Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Antimicrobial Susceptibility Testing
2.2. PCR and DNA Sequencing of Quinolone and Β-Lactamase Resistance Determination Genes
2.3. Pulsed-Field Gel Electrophoresis (PFGE)
2.4. Conjugation Experiment and Plasmid Analysis
2.5. Efflux Pump Inhibitor Test by Using Phe-Arg-β-Naphthylamide (PAβN)
2.6. Whole-Genome Sequencing of S. Enteritidis SJTUF12519
2.7. Data Availability
3. Results and Discussion
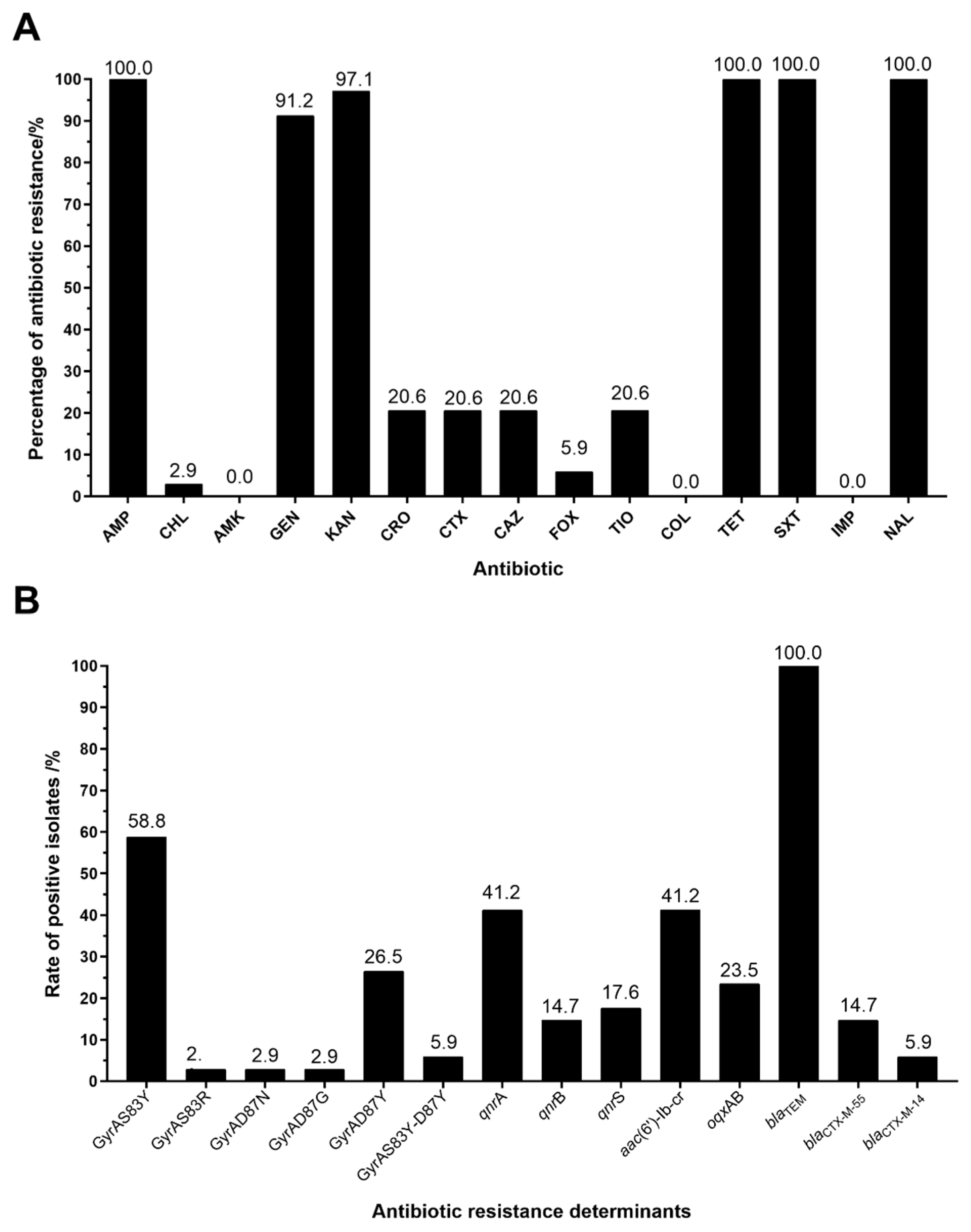
3.1. Antimicrobial Susceptibility Test Results
3.2. Prevalence and Distribution of Antibiotic Resistance Determinant
3.3. PFGE Analysis
3.4. Influence of Efflux Pump Inhibitors
3.5. The Analysis of Plasmid Transferability
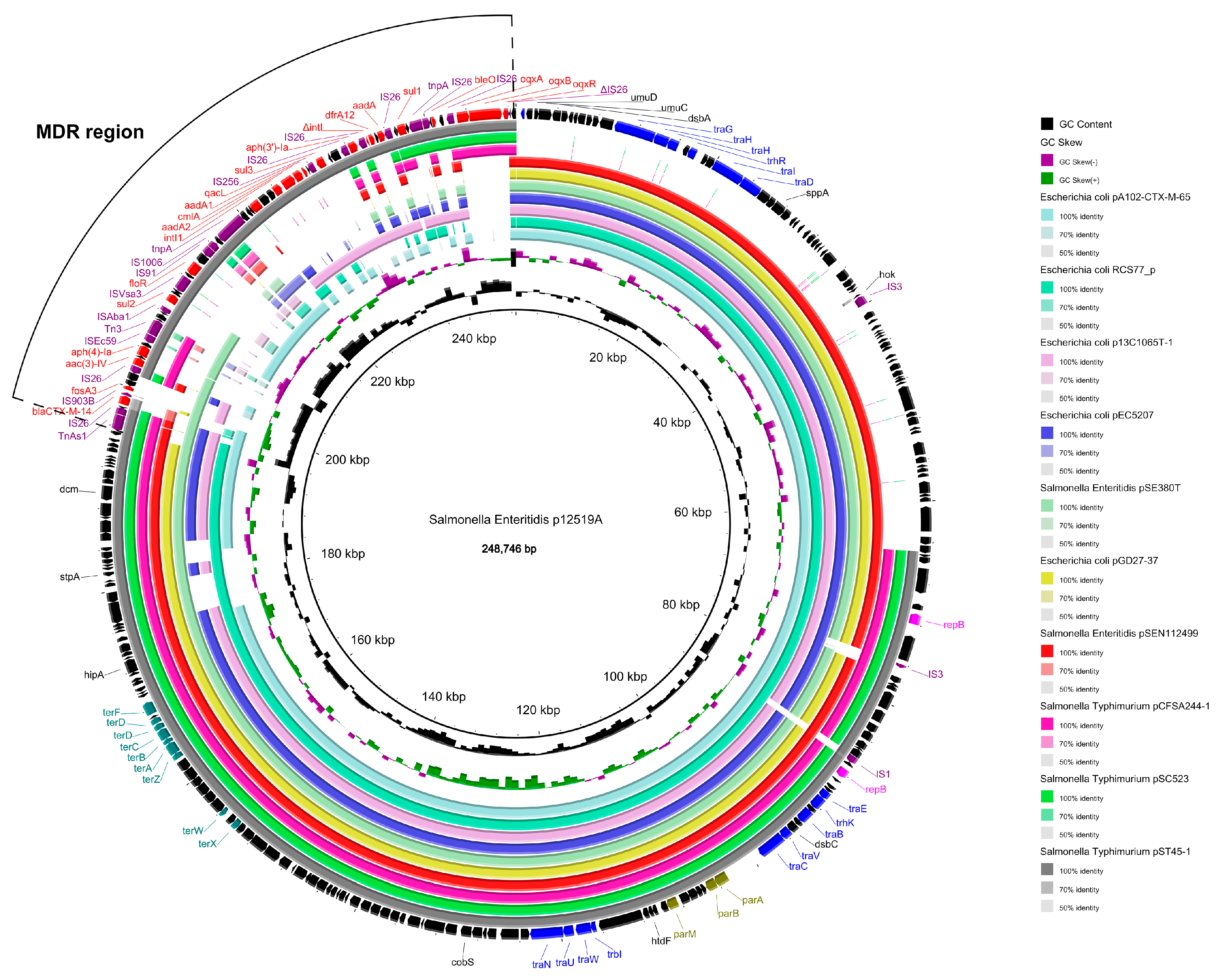
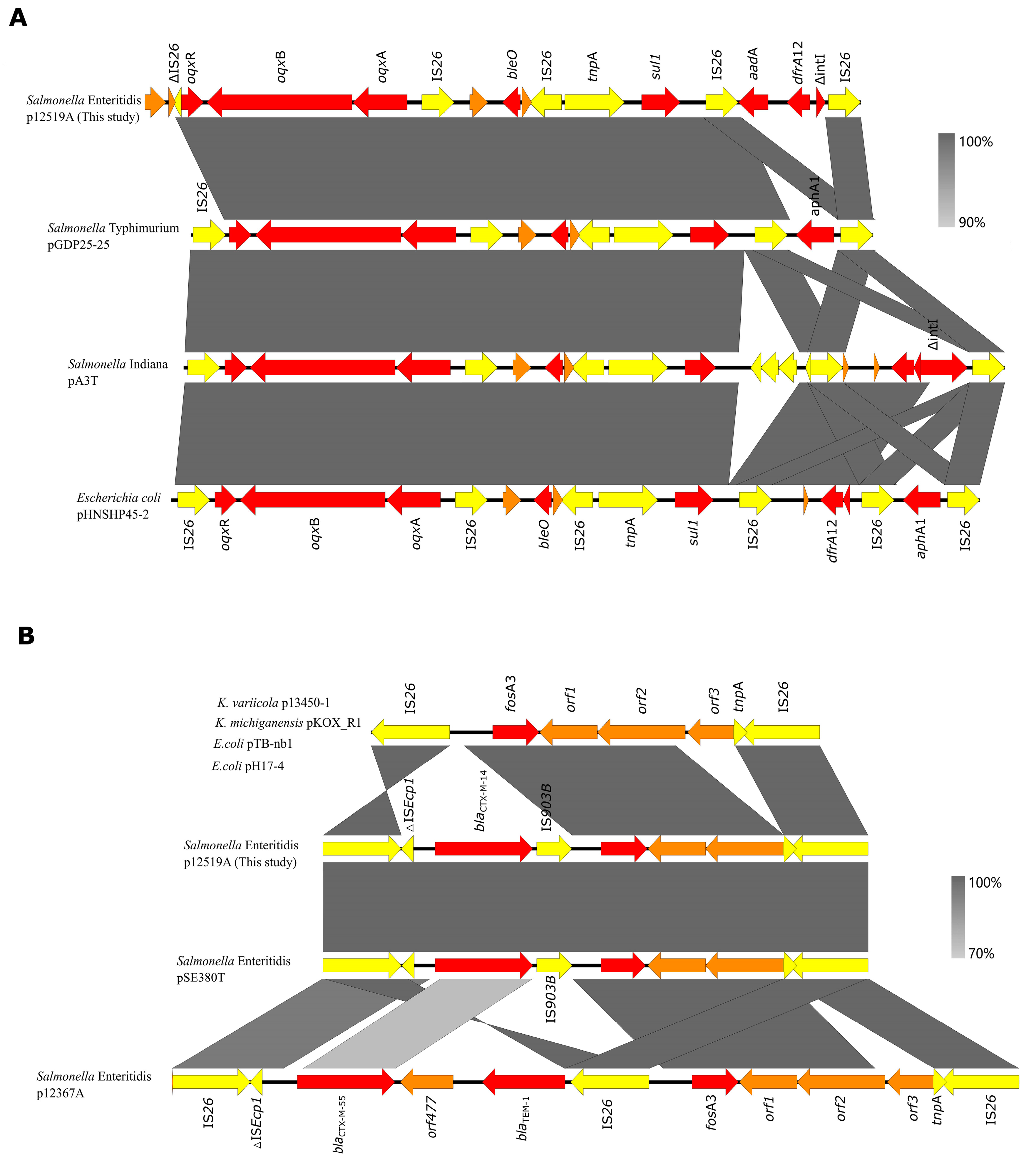
3.6. Complete Sequence of Plasmid p12519A
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-Analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, M.; Xu, X.; Fu, Y.; Xiong, Z.; Zhang, L.; Qu, X.; Zhang, H.; Wei, Y.; Zhan, Z.; et al. High-Levels of Resistance to Quinolone and Cephalosporin Antibiotics in MDR-ACSSuT Salmonella enterica Serovar Enteritidis Mainly Isolated from Patients and Foods in Shanghai, China. Int. J. Food Microbiol. 2018, 286, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Sher, A.A.; Mustafa, B.E.; Grady, S.C.; Gardiner, J.C.; Saeed, A.M. Outbreaks of Foodborne Salmonella enteritidis in the United States between 1990 and 2015: An Analysis of Epidemiological and Spatial-Temporal Trends. Int. J. Infect. Dis. 2021, 105, 54–61. [Google Scholar] [CrossRef] [PubMed]
- The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2017. EFSA J. 2018, 16, e05500. [CrossRef]
- Monte, D.F.M.; Sellera, F.P. Salmonella. Emerg. Infect. Dis. 2020, 26, 12. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Wang, Y.; Fanning, S.; Cui, S.; Chen, Q.; Liu, G.; Chen, Q.; Zhou, G.; Yang, B.; et al. Serovar Diversity and Antimicrobial Resistance of Non-Typhoidal Salmonella enterica Recovered from Retail Chicken Carcasses for Sale in Different Regions of China. Food Control 2017, 81, 46–54. [Google Scholar] [CrossRef]
- Li, Y.; Pei, X.; Zhang, X.; Wu, L.; Liu, Y.; Zhou, H.; Ma, G.; Chen, Q.; Liang, H.; Yang, D. A Surveillance of Microbiological Contamination on Raw Poultry Meat at Retail Markets in China. Food Control 2019, 104, 99–104. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Zhang, Y.; Liu, S.; Chen, L.; Xiao, C.; Zeng, H.; Wei, X.; Gu, Q.; Li, Y.; et al. Prevalence, Abundance, Serovars and Antimicrobial Resistance of Salmonella Isolated from Retail Raw Poultry Meat in China. Sci. Total Environ. 2020, 713, 136385. [Google Scholar] [CrossRef]
- Yu, X.; Zhu, H.; Bo, Y.; Li, Y.; Zhang, Y.; Liu, Y.; Zhang, J.; Jiang, L.; Chen, G.; Zhang, X. Prevalence and Antimicrobial Resistance of Salmonella enterica Subspecies Enterica Serovar Enteritidis Isolated from Broiler Chickens in Shandong Province, China, 2013–2018. Poult. Sci. 2021, 100, 1016–1023. [Google Scholar] [CrossRef]
- Hald, T.; Aspinall, W.; Devleesschauwer, B.; Cooke, R.; Corrigan, T.; Havelaar, A.H.; Gibb, H.J.; Torgerson, P.R.; Kirk, M.D.; Angulo, F.J.; et al. World Health Organization Estimates of the Relative Contributions of Food to the Burden of Disease Due to Selected Foodborne Hazards: A Structured Expert Elicitation. PLoS ONE 2016, 11, e0145839. [Google Scholar] [CrossRef]
- Harris, P.N.A.; Tambyah, P.A.; Paterson, D.L. Beta-Lactam and Beta-Lactamase Inhibitor Combinations in the Treatment of Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae: Time for a Reappraisal in the Era of Few Antibiotic Options? Lancet Infect. Dis. 2015, 15, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H. Safety Profile of the Fluoroquinolones Focus on Levofloxacin. Drug Saf. 2010, 33, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yang, C.; Dong, N.; Xie, M.; Ye, L.; Chan, E.W.C.; Chen, S. Evolution of Ciprofloxacin Resistance-Encoding Genetic Elements in Salmonella. mSystems 2020, 5, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.; Zhang, J.; Xu, X.; Shi, W.; Chen, S.; Yang, X.; Su, X.; Shi, X.; Meng, J. Emerging High-Level Ciprofloxacin Resistance and Molecular Basis of Resistance in Salmonella enterica from Humans, Food and Animals. Int. J. Food Microbiol. 2018, 280, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; You, L.; Wang, D.; Huang, H.; Li, S.; Wang, D. Antimicrobial Resistance and Molecular Genotyping of Salmonella enterica Serovar Enteritidis Clinical Isolates from Guizhou Province of Southwestern China. PLoS ONE 2019, 14, e0221492. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of Drug Resistance: Quinolone Resistance: Mechanisms of Quinolone Resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Chang, M.-X.; Zhang, J.-F.; Sun, Y.-H.; Li, R.-S.; Lin, X.-L.; Yang, L.; Webber, M.A.; Jiang, H.-X. Contribution of Different Mechanisms to Ciprofloxacin Resistance in Salmonella Spp. Front. Microbiol. 2021, 12, 663731. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Xu, X.; Zhou, X.; Shi, C.; Zhao, X.; Liu, Y.; Shi, X. Co-Existence of mphA, oqxAB and blaCTX-M-65 on the IncHI2 Plasmid in Highly Drug-Resistant Salmonella enterica Serovar Indiana ST17 Isolated from Retail Foods and Humans in China. Food Control 2020, 118, 107269. [Google Scholar] [CrossRef]
- Zhang, Z.; Meng, X.; Wang, Y.; Xia, X.; Wang, X.; Xi, M.; Meng, J.; Shi, X.; Wang, D.; Yang, B. Presence of qnr, aac(6’)-Ib, qepA, oqxAB, and Mutations in Gyrase and Topoisomerase in Nalidixic Acid-Resistant Salmonella Isolates Recovered from Retail Chicken Carcasses. Foodborne Pathog. Dis. 2014, 11, 698–705. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhang, J.; Yang, S.; Zhang, S.; Chen, M.; Xue, L.; Ding, Y.; Zeng, H.; Gu, Q.; et al. Molecular Characterisation of Antimicrobial Resistance Determinants and Class 1 Integrons of Salmonella enterica Subsp. Enterica Serotype Enteritidis Strains from Retail Food in China. Food Control 2021, 128, 108191. [Google Scholar] [CrossRef]
- Nolivos, S.; Cayron, J.; Dedieu, A.; Page, A.; Delolme, F.; Lesterlin, C. Role of AcrAB-TolC Multidrug Efflux Pump in Drug-Resistance Acquisition by Plasmid Transfer. Science 2019, 364, 778–782. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liao, X.-P.; Liu, Z.-Z.; Huang, T.; Li, X.; Sun, J.; Liu, B.-T.; Zhang, Q.; Liu, Y.-H. Co-Spread of oqxAB and blaCTX-M-9G in Non-Typhi Salmonella enterica Isolates Mediated by ST2-IncHI2 Plasmids. Int. J. Antimicrob. Agents 2014, 44, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chen, K.; Wai-Chi Chan, E.; Chen, S. Increasing Prevalence of Ciprofloxacin-Resistant Food-Borne Salmonella Strains Harboring Multiple PMQR Elements but Not Target Gene Mutations. Sci. Rep. 2015, 5, 14754. [Google Scholar] [CrossRef] [PubMed]
- Soares, F.B.; Camargo, C.H.; Cunha, M.P.V.; de Almeida, E.A.; de Jesus Bertani, A.M.; de Carvalho, E.; de Paiva, J.B.; Fernandes, S.A.; Tiba-Casas, M.R. Co-Occurrence of qnrE1 and blaCTX-M-8 in IncM1 Transferable Plasmids Contributing to MDR in Different Salmonella Serotypes. J. Antimicrob. Chemother. 2019, 74, 1155–1156. [Google Scholar] [CrossRef]
- Chen, K.; Dong, N.; Chan, E.W.-C.; Chen, S. Transmission of Ciprofloxacin Resistance in Salmonella Mediated by a Novel Type of Conjugative Helper Plasmids. Emerg. Microbes Infect. 2019, 8, 857–865. [Google Scholar] [CrossRef]
- Chen, K.; Dong, N.; Zhao, S.; Liu, L.; Li, R.; Xie, M.; Lin, D.; Wai-Chi Chan, E.; Meng, J.; McDermott, P.F.; et al. Identification and Characterization of Conjugative Plasmids That Encode Ciprofloxacin Resistance in Salmonella. Antimicrob. Agents Chemother. 2018, 62, e00575-18. [Google Scholar] [CrossRef]
- Thiyagarajan, Y.; Harish, B. Prevalence of Plasmid-Mediated Quinolone Resistance Genes among Ciprofloxacin-Resistant Clinical Isolates of Enterobacteriaceae over Four Years: A Descriptive Study. Int. J. Infect. Dis. 2016, 45, 119–120. [Google Scholar] [CrossRef][Green Version]
- Chen, K.; Chan, E.W.C.; Chen, S. Evolution and Transmission of a Conjugative Plasmid Encoding Both Ciprofloxacin and Ceftriaxone Resistance in Salmonella. Emerg. Microbes Infect. 2019, 8, 396–403. [Google Scholar] [CrossRef]
- CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- ECAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. 2019. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 13 January 2025).
- Qiao, J.; Zhang, Q.; Alali, W.Q.; Wang, J.; Meng, L.; Xiao, Y.; Yang, H.; Chen, S.; Cui, S.; Yang, B. Characterization of Extended-Spectrum β-Lactamases (ESBLs)-Producing Salmonella in Retail Raw Chicken Carcasses. Int. J. Food Microbiol. 2017, 248, 72–81. [Google Scholar] [CrossRef]
- Xu, L. Rapid and Simple Detection of blaCTX-M Genes by Multiplex PCR Assay. J. Med. Microbiol. 2005, 54, 1183–1187. [Google Scholar] [CrossRef]
- Zhang, Z.; He, S.; Xu, X.; Chang, J.; Zhan, Z.; Cui, Y.; Shi, C.; Shi, X. Antimicrobial Susceptibility and Molecular Characterization of Salmonella enterica Serovar Indiana from Foods, Patients, and Environments in China during 2007–2016. Food Control 2022, 131, 108427. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, M.; Xu, X.; Lv, C.; Shi, C. Dynamic Antimicrobial Resistance and Phylogenomic Structure of Salmonella Typhimurium from 2007 to 2019 in Shanghai, China. Microbiol. Spectr. 2024, 12, e00262-24. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Kehrenberg, C.; de Jong, A.; Friederichs, S.; Cloeckaert, A.; Schwarz, S. Molecular Mechanisms of Decreased Susceptibility to Fluoroquinolones in Avian Salmonella Serovars and Their Mutants Selected during the Determination of Mutant Prevention Concentrations. J. Antimicrob. Chemother. 2007, 59, 886–892. [Google Scholar] [CrossRef][Green Version]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Baucheron, S.; Imberechts, H.; Chaslus-Dancla, E.; Cloeckaert, A. The AcrB Multidrug Transporter Plays a Major Role in High-Level Fluoroquinolone Resistance in Salmonella enterica Serovar Typhimurium Phage Type DT204. Microb. Drug Resist. 2002, 8, 281–289. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, W.; Xu, X.; Zhou, X.; Shi, C. Transmissible ST3-IncHI2 Plasmids Are Predominant Carriers of Diverse Complex IS26-Class 1 Integron Arrangements in Multidrug-Resistant Salmonella. Front. Microbiol. 2018, 9, 2492. [Google Scholar] [CrossRef]
- Haenni, M.; Metayer, V.; Jarry, R.; Drapeau, A.; Puech, M.-P.; Madec, J.-Y.; Keck, N. Wide Spread of blaCTX-M-9/mcr-9 IncHI2/ST1 Plasmids and CTX-M-9-Producing Escherichia Coli and Enterobacter Cloacae in Rescued Wild Animals. Front. Microbiol. 2020, 11, 601317. [Google Scholar] [CrossRef]
- Shang, D.; Zhao, H.; Xu, X.; Arunachalam, K.; Chang, J.; Bai, L.; Shi, C. Conjugative IncHI2 Plasmid Harboring Novel Class 1 Integron Mediated Dissemination of Multidrug Resistance Genes in Salmonella Typhimurium. Food Control 2021, 122, 107810. [Google Scholar] [CrossRef]
- Du, P.; Liu, D.; Song, H.; Zhang, P.; Li, R.; Fu, Y.; Liu, X.; Jia, J.; Li, X.; Fanning, S.; et al. Novel IS26-Mediated Hybrid Plasmid Harboring Tet(X4) in Escherichia Coli. J. Glob. Antimicrob. Resist. 2020, 21, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hall, R.M. IS26-Mediated Precise Excision of the IS26-aphA1a Translocatable Unit. mBio 2015, 6, e01866-15. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hall, R.M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere 2016, 1, e00038-16. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Monno, R.; Addabbo, P.; Pesole, G.; Scrascia, M.; Calia, C.; Dionisi, A.M.; Chiara, M.; Horner, D.S.; Manzari, C.; et al. IS26 Mediated Antimicrobial Resistance Gene Shuffling from the Chromosome to a Mosaic Conjugative FII Plasmid. Plasmid 2018, 100, 22–30. [Google Scholar] [CrossRef]
- Partridge, S.R.; Zong, Z.; Iredell, J.R. Recombination in IS26 and Tn2 in the Evolution of Multiresistance Regions Carrying blaCTX-M-15 on Conjugative IncF Plasmids from Escherichia Coli. Antimicrob. Agents Chemother. 2011, 55, 4971–4978. [Google Scholar] [CrossRef]
- Wong, M.H.-Y.; Chan, E.W.-C.; Chen, S. IS26-Mediated Formation of a Virulence and Resistance Plasmid in Salmonella enteritidis . J. Antimicrob. Chemother. 2017, 72, 2750–2754. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Z.; Xu, X.; He, S.; Zhao, X.; Cui, Y.; Zhou, X.; Shi, C.; Liu, Y.; Zhou, M.; et al. Molecular Characterization of Cephalosporin-Resistant Salmonella enteritidis ST11 Isolates Carrying blaCTX-M from Children with Diarrhea. Foodborne Pathog. Dis. 2021, 18, 702–711. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, W.; Pan, W.; Yin, J.; Pan, Z.; Gao, S.; Jiao, X. Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 2012, 56, 3423–3427. [Google Scholar] [CrossRef]
- Eaves, D.J.; Randall, L.; Gray, D.T.; Buckley, A.; Woodward, M.J.; White, A.P.; Piddock, L.J.V. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 2004, 48, 4012–4015. [Google Scholar] [CrossRef]
- Park, C.H.; Robicsek, A.; Jacoby, G.A.; Sahm, D.; Hooper, D.C. Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006, 50, 3953–3955. [Google Scholar] [CrossRef]


| Isolate | Ciprofloxacin | PMQR Genes | GyrA Mutation | |
|---|---|---|---|---|
| MIC | MIC in the Presence of PaβN a | |||
| SJTUF11851 | 1 | 0.125 | aac(6′)-Ib-cr, qnrA | D87Y |
| SJTUF12065 | 1 | 0.06 | oqxAB | S83Y-D87Y |
| SJTUF12074 | 1 | 0.06 | oqxAB | D87Y |
| SJTUF12092 | 4 | 0.5 | aac(6′)-Ib-cr, oqxAB | D87Y |
| SJTUF12113 | 2 | 0.025 | aac(6′)-Ib-cr, qnrS | D87Y |
| SJTUF12122 | 2 | 0.025 | aac(6′)-Ib-cr, qnrA,qnrS | S83Y |
| SJTUF12519 | 1 | 0.06 | oqxAB | D87Y |
| Isolate a | MIC (µg/mL) | Other Resistant Profiles | PMQR Genes | ESBL Gene | Replicon Types b | Plasmid Sizes (kb) | |
|---|---|---|---|---|---|---|---|
| CRO | CIP | ||||||
| SJTUF11851 | ≥128 | 1 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT-NAL | aac(6′)-Ib-cr, qnrA | CTX-M-55 | IncHI2, NT | ~245, ~80 |
| SJTUF11851-TC | 64 | 0.25 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | aac(6′)-Ib-cr, qnrA | CTX-M-55 | IncHI2 | ~245 |
| SJTUF12065 | ≥128 | 1 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT-NAL | oqxAB | CTX-M-55 | IncHI2, NT | ~245, ~80 |
| SJTUF12065-TC | 64 | 0.06 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | oqxAB | CTX-M-55 | IncHI2 | ~245 |
| SJTUF12074 | ≥128 | 1 | AMP-TIO-CAZ-CTX-FOX-KAN-GEN-TET-SXT-NAL | oqxAB | CTX-M-55 | IncHI2, NT | ~245, ~80 |
| SJTUF12074-TC | 64 | 0.03 | AMP-TIO-CAZ-CTX-FOX-KAN-GEN-TET-SXT | oqxAB | CTX-M-55 | IncHI2 | ~245 |
| SJTUF12092 | ≥128 | 4 | AMP-TIO-CAZ-CTX-FOX-KAN-GEN-TET-SXT-NAL | aac(6′)-Ib-cr, oqxAB | CTX-M-55 | IncHI2, NT | ~245, ~80 |
| SJTUF12092-TC | 64 | 0.125 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | aac(6′)-Ib-cr,oqxAB | CTX-M-55 | IncHI2 | ~245 |
| SJTUF12113 | ≥128 | 2 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT-NAL | aac(6′)-Ib-cr, qnrS | CTX-M-55 | IncHI2, NT | ~245, ~80 |
| SJTUF12113-TC | 64 | 0.25 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | aac(6′)-Ib-cr, qnrS | CTX-M-55 | IncHI2 | ~245 |
| SJTUF12122 | ≥128 | 2 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT-NAL | aac(6′)-Ib-cr, qnrA,qnrS | CTX-M-14 | IncHI2, NT | ~245, ~80 |
| SJTUF12122-TC | 64 | 0.25 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | aac(6′)-Ib-cr, qnrA,qnrS | CTX-M-14 | IncHI2 | ~245 |
| SJTUF12519 | ≥128 | 1 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT-NAL | oqxAB | CTX-M-14 | IncHI2, NT | ~245, ~80 |
| SJTUF12519-TC | 64 | 0.03 | AMP-TIO-CAZ-CTX-KAN-GEN-TET-SXT | oqxAB | CTX-M-14 | IncHI2 | ~245 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhao, H.; Shi, C. Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis. Foods 2025, 14, 289. https://doi.org/10.3390/foods14020289
Zhang Z, Zhao H, Shi C. Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis. Foods. 2025; 14(2):289. https://doi.org/10.3390/foods14020289
Chicago/Turabian StyleZhang, Zengfeng, Hang Zhao, and Chunlei Shi. 2025. "Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis" Foods 14, no. 2: 289. https://doi.org/10.3390/foods14020289
APA StyleZhang, Z., Zhao, H., & Shi, C. (2025). Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis. Foods, 14(2), 289. https://doi.org/10.3390/foods14020289






