From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Identification of Isolates
2.2. PFGE
2.3. Reidentification of Listeria monocytogenes Isolates
2.4. WGS
2.5. Genomic Identification, Serotype, ST, and cgMLST of Listeria monocytogenes
2.6. Detection of Virulence and Antibiotic Resistance Genes
2.7. Plasmids and Mobile Genetic Element Detection
2.8. Bioinformatic Search for CRISPR/Cas Systems
3. Results
3.1. Isolation and Primary Species Identification of Listeria monocytogenes
3.2. PFGE
3.3. Genomic Identification, Serotype, ST, and cgMLST of Listeria monocytogenes
3.4. Detection of Virulence and Antibiotic Resistance Genes
3.5. Plasmids and Mobile Genetic Element Detection
3.6. Bioinformatic Search for CRISPR/Cas Systems
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monteiro, C. The big issue is ultra-processing. [Commentary]. World Nutr. 2010, 1, 237–269. [Google Scholar]
- Thienhirun, S.; Chung, S. Consumer Attitudes and Preferences Toward Cross-Cultural Ready-To-Eat (RTE) Food. J. Food Prod. Mark. 2018, 24, 56–79. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.; Roy, S.; Griffin, S. Foodborne Illness Acquired in the United States-Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Halbedel, S.; Wilking, H.; Holzer, A.; Kleta, S.; Fischer, M.A.; Lüth, S.; Pietzka, A.; Huhulescu, S.; Lachmann, R.; Krings, A.; et al. Large Nationwide Outbreak of Invasive Listeriosis Associated with Blood Sausage, Germany, 2018–2019. Emerg. Infect. Dis. 2020, 26, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Listeria Monocytogenes in Ready-to-Eat (RTE) Foods: Attribution, Characterization and Monitoring—Meeting Report; Microbiological Risk Assessment Series No. 38; Food and Agriculture Organization of the United Nations: Rome, Italy, 2022. [Google Scholar]
- Paduro, C.; Montero, D.A.; Chamorro, N.; Carreño, L.J.; Vidal, M.; Vidal, R. Ten years of molecular epidemiology surveillance of Listeria monocytogenes in Chile 2008–2017. Food Microbiol. 2020, 85, 103280. [Google Scholar] [CrossRef]
- Koopmans, M.M.; Brouwer, M.C.; Vázquez-Boland, J.A.; van de Beek, D. Human Listeriosis. Clin. Microbiol. Rev. 2023, 36, e0006019. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.; Hayman, M.; Jackson, T.; Whiting, R.C. A Review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Amajoud, N.; Leclercq, A.; Soriano, J.; Bracq-Dieye, H.; El Maadoudi, M.; Skalli, N.; Kounnoun, A.; Moura, A.; Lecuit, M.; Abrini, J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control 2018, 84, 436–441. [Google Scholar] [CrossRef]
- Gorski, L. Serotype assignment by sero-agglutination, ELISA, and PCR. Methods Mol. Biol. 2021, 2220, 57–78. [Google Scholar]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: A Comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Lopes-Luz, L.; Mendonça, M.; Bernardes Fogaça, M.; Kipnis, A.; Bhunia, A.K.; Bührer-Sékula, S. Listeria monocytogenes: Review of pathogenesis and virulence determinants-targeted immunological assays. Crit. Rev. Microbiol. 2021, 47, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Allam, M.; Tau, N.; Smouse, S.L.; Mtshali, P.S.; Mnyameni, F.; Khumalo, Z.T.; Ismail, A.; Govender, N.; Thomas, J.; Smith, A.M. Whole-genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc. 2018, 6, e00538-18. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Neoh, H.M.; Tan, X.E.; Sapri, H.F.; Tan, T.L. Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 2019, 74, 103935. [Google Scholar] [CrossRef]
- Barret, A.S.; Charron, M.; Mariani-Kurkdjian, P.; Gouali, M.; Loukiadis, E.; Poignet-Leroux, B.; Godron, A.; Gault, G.; Faure, M.; Mailles, A.; et al. Shopper cards data and storage practices for the investigation of an outbreak of Shiga-toxin producing Escherichia coli O157 infections. Med. Mal. Infect. 2013, 43, 368–373. [Google Scholar] [CrossRef]
- Lakicevic, B.; Jankovic, V.; Pietzka, A.; Ruppitsch, W. Wholegenome sequencing as the gold standard approach for control of Listeria monocytogenes in the food chain. J. Food Prot. 2023, 86, 100003. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Ortuño-Gutiérrez, N.; Sunyoto, T.; Gomes-Dias, C.A.; Bastidas-Caldes, C.; Ramírez, M.S.; Harries, A.D. Whole-genome sequencing for surveillance of antimicrobial resistance in Ecuador: Present and future implications. Rev. Panam. Salud Publica 2023, 47, e8. [Google Scholar] [CrossRef]
- Leopold, S.; Goering, R.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef]
- Cabal, A.; Prieto, B.; Raicevic, N.; Pietzka, A.; Chakeri, A.; Hyden, P.; Kundi, M.; Indra, A.; Mach, R.; Parra-Flores, J.; et al. Whole Genome Sequencing for Food Safety, Clinical and Public Health Microbiology. In MEDICON’23 and CMBEBIH’23; Badnjević, A., Gurbeta Pokvić, L., Eds.; IFMBE Proceedings Springer: Cham, Switzerland, 2023; Volume 93, pp. 865–873. [Google Scholar]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef]
- Kwon, H.J.; Chen, Z.; Evans, P.; Meng, J.; Chen, Y. Characterization of mobile genetic elements using long-read sequencing for tracking Listeria monocytogenes from food processing environments. Pathogens 2020, 9, 822. [Google Scholar] [CrossRef]
- Stessl, B.; Wagner, M.; Ruppitsch, W. Multilocus sequence typing (MLST) and whole genome sequencing (WGS) of Listeria monocytogenes and Listeria innocua. Methods Mol. Biol. 2021, 2220, 89–103. [Google Scholar] [PubMed]
- Cordero, N.; Maza, F.; Navea-Perez, H.; Aravena, A.; Marquez-Fontt, B.; Navarrete, P.; Figueroa, G.; González, M.; Latorre, M.; Reyes-Jara, A. Different transcriptional responses from slow and fast growth rate strains of Listeria monocytogenes adapted to low temperature. Front. Microbiol. 2016, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haesler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee:In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Lee, S.; Ward, T.J.; Graves, L.M.; Wolf, L.A.; Sperry, K.; Siletzky, R.M.; Kathariou, S. Atypical Listeria monocytogenes serotype 4b strains harboring a lineage II-specific gene cassette. Appl. Environ. Microbiol. 2012, 78, 660–667. [Google Scholar] [CrossRef]
- Salcedo, C.; Arreaza, L.; Alcalá, B.; de la Fuente, L.; Vázquez, J.A. Development of a multilocus sequence typing method for the analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 2003, 41, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.; Leclerq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.M.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.L.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 Years on. Nucleic Acids Res. 2016, 4, D694–D697. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Johansson, M.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Biswas, A.; Staals, R.; Morales, S.; Fineran, P.; Brown, C. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.C.; Naïtali, M.; Briandet, R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- Chowdhury, B.; Anand, S. Environmental persistence of Listeria monocytogenes and its implications in dairy processing plants. Compr. Rev. Food Sci. Food Saf. 2023, 22, 4573–4599. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.; NicAogáin, K.; Luque-Sastre, L.; McManamon, O.; Hunt, K.; Alvarez-Ordóñez, A.; Scollard, J.; Schmalenberger, A.; Fanning, S.; O’Byrne, C.; et al. A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 2017, 249, 18–26. [Google Scholar] [CrossRef]
- Ronholm, J. Editorial: Game Changer—Next Generation Sequencing and Its Impact on Food Microbiology. Front. Microbiol. 2018, 9, 363. [Google Scholar] [CrossRef]
- Toledo, V.; den Bakker, H.C.; Hormazábal, J.C.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A.I. Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef]
- Willis, C.; McLauchlin, J.; Aird, H.; Amar, C.; Barker, C.; Dallman, T.; Elviss, N.; Lai, S.; Sadler-Reeves, L. Occurrence of Listeria and Escherichia coli in frozen fruit and vegetables collected from retail and catering premises in England 2018–2019. Int. J. Food Microbiol. 2020, 334, 108849. [Google Scholar] [CrossRef]
- Daza-Prieto, B.; Pietzka, A.; Martinovic, A.; Ruppitsch, W.; Zuber Bogdanovic, I. Surveillance and genetic characterization of Listeria monocytogenes in the food chain in Montenegro during the period 2014–2022. Front. Microbiol. 2024, 15, 1418333. [Google Scholar] [CrossRef]
- Pyz-Łukasik, R.; Paszkiewicz, W.; Kiełbus, M.; Ziomek, M.; Gondek, M.; Domaradzki, P.; Michalak, K.; Pietras-Ożga, D. Genetic diversity and potential virulence of Listeria monocytogenes isolates originating from Polish artisanal cheeses. Foods 2022, 11, 2805. [Google Scholar] [CrossRef]
- Tricoli, M.R.; Massaro, C.; Arrigo, I.; Diquattro, O.; Di Bernardo, F.; Galia, E.; Palermo, M.; Fasciana, T.; Giammanco, A. Characterization of Listeria monocytogenes strains isolated in Palermo (Sicily and Italy) during the years 2018–2020 from severe cases of Listeriosis. Antibiotics 2024, 13, 57. [Google Scholar] [CrossRef]
- Pietzka, A.; Allerberger, F.; Murer, A.; Lennkh, A.; Stöger, A.; Cabal-Rosel, A.; Huhulescu, S.; Maritschnik, S.; Springer, B.; Lepuschitz, S.; et al. Whole genome sequencing based surveillance of L. monocytogenes for early detection and investigations of Listeriosis outbreaks. Front. Public Health 2019, 7, 139. [Google Scholar] [CrossRef]
- Halbedel, S.; Sperle, I.; Lachmann, R.; Kleta, S.; Fischer, M.A.; Wamp, S.; Holzer, A.; Lüth, S.; Murr, L.; Freitag, C.; et al. Large multicountry outbreak of invasive Listeriosis by a Listeria monocytogenes ST394 clone linked to smoked rainbow trout, 2020 to 2021. Microbiol. Spectr. 2023, 11, e03520-22. [Google Scholar] [CrossRef]
- Annabel, L.N.; Monika, D.; Martin, W.; Schmitz-Esser, S. Plasmids contribute to food processing environment-associated stress survival in three Listeria monocytogenes ST121, ST8, and ST5 strains. Int. J. Food Microbiol. 2019, 299, 39–46. [Google Scholar]
- Zhang, H.Z.; Wang, J.; Chang, Z.Y.; Liu, X.; Chen, W.; Yu, Y.; Wang, X.; Dong, Q.; Ye, Y.; Zhang, X. Listeria monocytogenes contamination characterization in two ready-to-eat meat plants from 2019 to 2020 in Shanghai. Front. Microbiol. 2021, 12, 729114. [Google Scholar]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and antibiotic resistance genes in Listeria monocytogenes strains isolated from ready-to-eat foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef] [PubMed]
- Pettengill, J.B.; Markell, A.; Conrad, A.; Carleton, H.A.; Beal, J.; Rand, H.; Musser, S.; Brown, E.W.; Allard, M.W.; Huffman, J.; et al. A multinational listeriosis outbreak and the importance of sharing genomic data. Lancet Microbe 2020, 1, e233–e234. [Google Scholar] [CrossRef] [PubMed]
- Tuytschaever, T.; Raes, K.; Sampers, I. Listeria monocytogenes in food businesses: From persistence strategies to intervention/prevention strategies—A review. Compr. Rev. Food Sci. Food Saf. 2023, 22, 3910–3950. [Google Scholar] [CrossRef]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-Del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Lee, S.; Ward, T.J.; Jima, D.D.; Parsons, C.; Kathariou, S. The arsenic resistance-associated Listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 2017, 83, e01189-17. [Google Scholar] [CrossRef]
- Gelbicova, T.; Florianova, M.; Hluchanova, L.; Kalova, A.; Korena, K.; Strakova, N.; Karpiskova, R. Comparative analysis of genetic determinants encoding cadmium, arsenic, and benzalkonium chloride resistance in Listeria monocytogenes of human, food, and environmental origin. Front. Microbiol. 2021, 11, 599882. [Google Scholar] [CrossRef]
- Hingston, P.; Brenner, T.; Hansen, L.T.; Wang, S. Comparative analysis of Listeria monocytogenes plasmids and expression levels of plasmid-encoded genes during growth under salt and acid stress conditions. Toxins 2019, 11, 426. [Google Scholar] [CrossRef]
- Tebano, G.; Zaghi, I.; Baldasso, F.; Calgarini, C.; Capozzi, R.; Salvadori, C.; Cricca, M.; Cristini, F. Antibiotic resistance to molecules commonly prescribed for the treatment of antibiotic-resistant gram-positive pathogens: What is relevant for the clinician? Pathogens 2024, 13, 88. [Google Scholar] [CrossRef]
- Mohr, K.I. History of Antibiotics Research. In How to Overcome the Antibiotic Crisis. Current Topics in Microbiology and Immunology; Stadler, M., Dersch, P., Eds.; Springer: Cham, Switzerland, 2016; Volume 398. [Google Scholar]
- Hussain, I.; Jabbar, T.; Naureen, A.; Hussain, A.; Gilani, M.R.H.S.; Abbas, N.; Naqvi, S.A.R. Lincosamide and Glycopeptide Antibiotics. In Antibiotics-Therapeutic Spectrum and Limitations; Academic Press: Cambridge, MA, USA, 2023; Volume 398, pp. 183–202. [Google Scholar]
- Mota, M.I.; Vázquez, S.; Cornejo, C.; D’Alessandro, B.; Braga, V.; Caetano, A.; Betancor, L.; Varela, G. Does Shiga Toxin-Producing Escherichia coli and Listeria monocytogenes contribute significantly to the burden of antimicrobial resistance in Uruguay? Front. Vet. Sci. 2020, 7, 583930. [Google Scholar] [CrossRef] [PubMed]
- Haubert, L.; Zehetmeyr, M.L.; da Silva, W.P. Resistance to benzalkonium chloride and cadmium chloride in Listeria monocytogenes isolates from food and food-processing environments in southern Brazil. Can. J. Microbiol. 2019, 65, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Deng, Y.; Fan, R.; Shi, L.; Bai, J.; Yan, H. Coresistance to benzalkonium chloride disinfectant and heavy metal ions in Listeria monocytogenes and Listeria innocua swine isolates from China. Foodborne Pathog. Dis. 2019, 16, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.L.; Carrillo, C.D.; DeschÊnes, M.; Blais, B.W. Genomic markers for quaternary ammonium compound resistance as a persistence indicator for Listeria monocytogenes contamination in food manufacturing environments. J. Food Prot. 2021, 84, 389–398. [Google Scholar] [CrossRef]
- Silva, A.; Silva, V.; Gomes, J.P.; Coelho, A.; Batista, R.; Saraiva, C.; Esteves, A.; Martins, Â.; Contente, D.; Diaz-Formoso, L.; et al. Listeria monocytogenes from food products and food associated environments: Antimicrobial resistance, genetic clustering and biofilm insights. Antibiotics 2024, 13, 447. [Google Scholar] [CrossRef]
- Cherifi, T.; Carrillo, C.; Lambert, D.; Miniaï, I.; Quessy, S.; Larivière-Gauthier, G.; Blais, B.; Fravalo, P. Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 2018, 18, 220. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, Y.; Carleton, H.; Timme, R.; Melka, D.; Muruvanda, T.; Wang, C.; Kastanis, G.; Katz, L.S.; Turner, L.; et al. Whole Genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes isolates associated with an outbreak linked to cheese, United States, 2013. Appl. Environ. Microbiol. 2017, 83, e00633-17. [Google Scholar] [CrossRef]
- Chmielowska, C.; Korsak, D.; Chapkauskaitse, E.; Decewicz, P.; Lasek, R.; Szuplewska, M.; Bartosik, D. Plasmidome of Listeria spp.—The repA-Family Business. Int. J. Mol. Sci. 2021, 22, 10320. [Google Scholar] [CrossRef]
- Ibrach, I.M.; Seth, R.K. Human listerial meningitis. J. Clin. Pathol. 1961, 14, 193–195. [Google Scholar] [CrossRef]
- Hupfeld, M.; Trasanidou, D.; Ramazzini, L.; Klumpp, J.; Loessner, M.J.; Kilcher, S. A functional type II-A CRISPR-Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res. 2018, 46, 6920–6933. [Google Scholar] [CrossRef]
- Di, H.; Ye, L.; Yan, H.; Meng, H.; Yamasak, S.; Shi, L. Comparative analysis of CRISPR loci in different Listeria monocytogenes lineages. Biochem. Biophys. Res. Commun. 2014, 454, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J.; Inman, R.B.; Lauer, P.; Calendar, R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: Implications for phage evolution. Mol. Microbiol. 2000, 35, 324–340. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Comparative genomics of Listeria species recovered from meat and food processing facilities. Microbiol. Spectr. 2022, 10, e0118922. [Google Scholar] [CrossRef] [PubMed]

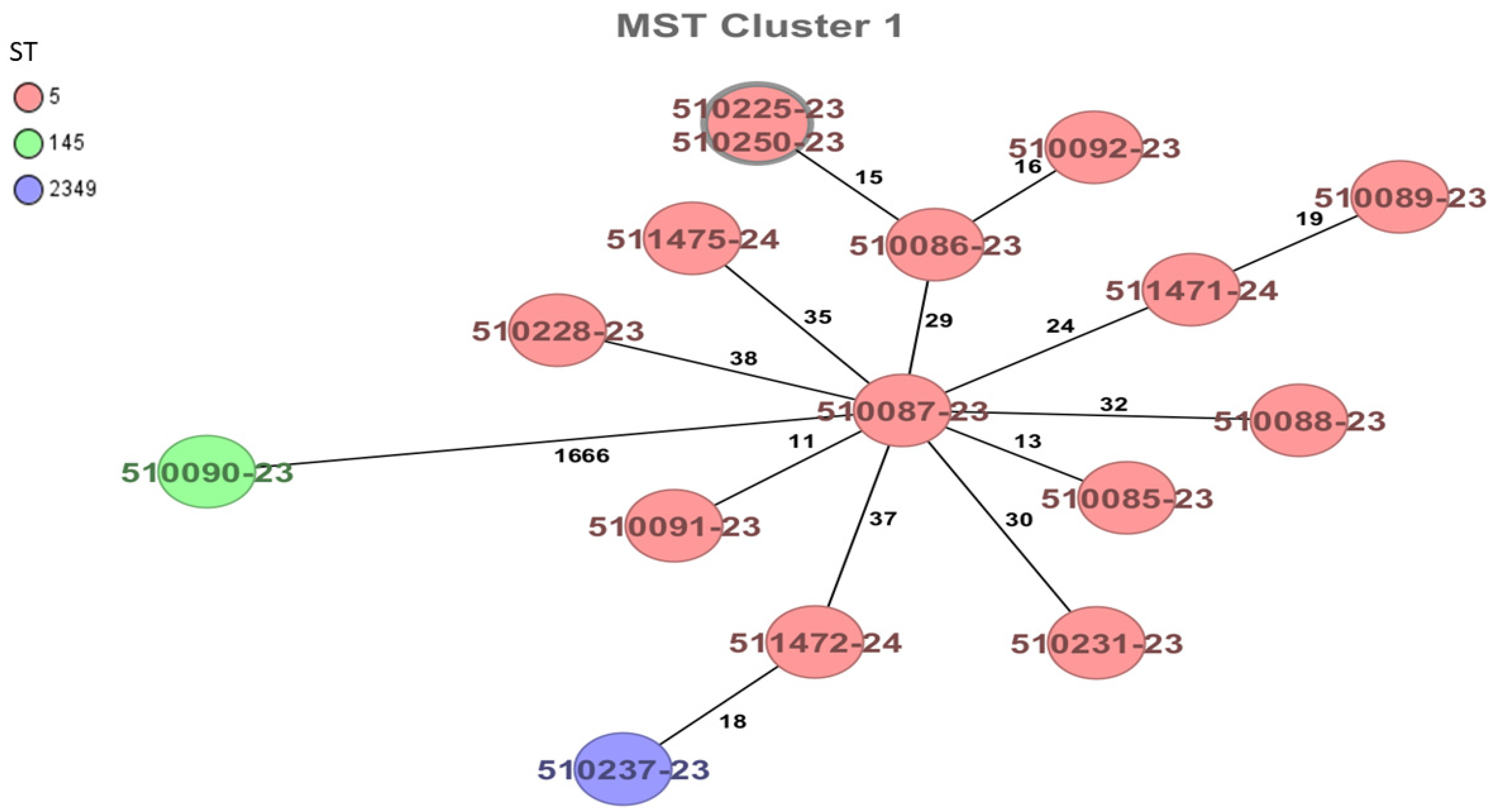
| Source | Number of Samples n | Positivity n (%) |
|---|---|---|
| Surfaces and equipment | 103 | 12 (11.6) |
| Raw material samples | 18 | 0 (0) |
| Finished product samples | 105 | 4 (3.8) |
| Total | 216 | 16 (4.4) |
| Sample ID | Food | MALDI-TOF MS | rMLST | ST | CC | CT | Serotype |
|---|---|---|---|---|---|---|---|
| 510085-23 | Finished product | L. monocytogenes | L. monocytogenes | 5 | CC5 | 9152 | 1/2b |
| 510086-23 | Surfaces and equipment (floor) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 9170 | 1/2b |
| 510087-23 | Surfaces and equipment (worktable) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 18381 | 1/2b |
| 510088-23 | Surfaces and equipment (preparation tank) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 18382 | 1/2b |
| 510089-23 | Surfaces and equipment (drain) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 18384 | 1/2b |
| 510091-23 | Surfaces and equipment (worktable) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 18381 | 1/2b |
| 510092-23 | Surfaces and equipment (transport cart wheel) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 9170 | 1/2b |
| 511471-24 | Finished product | L. monocytogenes | L. monocytogenes | 5 | CC5 | 10192 | 1/2b |
| 510225-23 | Surfaces and equipment (preparation tank) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 9170 | 1/2b |
| 510228-23 | Surfaces and equipment (mixing paddle) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 18386 | 1/2b |
| 511472-24 | Finished product | L. monocytogenes | L. monocytogenes | 5 | CC5 | 21278 | 1/2b |
| 510231-23 | Surfaces and equipment (grate) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 8061 | 1/2b |
| 510250-23 | Surfaces and equipment (floor) | L. monocytogenes | L. monocytogenes | 5 | CC5 | 9170 | 1/2b |
| 510237-23 | Surfaces and equipment (grate) | L. monocytogenes | L. monocytogenes | 2349 | CC5 | 18391 | 1/2b |
| 510090-23 | Surfaces and equipment (worktable) | L. monocytogenes | L. monocytogenes | 145 | CC2 | 66 | 4b |
| 511475-24 | Finished product | L. monocytogenes | L. monocytogenes | 5 | CC5 | 21280 | 1/2b |
| ST | CC | Virulence Genes | Resistance Genes |
|---|---|---|---|
| 145 | CC2 | bsh, clpC, clpE, clpP, fbpA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlP, lap, lntA, lpeA, lplA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 | fosX, vga(G) |
| 2349 | CC5 | ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inl, inlK, inlP, lap, lapB, lntA, lpeA, lplA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 | bcrB/bcrC, fosX, vga(G) |
| 5 | CC5 | ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlK, inlP, lap, lapB, lntA, lpeA, lplA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 | bcrB/bcrC, qacJ, fosX, vga(G) |
| ID Strain | ST | Plasmid | Plasmid Accession Number | Size (Kb) | Mobile Genetic Elements |
|---|---|---|---|---|---|
| 510085-23 | 5 | P40.3_510085 | CP014251 | 40,325 | cn_8427_ISLmo3, ISLmo3, cn_17195_ISLmo3, ISLmo4, ISLmo5, ISLmo19 |
| 510086-23 | 5 | P68.0_510086 | CP014251 | 68,042 | cn_8427_ISLmo3, ISLmo3, cn_17195_ISLmo3, ISLmo4, cn_12275_ISLmo3, ISLmo19, cn_13026_ISLmo19, ISLmo8, ISLmo4, ISLmo5 |
| 510087-23 | 5 | p34.8_510087 | CP014251 | 34,816 | cn_8427_ISLmo3, ISLmo3, ISLmo4, ISLmo5, ISLmo19, |
| 510088-23 | 5 | No found | |||
| 510089-23 | 5 | p34.8_510089 | CP014251 | 34,816 | ISLmo3, ISLmo4, ISLmo5, ISLmo19, |
| 510091-23 | 5 | p34.8_510091 | CP014251 | 34,816 | ISLmo3, ISLmo4, ISLmo5, ISLmo19, Cn_8427_ISLmo3 |
| 510092-23 | 5 | P69.1_510092 | CP014251 | 69,077 | ISLmo5, cn_13026_ISLmo4, ISLmo4, cn_12275_ISLmo19, ISLmo8, cn_17195_ISLmo19, ISLmo19, cn_17195_ISLmo3, cn_8427_ISLmo3 |
| 511471-24 | 5 | P406.7_511471 P50.7_511471 | LR134399 CP014251 | 406,655 50,672 | ISLmo3, ISLm19, ISLmo5, ISLmo4 |
| 510225-23 | 5 | P69.1_510092 | CP014251 | 69,077 | ISLmo5, cn_13026_ISLmo4, ISLmo4, cn_12275_ISLmo19, ISLmo8, cn_17195_ISLmo19, ISLmo19, cn_17195_ISLmo3, cn_8427_ISLmo3, ISLmo3 |
| 510228-23 | 5 | P34.6_510228 | CP014251 | 34,626 | cn_8427_ISLmo3, ISLmo3, ISLmo4, ISLmo5, ISLmo19, ISLmo8 |
| 511472-24 | 5 | P51.1_511472 | CP014251 | 51,100 | cn_7697_ISLmo3, ISLmo3, ISLmo4, ISLmo5, ISLmo19, ISLmo8 |
| 510231-23 | 5 | P37.2_510231 | CP014251 | cn_8427_ISLmo3, ISLmo3, ISLmo4, ISLmo5, ISLmo19, ISLmo8 | |
| 510250-23 | 5 | P69.1_510092 | CP014251 | 69,077 | cn_8427_ISLmo3, ISLmo3, cn_17195_ISLmo3, ISLmo4, cn_12275_ISLmo3, ISLmo19, cn_13026_ISLmo19, ISLmo8, ISLmo4, ISLmo5 |
| 510237-23 | 2349 | P40.3_510237 | CP014251 | 40,325 | cn_8427_ISLmo3, ISLmo3, ISLmo4, ISLmo5, ISLmo19, ISLmo8 |
| 510090-23 | 145 | No found | |||
| 511475-24 | 5 | P426_511475 | LR134399 | 425,974 | Not found |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra-Flores, J.; Daza-Prieto, B.; Chavarria, P.; Troncoso, M.; Stöger, A.; Figueroa, G.; Mancilla-Rojano, J.; Cruz-Córdova, A.; Martinovic, A.; Ruppitsch, W. From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile. Foods 2025, 14, 290. https://doi.org/10.3390/foods14020290
Parra-Flores J, Daza-Prieto B, Chavarria P, Troncoso M, Stöger A, Figueroa G, Mancilla-Rojano J, Cruz-Córdova A, Martinovic A, Ruppitsch W. From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile. Foods. 2025; 14(2):290. https://doi.org/10.3390/foods14020290
Chicago/Turabian StyleParra-Flores, Julio, Beatriz Daza-Prieto, Pamela Chavarria, Miriam Troncoso, Anna Stöger, Guillermo Figueroa, Jetsi Mancilla-Rojano, Ariadnna Cruz-Córdova, Aleksandra Martinovic, and Werner Ruppitsch. 2025. "From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile" Foods 14, no. 2: 290. https://doi.org/10.3390/foods14020290
APA StyleParra-Flores, J., Daza-Prieto, B., Chavarria, P., Troncoso, M., Stöger, A., Figueroa, G., Mancilla-Rojano, J., Cruz-Córdova, A., Martinovic, A., & Ruppitsch, W. (2025). From Traditional Typing to Genomic Precision: Whole-Genome Sequencing of Listeria monocytogenes Isolated from Refrigerated Foods in Chile. Foods, 14(2), 290. https://doi.org/10.3390/foods14020290









