.1 | 120.1 | 50 | Cys-2 | 33 | 24 |
| Relative ion abundance/% | >50 | >20–50 | >20–50 | ≤10 |
| Allowable relative deviation/% | ±20 | ±25 | ±30 | ±50 |
| Amino Acid | CAS No. | Molecular Weight | LOQ (μg/kg) | |
|---|---|---|---|---|
| Alanine | Ala | 56-41-7 | 89.09 | 25 |
| Arginine | Arg | 1119-34-2 | 174.2 | 12.5 |
| Asparagine | Asn | 70-47-3 | 132.12 | 50 |
| Aspartic acid | Asp | 56-84-8 | 133.1 | 50 |
| Cystine | Cys | 56-89-3 | 240.3 | 50 |
| Glutamine | Gln | 56-85-9 | 146.15 | 25 |
| Glutamic acid | Glu | 56-86-0 | 147.13 | 25 |
| Glycine | Gly | 56-40-6 | 75.07 | 50 |
| Histidine | His | 5934-29-2 | 155.16 | 12.5 |
| Isoleucine | Ile | 73-32-5 | 131.18 | 12.5 |
| Leucine | Leu | 61-90-5 | 131.18 | 12.5 |
| Lysine | Lys | 657-27-2 | 146.19 | 25 |
| Methionine | Met | 63-68-3 | 149.2 | 12.5 |
| Phenylalanine | Phe | 63-91-2 | 165.19 | 12.5 |
| Proline | Pro | 147-85-3 | 115.13 | 12.5 |
| Serine | Ser | 56-45-1 | 105.09 | 25 |
| Threonine | Thr | 72-19-5 | 119.13 | 25 |
| Tyrosine | Tyr | 60-18-4 | 181.19 | 25 |
| Valine | Val | 72-18-4 | 117.15 | 12.5 |
| Tryptophan | Trp | 73-22-3 | 204.23 | 25 |
| Time (min) | A (%) | B (%) |
|---|---|---|
| 0 | 95 | 5 |
| 5 | 35 | 65 |
| 6 | 1 | 99 |
| 7.5 | 1 | 99 |
| 7.6 | 95 | 5 |
| 10 | 95 | 5 |
| ESI+ | ESI− | |
|---|---|---|
| Duration (min) | 10 | 10 |
| IonSpray Voltage (V) | 5000 | −4000 |
| Temperature (°C) | 550 | 550 |
| Ion Source Gas1 (psi) | 50 | 50 |
| Ion Source Gas2 (psi) | 60 | 60 |
| Curtain Gas (psi) | 35 | 35 |
| De-clustering Potential (V) | 60 | −60 |
| MS1 Collision Energy (V) | 10 | −10 |
| MS2 Collision Energy (V) | 30 | −30 |
| Collision Energy Spread (V) | 15 | 15 |
| MS1 TOF Masses (Da) | 50~1250 | 50~1250 |
| MS2 TOF Masses (Da) | 25~1250 | 25~1250 |
| Genes IDs | Binding Motifs Numbers (bZIPs) | Gene Annotations |
|---|---|---|
| LOC542214/Zm00001eb253820 | 25 | glutamine synthetase 4(gln4) |
| LOC542401/Zm00001eb190340 | 32 | glutamine synthetase 5 (gln5) |
| LOC103651348/Zm00001eb156610 | 19 | glutamate synthase2/glutamine oxoglutarate aminotransferase2 (GS2/gogat2) |
| LOC542710/Zm00001eb329710 | 26 | ferredoxin-dependent glutamate synthase (Fd-gogat) |
| LOC100274119/Zm00001eb238900 | 20 | aspartate aminotransferase (AspAT) |
| LOC100273311/Zm00001eb152450 | 17 | glutamate-oxaloacetate transaminase 1 (got1) |
| LOC100276531/Zm00001eb146400 | 17 | glutamate-oxaloacetate transaminase 4 (gogat4) |
| LOC111590387 | 12 | small nucleolar RNA U3-2 |
| LOC11847246 | 23 | 28S ribosomal RNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Huang, K.; Luo, L.; Yu, X.; Shen, N.; Wu, Y.; Wu, J.; Shi, J.; Yue, E. Regulatory Mechanisms of Free Umami Amino Acid Accumulation in Fresh Waxy Kernels: Insights from Transcriptome and Metabolomics Analyses. Foods 2025, 14, 3628. https://doi.org/10.3390/foods14213628
Zhao L, Huang K, Luo L, Yu X, Shen N, Wu Y, Wu J, Shi J, Yue E. Regulatory Mechanisms of Free Umami Amino Acid Accumulation in Fresh Waxy Kernels: Insights from Transcriptome and Metabolomics Analyses. Foods. 2025; 14(21):3628. https://doi.org/10.3390/foods14213628
Chicago/Turabian StyleZhao, Lin, Kaimei Huang, Letan Luo, Xiangqun Yu, Ning Shen, Yifan Wu, Jianguo Wu, Jiang Shi, and Erkui Yue. 2025. "Regulatory Mechanisms of Free Umami Amino Acid Accumulation in Fresh Waxy Kernels: Insights from Transcriptome and Metabolomics Analyses" Foods 14, no. 21: 3628. https://doi.org/10.3390/foods14213628
APA StyleZhao, L., Huang, K., Luo, L., Yu, X., Shen, N., Wu, Y., Wu, J., Shi, J., & Yue, E. (2025). Regulatory Mechanisms of Free Umami Amino Acid Accumulation in Fresh Waxy Kernels: Insights from Transcriptome and Metabolomics Analyses. Foods, 14(21), 3628. https://doi.org/10.3390/foods14213628







