Lacticaseibacillus paracasei subsp. paracasei 2LB: Identification of Genes to Assess the Safety and Probiotic Potential of the Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. L. paracasei 2LB Isolation and Cultivation Conditions
2.2. Whole-Genome Sequencing and De Novo Assembly
2.3. Phylogenomic Analysis of the Core- and Pan-Genome and Functional Annotation
2.4. In Silico Safety and Stability Assessment
2.5. Data Visualization
3. Results and Discussion
3.1. Genome Features of the L. paracasei 2LB
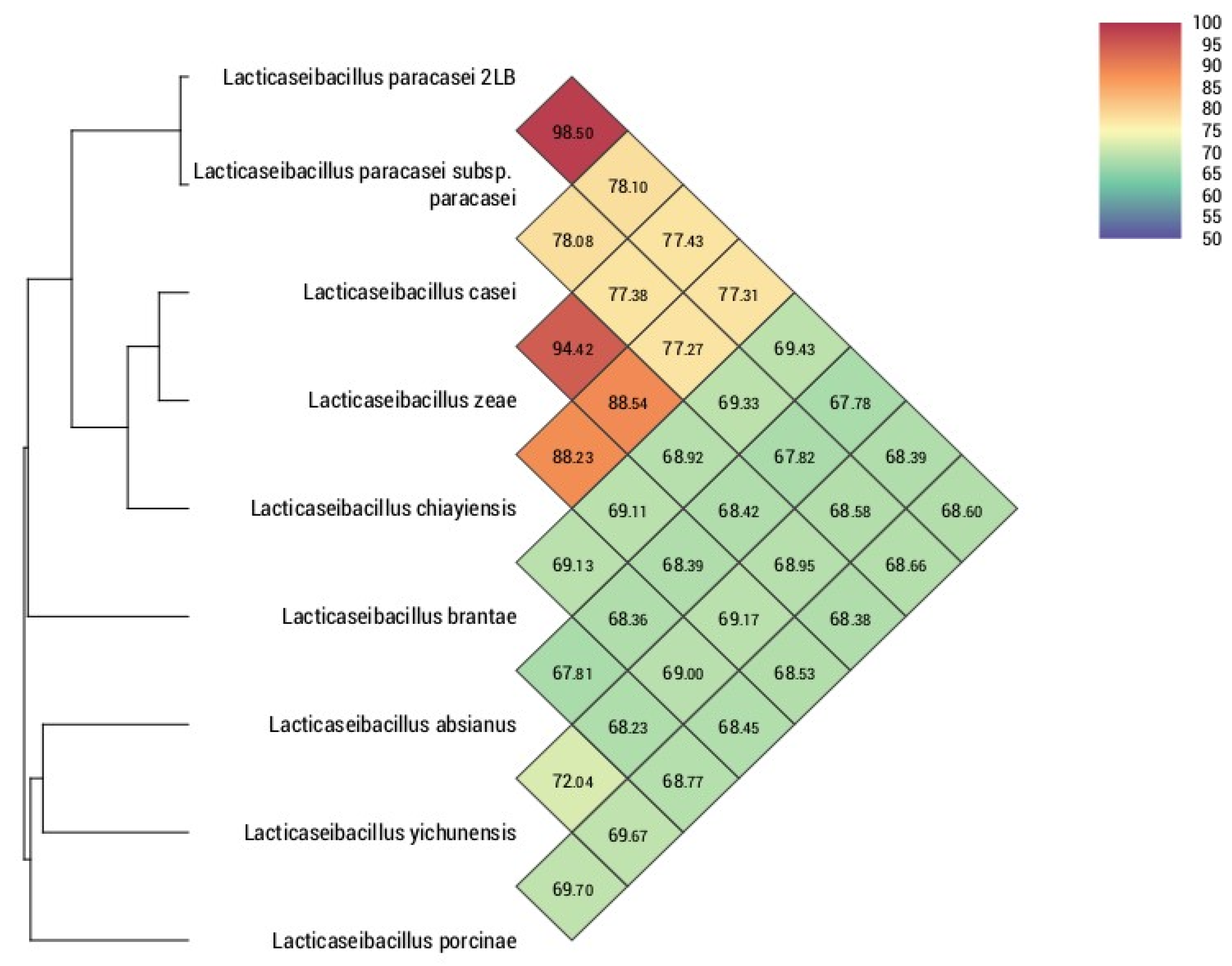
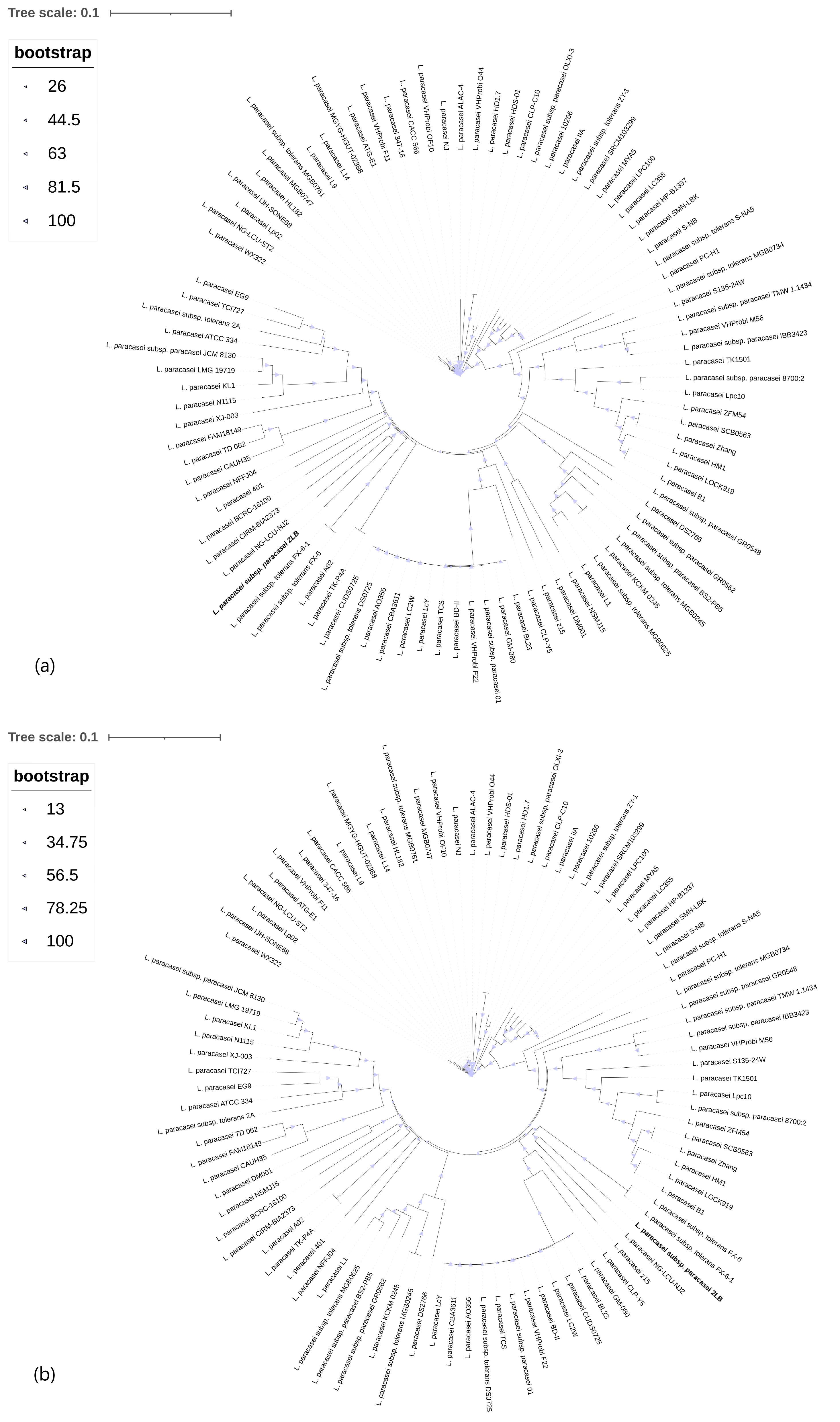
3.2. Phylogenomic Analysis of the Core- and Pan-Genome
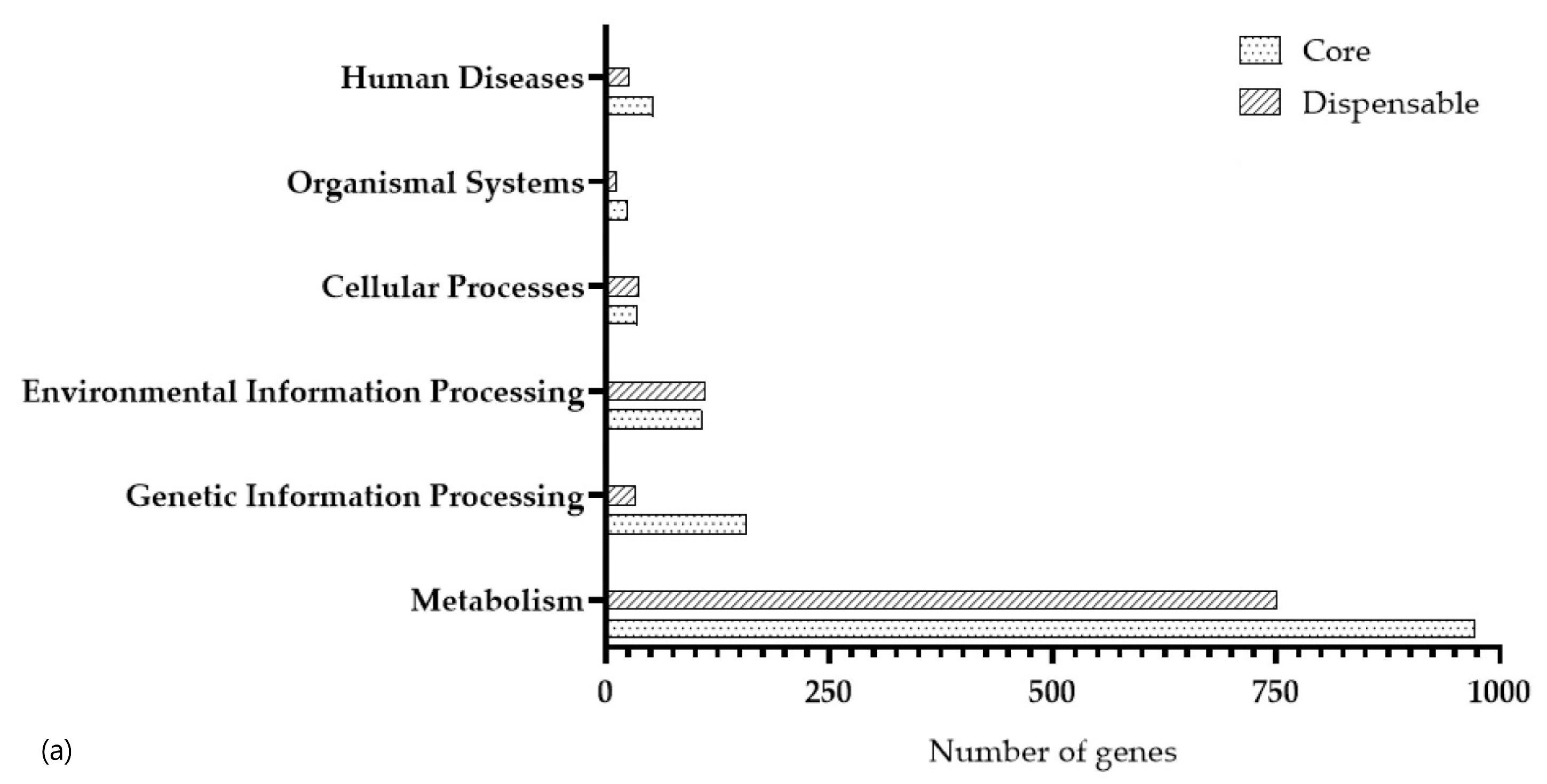
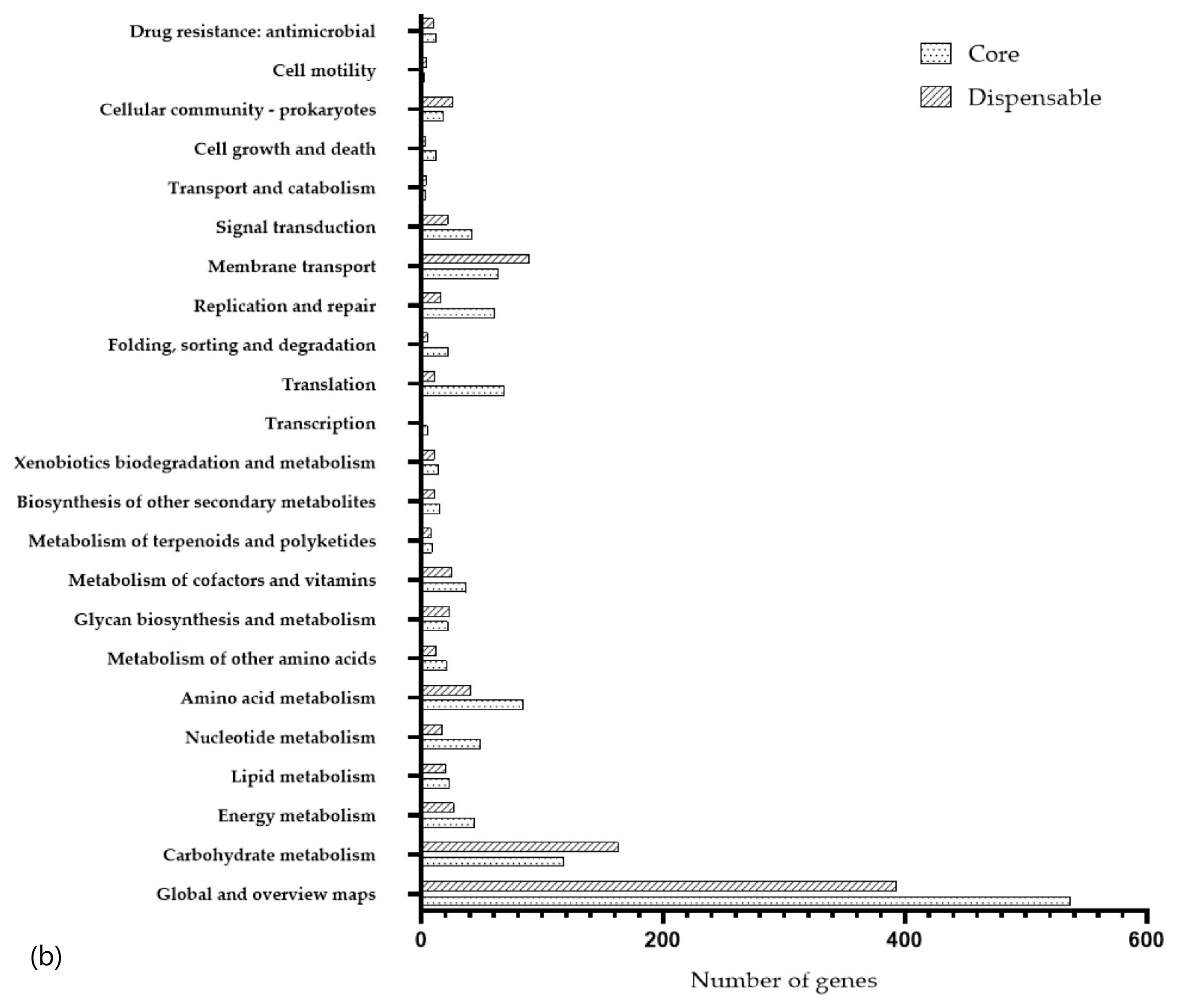
3.3. Functional Annotation
3.4. CAZymes Carbohydrate-Active Enzymes Analysis
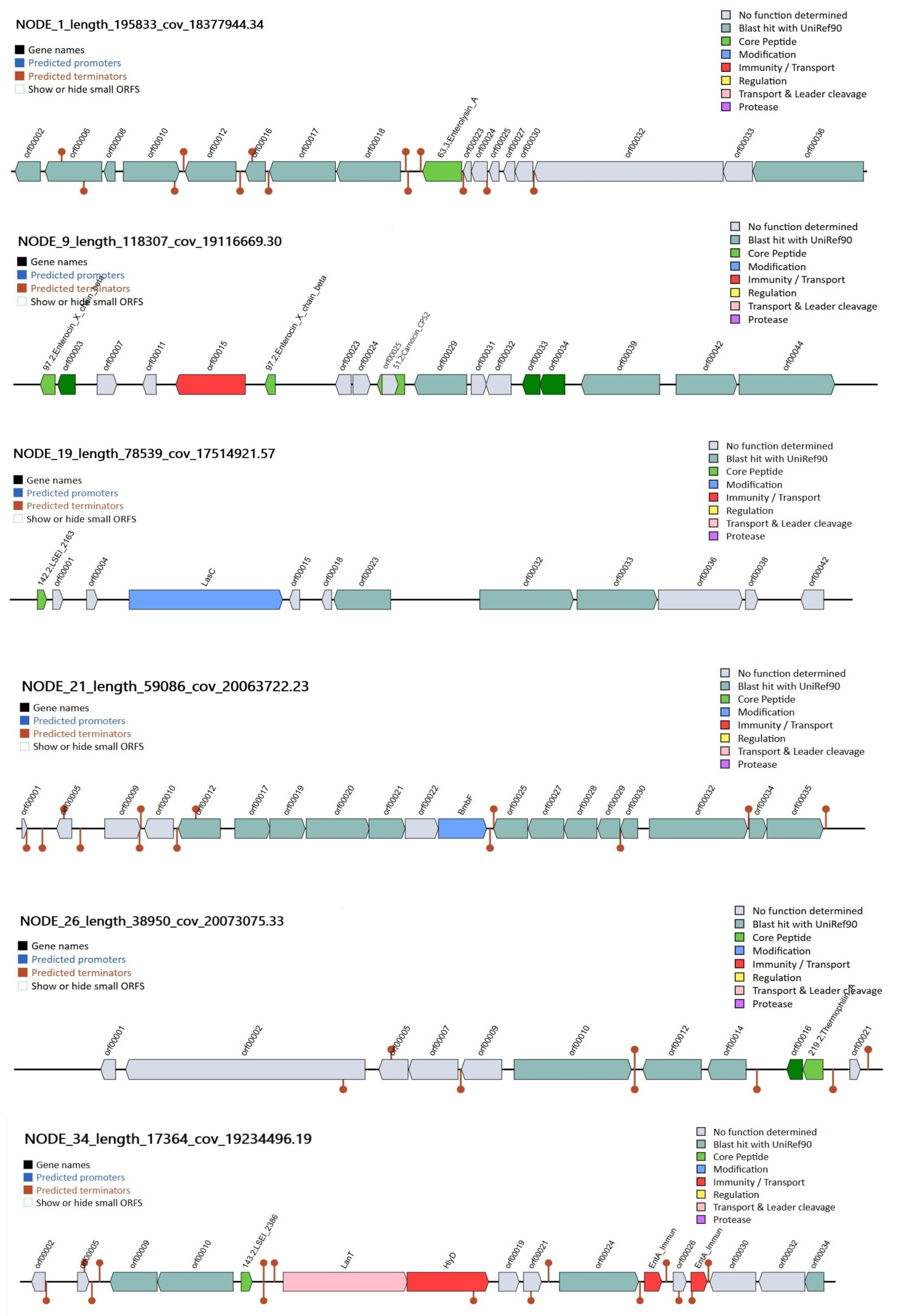
3.5. Genomic Analysis of Bacteriocins
3.6. Probiotic Related Multiple Genes Presented of the L. paracasei 2LB Strain
3.7. In Silico Safety and Stability Assessment
3.8. Analysis of CRISPR/Cas Systems
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, M.; Lu, M.; Chen, K.; Xu, R.; Xia, Y.; Liu, X.; Liu, Z.; Liu, Q. Akkermansia muciniphila and Lactobacillus plantarum Ameliorate Systemic Lupus Erythematosus by Possibly Regulating Immune Response and Remodeling Gut Microbiota. mSphere 2023, 8, e00070-23. [Google Scholar] [CrossRef]
- Meng, Y.; Qiu, X.; Tang, Z.; Mao, Y.; Tan, Y. Lactobacillus paracasei L9 Affects Disease Progression in Experimental Autoimmune Neuritis by Regulating Intestinal Flora Structure and Arginine Metabolism. J. Neuroinflammation 2023, 20, 122. [Google Scholar] [CrossRef]
- Torres-Miranda, A.; Melis-Arcos, F.; Garrido, D. Characterization and Identification of Probiotic Features in Lacticaseibacillus paracasei Using a Comparative Genomic Analysis Approach. Probiotics Antimicrob. Proteins 2022, 14, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Peng, K.; Xiao, S.; Long, Y.; Yu, Q. The Role of Lactobacillus in Inflammatory Bowel Disease: From Actualities to Prospects. Cell Death Discov. 2023, 9, 361. [Google Scholar] [CrossRef]
- Chen, Z.; Leng, X.; Zhou, F.; Shen, W.; Zhang, H.; Yu, Q.; Meng, X.; Fan, H.; Qin, M. Screening and Identification of Probiotic Lactobacilli from the Infant Gut Microbiota to Alleviate Lead Toxicity. Probiotics Antimicrob. Proteins 2023, 15, 821–831. [Google Scholar] [CrossRef]
- Xiao, L.; Yang, Y.; Han, S.; Rui, X.; Ma, K.; Zhang, C.; Wang, G.; Li, W. Effects of Genes Required for Exopolysaccharides Biosynthesis in Lacticaseibacillus paracasei S-NB on Cell Surface Characteristics and Probiotic Properties. Int. J. Biol. Macromol. 2023, 224, 292–305. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Chee, W.J.Y.; Chew, S.Y.; Than, L.T.L. Vaginal Microbiota and the Potential of Lactobacillus Derivatives in Maintaining Vaginal Health. Microb. Cell Factories 2020, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- Dehghani Champiri, I.; Bamzadeh, Z.; Rahimi, E.; Rouhi, L. Lacticaseibacillus paracasei LB12, a Potential Probiotic Isolated from Traditional Iranian Fermented Milk (Doogh). Curr. Microbiol. 2023, 80, 333. [Google Scholar] [CrossRef] [PubMed]
- Lazarenko, L.M.; Babenko, L.P.; Gichka, S.G.; Sakhno, L.O.; Demchenko, O.M.; Bubnov, R.V.; Sichel, L.M.; Spivak, M.Y. Assessment of the Safety of Lactobacillus casei IMV B-7280 Probiotic Strain on a Mouse Model. Probiotics Antimicrob. Proteins 2021, 13, 1644–1657. [Google Scholar] [CrossRef]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef]
- Castro-González, J.M.; Castro, P.; Sandoval, H.; Castro-Sandoval, D. Probiotic Lactobacilli Precautions. Front. Microbiol. 2019, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sarangi, A.N.; Mukherjee, M.; Bhowmick, S.; Tripathy, S. Reanalysis of Lactobacillus paracasei Lbs2 Strain and Large-Scale Comparative Genomics Places Many Strains into Their Correct Taxonomic Position. Microorganisms 2019, 7, 487. [Google Scholar] [CrossRef]
- Poon, T.; Juana, J.; Noori, D.; Jeansen, S.; Pierucci-Lagha, A.; Musa-Veloso, K. Effects of a Fermented Dairy Drink Containing Lacticaseibacillus paracasei subsp. paracasei CNCM I-1518 (Lactobacillus casei CNCM I-1518) and the Standard Yogurt Cultures on the Incidence, Duration, and Severity of Common Infectious Diseases: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2020, 12, 3443. [Google Scholar] [CrossRef]
- Senda-Sugimoto, Y.; Mihara, T.; Higuchi, Y.; Uchiyama, K.; Takara, T.; Takahashi, H. Effects of Lacticaseibacillus paracasei 327 Intake on the Intestinal Environment in Healthy Adult Japanese: A Randomized, Double-Blind, Placebo-Controlled, Parallel-Group Trial. Funct. Foods Health Dis. 2024, 14, 184–206. [Google Scholar] [CrossRef]
- Moiseenko, K.V.; Glazunova, O.A.; Savinova, O.S.; Shabaev, A.V.; Fedorova, T.V. Changes in Composition of Some Bioactive Molecules upon Inclusion of Lacticaseibacillus paracasei Probiotic Strains into a Standard Yogurt Starter Culture. Foods 2023, 12, 4238. [Google Scholar] [CrossRef]
- Khushboo; Karnwal, A.; Malik, T. Characterization and Selection of Probiotic Lactic Acid Bacteria from Different Dietary Sources for Development of Functional Foods. Front. Microbiol. 2023, 14, 1170725. [Google Scholar] [CrossRef] [PubMed]
- Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Alitheen Isolation and Characterization of Lactobacillus Spp. from Kefir Samples in Malaysia. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef] [PubMed]
- Tarrah, A.; Pakroo, S.; Corich, V.; Giacomini, A. Whole-Genome Sequence and Comparative Genome Analysis of Lactobacillus paracasei DTA93, a Promising Probiotic Lactic Acid Bacterium. Arch. Microbiol. 2020, 202, 1997–2003. [Google Scholar] [CrossRef]
- Wang, J.; Wang, T.; Li, Y.; Fan, Z.; Lv, Z.; Liu, L.; Li, X.; Li, B. Comparative Genomic Analysis of Lacticaseibacillus paracasei SMN-LBK from Koumiss. Front. Microbiol. 2022, 13, 1042117. [Google Scholar] [CrossRef]
- Barreto Pinilla, C.M.; Guzman Escudero, F.; Torres Silva E Alves, A.; Spadoti, L.M.; Brandelli, A. Draft Genome Sequence and Comparative Genome Analysis Reveal Potential Functional Properties in Lacticaseibacillus paracasei ItalPN16. Curr. Microbiol. 2023, 80, 399. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Chen, J.; Wu, H.; Wang, N. KEGG Orthology Prediction of Bacterial Proteins Using Natural Language Processing. BMC Bioinform. 2024, 25, 146. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated Carbohydrate-Active Enzyme and Substrate Annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Mahram, A.; Herbordt, M.C. NCBI BLASTP on High-Performance Reconfigurable Computing Systems. ACM Trans. Reconfigurable Technol. Syst. 2015, 7, 1–20. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia Coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Guan, J.; Tai, C.; Weng, Y.; Chen, X.; Ou, H.-Y. oriTDB: A Database of the Origin-of-Transfer Regions of Bacterial Mobile Genetic Elements. Nucleic Acids Res. 2025, 53, D163–D168. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, Better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to Qualify Microorganisms as “Probiotic” in Foods and Dietary Supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Allende, A.; Alvarez-Ordóñez, A.; Bortolaia, V.; Bover-Cid, S.; De Cesare, A.; Dohmen, W.; Guillier, L.; Jacxsens, L.; Nauta, M.; et al. Update of the List of Qualified Presumption of Safety (QPS) Recommended Microbiological Agents Intentionally Added to Food or Feed as Notified to EFSA 21: Suitability of Taxonomic Units Notified to EFSA until September 2024. EFSA J. 2025, 23, e9169. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Hall, B.G.; Nisbet, J. Building Phylogenetic Trees from Genome Sequences with kSNP4. Mol. Biol. Evol. 2023, 40, msad235. [Google Scholar] [CrossRef]
- Inglin, R.C.; Meile, L.; Stevens, M.J.A. Clustering of Pan- and Core-Genome of Lactobacillus Provides Novel Evolutionary Insights for Differentiation. BMC Genom. 2018, 19, 284. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Zhang, C.; Li, T.; Jin, F.-J.; Sung, Y.-J.; Oh, H.-M.; Lee, H.-G.; Jin, L. Development and Applications of CRISPR/Cas9-Based Genome Editing in Lactobacillus. Int. J. Mol. Sci. 2022, 23, 12852. [Google Scholar] [CrossRef]
- Jarocki, P.; Komoń-Janczara, E.; Podleśny, M.; Kholiavskyi, O.; Pytka, M.; Kordowska-Wiater, M. Genomic and Proteomic Characterization of Bacteriophage BH1 Spontaneously Released from Probiotic Lactobacillus rhamnosus Pen. Viruses 2019, 11, 1163. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, S.; Lee, K.-H.; Yoo, J.; Yun, J.; Kang, H.B.; Kim, J.E.; Oh, M.-H.; Ham, J.-S. Whole Genome Sequencing of Lacticaseibacillus Casei KACC92338 Strain with Strong Antioxidant Activity, Reveals Genes and Gene Clusters of Probiotic and Antimicrobial Potential. Front. Microbiol. 2024, 15, 1458221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ji, H.; Zhang, D.; Liu, H.; Wang, S.; Wang, J.; Wang, Y. Complete Genome Sequencing of Lactobacillus plantarum ZLP001, a Potential Probiotic That Enhances Intestinal Epithelial Barrier Function and Defense Against Pathogens in Pigs. Front. Physiol. 2018, 9, 1689. [Google Scholar] [CrossRef]
- Kuo, Y.-C.; Liu, C.-F.; Lin, J.-F.; Li, A.-C.; Lo, T.-C.; Lin, T.-H. Characterization of Putative Class II Bacteriocins Identified from a Non-Bacteriocin-Producing Strain Lactobacillus casei ATCC 334. Appl. Microbiol. Biotechnol. 2013, 97, 237–246. [Google Scholar] [CrossRef]
- Chintakovid, N.; Singkhamanan, K.; Yaikhan, T.; Nokchan, N.; Wonglapsuwan, M.; Jitpakdee, J.; Kantachote, D.; Surachat, K. Probiogenomic Analysis of Lactiplantibacillus Plantarum SPS109: A Potential GABA-Producing and Cholesterol-Lowering Probiotic Strain. Heliyon 2024, 10, e33823. [Google Scholar] [CrossRef]
- Ali, A.; Happel, D.; Habermann, J.; Schoenfeld, K.; Macarrón Palacios, A.; Bitsch, S.; Englert, S.; Schneider, H.; Avrutina, O.; Fabritz, S.; et al. Sactipeptide Engineering by Probing the Substrate Tolerance of a Thioether-Bond-Forming Sactisynthase. Angew. Chem. Int. Ed. 2022, 61, e202210883. [Google Scholar] [CrossRef]
- Sornsenee, P.; Surachat, K.; Kang, D.-K.; Mendoza, R.; Romyasamit, C. Probiotic Insights from the Genomic Exploration of Lacticaseibacillus paracasei Strains Isolated from Fermented Palm Sap. Foods 2024, 13, 1773. [Google Scholar] [CrossRef] [PubMed]
- Romyasamit, C.; Surachat, K.; Pattaranggoon, N.C.; Suksabay, P.; Permpoon, U.; Nam, T.-G.; Sornsenee, P. Phenotypic and Genomic Insights into Schleiferilactobacillus harbinensis WU01, a Candidate Probiotic with Broad-Spectrum Antimicrobial Activity Against ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter) Pathogens. Foods 2025, 14, 1161. [Google Scholar] [CrossRef] [PubMed]
- Salini, F.; Iacumin, L.; Comi, G.; Dicks, L. Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production. Microorganisms 2023, 11, 611. [Google Scholar] [CrossRef]
- Santarelli, G.; Sanguinetti, M.; Delogu, G.; De Maio, F. Comparative Genomic Analysis of Bacteriocin Genes in Lactobacillus crispatus Strains. Sci. Rep. 2025, 15, 20798. [Google Scholar] [CrossRef]
- Moiseenko, K.V.; Begunova, A.V.; Savinova, O.S.; Glazunova, O.A.; Rozhkova, I.V.; Fedorova, T.V. Biochemical and Genomic Characterization of Two New Strains of Lacticaseibacillus paracasei Isolated from the Traditional Corn-Based Beverage of South Africa, Mahewu, and Their Comparison with Strains Isolated from Kefir Grains. Foods 2023, 12, 223. [Google Scholar] [CrossRef]
- Dishan, A.; Gönülalan, Z. Lacticaseibacillus paracasei AD22 Stress Response in Brined White Cheese Matrix: In Vitro Probiotic Profiles and Molecular Characterization. Probiotics Antimicrob. Proteins 2025, 17, 1725–1738. [Google Scholar] [CrossRef]
- El-Arabi, N.; Salim, R.; Abosereh, N.; Abdelhadi, A. Molecular Characterization of Some Antilisterial Bacteriocin Genes from Enterococcus faecium and Pediococcus pentosaceus. Microbiol. Biotechnol. Lett. 2018, 46, 288–299. [Google Scholar] [CrossRef]
- Surachat, K.; Sangket, U.; Deachamag, P.; Chotigeat, W. In Silico Analysis of Protein Toxin and Bacteriocins from Lactobacillus paracasei SD1 Genome and Available Online Databases. PLoS ONE 2017, 12, e0183548. [Google Scholar] [CrossRef]
- Liu, Z.-S.; Lin, C.-F.; Chen, P.-W. Transcriptome Analysis of Lactobacillus rhamnosus GG Strain Treated with Prebiotic—Bovine Lactoferrin under a Cold Environment. J. Food Drug Anal. 2021, 29, 402–418. [Google Scholar] [CrossRef]
- Zhang, Y.; Bao, C.; Shen, L.; Tian, C.; Zang, X.; Chen, G.; Zhang, S. Microbial Cold Shock Proteins: Overview of Their Function and Mechanism of Action. Protein Pept. Lett. 2022, 29, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Responses of Lactic Acid Bacteria to Cold Stress. In Stress Responses of Lactic Acid Bacteria; Springer: Boston, MA, USA, 2011; pp. 91–110. ISBN 978-0-387-92770-1. [Google Scholar]
- Gao, X.; Kong, J.; Zhu, H.; Mao, B.; Cui, S.; Zhao, J. Lactobacillus, Bifidobacterium and Lactococcus Response to Environmental Stress: Mechanisms and Application of Cross-protection to Improve Resistance against Freeze-drying. J. Appl. Microbiol. 2022, 132, 802–821. [Google Scholar] [CrossRef]
- Kim, M.-S.; Min, B.; Kim, I.; Kwon, Y.-J.; Park, S.-Y.; Kim, B.-Y.; Kim, B.-K.; Park, S.S. Complete Genome Sequencing and Comparative Genomic Analysis of Streptococcus thermophilus CKDB027, a Promising Probiotic Bacterial Strain. Food Suppl. Biomater. Health 2021, 1, e28. [Google Scholar] [CrossRef]
- Duru, I.C.; Ylinen, A.; Belanov, S.; Pulido, A.A.; Paulin, L.; Auvinen, P. Transcriptomic Time-Series Analysis of Cold- and Heat-Shock Response in Psychrotrophic Lactic Acid Bacteria. BMC Genom. 2021, 22, 28. [Google Scholar] [CrossRef]
- Michaux, C.; Martini, C.; Shioya, K.; Ahmed Lecheheb, S.; Budin-Verneuil, A.; Cosette, P.; Sanguinetti, M.; Hartke, A.; Verneuil, N.; Giard, J.-C. CspR, a Cold Shock RNA-Binding Protein Involved in the Long-Term Survival and the Virulence of Enterococcus Faecalis. J. Bacteriol. 2012, 194, 6900–6908. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Giard, J.-C. New Insight into Cold Shock Proteins: RNA-binding Proteins Involved in Stress Response and Virulence. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; Wiley: Hoboken, NJ, USA, 2016; pp. 873–880. ISBN 978-1-119-00488-2. [Google Scholar]
- Adu, K.T.; Wilson, R.; Nichols, D.S.; Baker, A.L.; Bowman, J.P.; Britz, M.L. Proteomic Analysis of Lactobacillus casei GCRL163 Cell-Free Extracts Reveals a SecB Homolog and Other Biomarkers of Prolonged Heat Stress. PLoS ONE 2018, 13, e0206317. [Google Scholar] [CrossRef]
- Alcántara, C.; Zúñiga, M. Proteomic and Transcriptomic Analysis of the Response to Bile Stress of Lactobacillus casei BL23. Microbiology 2012, 158, 1206–1218. [Google Scholar] [CrossRef]
- Heydari, A.; Parvini, F.; Allahyari Fard, N. Transcriptomic Analysis of Probiotic Oxidative Stress Resistance in Anti-Inflammatory Pathways. J. Genet. Resour. 2025, 11, 146–159. [Google Scholar] [CrossRef]
- Diaz, M.; Sayavedra, L.; Atter, A.; Mayer, M.J.; Saha, S.; Amoa-Awua, W.; Narbad, A. Lactobacillus garii Sp. Nov., Isolated from a Fermented Cassava Product. Int. J. Syst. Evol. Microbiol. 2020, 70, 3012–3017. [Google Scholar] [CrossRef]
- Noirot-Gros, M.-F.; Velten, M.; Yoshimura, M.; McGovern, S.; Morimoto, T.; Ehrlich, S.D.; Ogasawara, N.; Polard, P.; Noirot, P. Functional Dissection of YabA, a Negative Regulator of DNA Replication Initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2006, 103, 2368–2373. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Ogasawara, N.; Ishikawa, S. The Functional Analysis of YabA, Which Interacts with DnaA and Regulates Initiation of Chromosome Replication in Bacillus subtils. Genes Genet. Syst. 2008, 83, 111–125. [Google Scholar] [CrossRef]
- Balakrishnan, R.; Oman, K.; Shoji, S.; Bundschuh, R.; Fredrick, K. The Conserved GTPase LepA Contributes Mainly to Translation Initiation in Escherichia Coli. Nucleic Acids Res. 2014, 42, 13370–13383. [Google Scholar] [CrossRef] [PubMed]
- Pech, M.; Karim, Z.; Yamamoto, H.; Kitakawa, M.; Qin, Y.; Nierhaus, K.H. Elongation Factor 4 (EF4/LepA) Accelerates Protein Synthesis at Increased Mg2+ Concentrations. Proc. Natl. Acad. Sci. USA 2011, 108, 3199–3203. [Google Scholar] [CrossRef]
- Huang, C.-H.; Chen, C.-C.; Liou, J.-S.; Lee, A.-Y.; Blom, J.; Lin, Y.-C.; Huang, L.; Watanabe, K. Genome-Based Reclassification of Lactobacillus casei: Emended Classification and Description of the Species Lactobacillus zeae. Int. J. Syst. Evol. Microbiol. 2020, 70, 3755–3762. [Google Scholar] [CrossRef]
- Arena, M.P.; Capozzi, V.; Longo, A.; Russo, P.; Weidmann, S.; Rieu, A.; Guzzo, J.; Spano, G.; Fiocco, D. The Phenotypic Analysis of Lactobacillus plantarum Shsp Mutants Reveals a Potential Role for Hsp1 in Cryotolerance. Front. Microbiol. 2019, 10, 838. [Google Scholar] [CrossRef] [PubMed]
- Longo, A.; Russo, P.; Capozzi, V.; Spano, G.; Fiocco, D. Knock out of sHSP Genes Determines Some Modifications in the Probiotic Attitude of Lactiplantibacillus Plantarum. Biotechnol. Lett. 2021, 43, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Mora, D.; Eijlander, R.T.; Wels, M.; De Vos, W.M. The PLoS ONE Editors Correction: Comparative Genomic Analysis of the Multispecies Probiotic-Marketed Product VSL#3. PLoS ONE 2018, 13, e0203548. [Google Scholar] [CrossRef]
- Cheng, Z.; Chen, J.; Zhang, Y.; Li, X.; Zhang, N.; Liu, F.; Jiao, Y. In Vitro Hypoglycemic Activities of Lactobacilli and Bifidobacterium Strains from Healthy Children’s Sources and Their Effect on Stimulating GLP-1 Secretion in STC-1 Cells. Foods 2024, 13, 519. [Google Scholar] [CrossRef]
- Rivas-Sendra, A.; Landete, J.M.; Alcántara, C.; Zúñiga, M. Response of Lactobacillus casei BL23 to Phenolic Compounds: Response to Phenolic Compounds. J. Appl. Microbiol. 2011, 111, 1473–1481. [Google Scholar] [CrossRef]
- Backert, S.; Bernegger, S.; Skórko-Glonek, J.; Wessler, S. Extracellular HtrA Serine Proteases: An Emerging New Strategy in Bacterial Pathogenesis. Cell. Microbiol. 2018, 20, e12845. [Google Scholar] [CrossRef]
- Bove, P.; Capozzi, V.; Garofalo, C.; Rieu, A.; Spano, G.; Fiocco, D. Inactivation of the ftsH Gene of Lactobacillus plantarum WCFS1: Effects on Growth, Stress Tolerance, Cell Surface Properties and Biofilm Formation. Microbiol. Res. 2012, 167, 187–193. [Google Scholar] [CrossRef]
- Kapse, N.G.; Engineer, A.S.; Gowdaman, V.; Wagh, S.; Dhakephalkar, P.K. Genome Profiling for Health Promoting and Disease Preventing Traits Unraveled Probiotic Potential of Bacillus Clausii B106. Microbiol. Biotechnol. Lett. 2018, 46, 334–345. [Google Scholar] [CrossRef]
- Marrec, C.L. Responses of Lactic Acid Bacteria to Osmotic Stress. In Stress Responses of Lactic Acid Bacteria; Springer: Boston, MA, USA, 2011; pp. 67–90. ISBN 978-0-387-92770-1. [Google Scholar]
- Mollova, D.; Gozmanova, M.; Apostolova, E.; Yahubyan, G.; Iliev, I.; Baev, V. Illuminating the Genomic Landscape of Lactiplantibacillus Plantarum PU3—A Novel Probiotic Strain Isolated from Human Breast Milk, Explored through Nanopore Sequencing. Microorganisms 2023, 11, 2440. [Google Scholar] [CrossRef]
- Fan, X.; Bao, T.; Yi, H.; Zhang, Z.; Zhang, K.; Liu, X.; Lin, X.; Zhang, Z.; Feng, Z. Ribosome Profiling and RNA Sequencing Reveal Genome-Wide Cellular Translation and Transcription Regulation Under Osmotic Stress in Lactobacillus rhamnosus ATCC 53103. Front. Microbiol. 2021, 12, 781454. [Google Scholar] [CrossRef]
- Sevillano, E.; Lafuente, I.; Peña, N.; Cintas, L.M.; Muñoz-Atienza, E.; Hernández, P.E.; Borrero, J. Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S. Foods 2023, 13, 107. [Google Scholar] [CrossRef]
- Yogeswara, I.B.A.; Mariyatun; Pramesi, P.C.; Rahayu, E.S. Whole-Genome Sequence of Lactiplantibacillus plantarum Mut-3, Isolated from Indonesian Fermented Soybean (Tempeh). Microbiol. Resour. Announc. 2023, 12, e0051322. [Google Scholar] [CrossRef] [PubMed]
- Sałański, P.; Kowalczyk, M.; Bardowski, J.K.; Szczepankowska, A.K. Health-Promoting Nature of Lactococcus Lactis IBB109 and Lactococcus Lactis IBB417 Strains Exhibiting Proliferation Inhibition and Stimulation of Interleukin-18 Expression in Colorectal Cancer Cells. Front. Microbiol. 2022, 13, 822912. [Google Scholar] [CrossRef]
- Ma, X.; Wang, G.; Zhai, Z.; Zhou, P.; Hao, Y. Global Transcriptomic Analysis and Function Identification of Malolactic Enzyme Pathway of Lactobacillus paracasei L9 in Response to Bile Stress. Front. Microbiol. 2018, 9, 1978. [Google Scholar] [CrossRef] [PubMed]
- Kusada, H.; Arita, M.; Tohno, M.; Tamaki, H. Bile Salt Hydrolase Degrades β-Lactam Antibiotics and Confers Antibiotic Resistance on Lactobacillus paragasseri. Front. Microbiol. 2022, 13, 858263. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Ferrer, S.; Monedero, V.; Zúñiga, M. Malic Enzyme and Malolactic Enzyme Pathways Are Functionally Linked but Independently Regulated in Lactobacillus casei BL23. Appl. Environ. Microbiol. 2013, 79, 5509–5518. [Google Scholar] [CrossRef]
- Horn, N.; Wegmann, U.; Dertli, E.; Mulholland, F.; Collins, S.R.A.; Waldron, K.W.; Bongaerts, R.J.; Mayer, M.J.; Narbad, A. Spontaneous Mutation Reveals Influence of Exopolysaccharide on Lactobacillus johnsonii Surface Characteristics. PLoS ONE 2013, 8, e59957. [Google Scholar] [CrossRef]
- Deo, D.; Davray, D.; Kulkarni, R. A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms 2019, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liang, Q.; Song, X.; Zhang, Y. Whole Genome Sequence of Lactiplantibacillus Plantarum MC5 and Comparative Analysis of Eps Gene Clusters. Front. Microbiol. 2023, 14, 1146566. [Google Scholar] [CrossRef]
- Muñoz-Provencio, D.; Pérez-Martínez, G.; Monedero, V. Characterization of a Fibronectin-binding Protein from Lactobacillus casei BL23. J. Appl. Microbiol. 2010, 108, 1050–1059. [Google Scholar] [CrossRef]
- Von Ossowski, I.; Satokari, R.; Reunanen, J.; Lebeer, S.; De Keersmaecker, S.C.J.; Vanderleyden, J.; De Vos, W.M.; Palva, A. Functional Characterization of a Mucus-Specific LPXTG Surface Adhesin from Probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2011, 77, 4465–4472. [Google Scholar] [CrossRef]
- Souza, R.F.S.; Jardin, J.; Cauty, C.; Rault, L.; Bouchard, D.S.; Bermúdez-Humarán, L.G.; Langella, P.; Monedero, V.; Seyffert, N.; Azevedo, V.; et al. Contribution of Sortase SrtA2 to Lactobacillus casei BL23 Inhibition of Staphylococcus aureus Internalization into Bovine Mammary Epithelial Cells. PLoS ONE 2017, 12, e0174060. [Google Scholar] [CrossRef]
- Van Baarlen, P.; Wells, J.M.; Kleerebezem, M. Regulation of Intestinal Homeostasis and Immunity with Probiotic Lactobacilli. Trends Immunol. 2013, 34, 208–215. [Google Scholar] [CrossRef]
- Rossi, F.; Amadoro, C.; Pallotta, M.L.; Colavita, G. Variability of Genetic Characters Associated with Probiotic Functions in Lacticaseibacillus Species. Microorganisms 2022, 10, 1023. [Google Scholar] [CrossRef]
- Bäuerl, C.; Abitayeva, G.; Sosa-Carrillo, S.; Mencher-Beltrán, A.; Navarro-Lleó, N.; Coll-Marqués, J.M.; Zúñiga-Cabrera, M.; Shaikhin, S.; Pérez-Martinez, G. P40 and P75 Are Singular Functional Muramidases Present in the Lactobacillus casei/paracasei/rhamnosus Taxon. Front. Microbiol. 2019, 10, 1420. [Google Scholar] [CrossRef]
- Salzillo, M.; Vastano, V.; Capri, U.; Muscariello, L.; Marasco, R. Pyruvate Dehydrogenase Subunit β of Lactobacillus plantarum Is a Collagen Adhesin Involved in Biofilm Formation. J. Basic Microbiol. 2017, 57, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Sotoodehnejadnematalahi, F.; Amiri, M.M.; Pourshafie, M.R.; Rohani, M. Decreased Mucosal Adhesion of Lactobacillus Species in Patients with Inflammatory Bowel Disease. Casp. J. Intern. Med. 2022, 13, 713–720. [Google Scholar] [CrossRef]
- Broadbent, J.R.; Larsen, R.L.; Deibel, V.; Steele, J.L. Physiological and Transcriptional Response of Lactobacillus casei ATCC 334 to Acid Stress. J. Bacteriol. 2010, 192, 2445–2458. [Google Scholar] [CrossRef]
- Terán, L.C.; Mortera, P.; Tubio, G.; Alarcón, S.H.; Blancato, V.S.; Espariz, M.; Esteban, L.; Magni, C. Genomic Analysis Revealed Conserved Acid Tolerance Mechanisms from Native Micro-organisms in Fermented Feed. J. Appl. Microbiol. 2022, 132, 1152–1165. [Google Scholar] [CrossRef]
- Najar, I.N.; Sharma, P.; Das, R.; Mondal, K.; Singh, A.K.; Radha, A.; Sharma, V.; Sharma, S.; Thakur, N.; Gandhi, S.G.; et al. Exploring Probiotic Potential: A Comparative Genomics and In Silico Assessment of Genes within the Genus Geobacillus. bioRxiv 2024. [Google Scholar] [CrossRef]
- Duary, R.K.; Batish, V.K.; Grover, S. Expression of the atpD Gene in Probiotic Lactobacillus plantarum Strains under in Vitro Acidic Conditions Using RT-qPCR. Res. Microbiol. 2010, 161, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Senan, S.; Prajapati, J.B.; Joshi, C.G. Comparative Genome-Scale Analysis of Niche-Based Stress-Responsive Genes in Lactobacillus helveticus Strains. Genome 2014, 57, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Kamarinou, C.S.; Kiousi, D.E.; Repanas, P.; Argyri, A.A.; Chorianopoulos, N.G.; Galanis, A. Dissecting the Genetic Basis of the Technological, Functional, and Safety Characteristics of Lacticaseibacillus paracasei SRX10. Microorganisms 2024, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, J.; Zhang, D.; Liu, H.; Wang, S.; Wang, Y.; Ji, H. Complete Genome Sequencing and Comparative Genome Characterization of Lactobacillus johnsonii ZLJ010, a Potential Probiotic with Health-Promoting Properties. Front. Genet. 2019, 10, 812. [Google Scholar] [CrossRef]
- Park, S.; Kwon, H.; Tran, T.D.; Choi, M.; Jung, S.; Lee, S.; Briles, D.E.; Rhee, D. ClpL Is a Chaperone without Auxiliary Factors. FEBS J. 2015, 282, 1352–1367. [Google Scholar] [CrossRef]
- Chen, M.-J.; Tang, H.-Y.; Chiang, M.-L. Effects of Heat, Cold, Acid and Bile Salt Adaptations on the Stress Tolerance and Protein Expression of Kefir-Isolated Probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 2017, 66, 20–27. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Efstathiou, C.; Tegopoulos, K.; Mantzourani, I.; Alexopoulos, A.; Plessas, S.; Kolovos, P.; Koffa, M.; Galanis, A. Genomic Insight Into Lacticaseibacillus paracasei SP5, Reveals Genes and Gene Clusters of Probiotic Interest and Biotechnological Potential. Front. Microbiol. 2022, 13, 922689. [Google Scholar] [CrossRef]
- Nakano, A.; Kishikawa, J.; Nakanishi, A.; Mitsuoka, K.; Yokoyama, K. Structural Basis of Unisite Catalysis of Bacterial F0F1-ATPase. PNAS Nexus 2022, 1, pgac116. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Mu, G.; Tuo, Y. Interpretation of Safety and Potential Probiotic Properties of Lactiplantibacillus plantarum Y42 Based on Genome-Wide Sequencing. Food Biosci. 2023, 56, 103249. [Google Scholar] [CrossRef]
- Bäuerl, C.; Pérez-Martínez, G.; Yan, F.; Polk, D.B.; Monedero, V. Functional Analysis of the P40 and P75 Proteins from Lactobacillus casei BL23. Microb. Physiol. 2010, 19, 231–241. [Google Scholar] [CrossRef]
- Singh, T.P.; Tehri, N.; Kaur, G.; Malik, R.K. Cell Surface and Extracellular Proteins of Potentially Probiotic Lactobacillus reuteri as an Effective Mediator to Regulate Intestinal Epithelial Barrier Function. Arch. Microbiol. 2021, 203, 3219–3228. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain. EFSA J. 2024, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Joint FAO/WHO Food Standards Programme; Codex Committee on Nutrition and Foods for Special Dietary Uses. Discussion Paper on Harmonized Probiotic Guidelines for Use in Foods and Food Supplements; FAO/WHO: Dresden, Germany, 2024.
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; De La Cruz, F. Mobility of Plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef]
- Park, C.-H.; Yang, H.; Kim, S.; Yun, C.-S.; Jang, B.-C.; Hong, Y.-J.; Park, D.-S. Comparison of Plasmid Curing Efficiency across Five Lactic Acid Bacterial Species. J. Microbiol. Biotechnol. 2024, 34, 2385–2395. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal Transfer of Antibiotic Resistance Genes in Clinical Environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrzak-Piekarczyk, T.; Koryszewska-Bagińska, A.; Grynberg, M.; Nowak, A.; Cukrowska, B.; Kozakova, H.; Bardowski, J. Genomic and Functional Characterization of the Unusual pLOCK 0919 Plasmid Harboring the spaCBA Pili Cluster in Lactobacillus casei LOCK 0919. Genome Biol. Evol. 2016, 8, 202–217. [Google Scholar] [CrossRef]
- Fernández-López, C.; Bravo, A.; Ruiz-Cruz, S.; Solano-Collado, V.; Garsin, D.A.; Lorenzo-Díaz, F.; Espinosa, M. Mobilizable Rolling-Circle Replicating Plasmids from Gram-Positive Bacteria: A Low-Cost Conjugative Transfer. In Plasmids; Tolmasky, M.E., Alonso, J.C., Eds.; ASM Press: Washington, DC, USA, 2015; pp. 257–276. ISBN 978-1-68367-097-1. [Google Scholar]
- Zhang, H.; Hao, Y.; Zhang, D.; Luo, Y. Characterization of the Cryptic Plasmid pTXW from Lactobacillus paracasei TXW. Plasmid 2011, 65, 1–7. [Google Scholar] [CrossRef]
- Belloso Daza, M.V.; Milani, G.; Cortimiglia, C.; Pietta, E.; Bassi, D.; Cocconcelli, P.S. Genomic Insights of Enterococcus faecium UC7251, a Multi-Drug Resistant Strain from Ready-to-Eat Food, Highlight the Risk of Antimicrobial Resistance in the Food Chain. Front. Microbiol. 2022, 13, 894241. [Google Scholar] [CrossRef] [PubMed]
- Aziz, G.; Zaidi, A.; Sullivan, D.J. O’. Insights from Metagenome-Assembled Genomes on the Genetic Stability and Safety of over-the-Counter Probiotic Products. Curr. Genet. 2023, 69, 213–234. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.K.; Shah, N.P.; Mishra, V. Conjugal Transfer of Antibiotic Resistances in Lactobacillus spp. Curr. Microbiol. 2021, 78, 2839–2849. [Google Scholar] [CrossRef]
- Scardaci, R.; Bietto, F.; Racine, P.-J.; Boukerb, A.M.; Lesouhaitier, O.; Feuilloley, M.G.J.; Scutera, S.; Musso, T.; Connil, N.; Pessione, E. Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved? Microorganisms 2022, 10, 487. [Google Scholar] [CrossRef]
- Kong, L.; Su, M.; Sang, J.; Huang, S.; Wang, M.; Cai, Y.; Xie, M.; Wu, J.; Wang, S.; Foster, S.J.; et al. The W-Acidic Motif of Histidine Kinase Walk Is Required for Signaling and Transcriptional Regulation in Streptococcus Mutans. Front. Microbiol. 2022, 13, 820089. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Chen, Y.; Wang, L.; Li, L.; Zhang, X.; Ma, Z.; Liu, J. Screening, Identification, and Whole-Genome Sequencing of Ferulic Acid Esterase-Producing Lactic Acid Bacteria from Sheep Rumen. Microorganisms 2025, 13, 1295. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. Origins and Evolution of CRISPR-Cas Systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Kirchner, M.; Schneider, S. CRISPR-Cas: From the Bacterial Adaptive Immune System to a Versatile Tool for Genome Engineering. Angew. Chem. Int. Ed. 2015, 54, 13508–13514. [Google Scholar] [CrossRef]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas System and Its Role in Phage-Bacteria Interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Richter, H.; Charpentier, E.; Qimron, U. Adaptation in CRISPR-Cas Systems. Mol. Cell 2016, 61, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Abitaeva, G.K.; Sarmurzina, Z.S.; Bisenova, G.N.; Musabayeva, B.K.; Tultabayeva , C.T. Characteristics of probiotic strains for the development of preventive drinks. Microbiol. Virol. 2022, 4, 142–158. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matošić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]


| Sequencing Assembly | Structural Annotation | ||
|---|---|---|---|
| Assembled genome size, bp | 3,066,038 | G+C content (%) | 46.25% |
| Number of contigs | 219 | CDS | 2932 |
| Longest contig, bp | 195,833 | tRNAs | 59 |
| Shortest contig, bp | 129 | tmRNAs | 1 |
| Contig number (>1000 bp) | 75 | rRNAs | 6 |
| Contig number (>5000 bp) | 49 | CRISPR number | 2 |
| Contig number (>10,000 bp) | 40 | Prophages | 3 intact and 1 incomplete |
| Contig number (>25,000 bp) | 32 | ||
| Contig number (>50,000 bp) | 22 | ||
| Metric N50 | 105,408 | ||
| Metric N90 | 27,962 | ||
| CAZy Property | Numbers of Genes | Percentage (%) |
|---|---|---|
| AA | 3 | 3.53 |
| CBM | 2 | 2.35 |
| CE | 5 | 5.88 |
| GH | 40 | 47.06 |
| GT | 33 | 38.82 |
| PL | 2 | 2.35 |
| Total | 85 |
| Category | Role/Function | Genes | References |
|---|---|---|---|
| Cold shock | Cold shock protein | cspA | [59,60] |
| Cold shock protein | cspB | [61] | |
| Cold shock protein | cspC | [62] | |
| Cold shock protein | cspG | [63,64] | |
| Cold shock protein | cspR | [65,66] | |
| Heat shock | Chaperone protein | DnaK | [67,68] |
| Heat shock protein | GrpE | [69] | |
| tmRNA-binding protein | SmpB | [70] | |
| DNA replication intiation control protein | YabA | [71,72] | |
| Translation elongation factor | LepA | [73,74] | |
| Chaperone protein | DnaJ | [75] | |
| Belongs to the small heat shock protein (HSP20) family | hsp1 | [76] | |
| Belongs to the small heat shock protein (HSP20) family | hsp3 | [76,77] | |
| Part of a stress-induced multi-chaperone system, it is involved in the recovery of the cell from heat-induced damage, in cooperation with DnaK, DnaJ and GrpE | clpC | [78,79] | |
| It cleaves misfolded, damaged, or unnecessary proteins and also plays a role in regulating cellular processes. | Clp protease | [80] | |
| serine protease | htrA | [80,81] | |
| Acts as a processive, ATP-dependent zinc metallopeptidase for both cytoplasmic and membrane proteins. Plays a role in the quality control of integral membrane proteins | ftsH | [82] | |
| Osmotic stress | Glycine betaine ABC transport system, ATP-binding protein | OpuAA (EC 3.6.3.32) | [83] |
| Glycine betaine ABC transport system, glycine betaine-binding protein | OpuAC | [84] | |
| Osmotically activated L-carnitine/choline ABC transporter, ATP-binding protein | OpuCA | [85] | |
| Osmotically activated L-carnitine/choline ABC transporter, permease protein | OpuCB | [86] | |
| L-proline glycine betaine ABC transport system permease protein | ProV (TC 3.A.1.12.1) | [87] | |
| Osmotically activated L-carnitine/choline ABC transporter, substrate-binding protein | OpuCC | [88] | |
| Glycine betaine ABC transport system, permease protein | OpuAB | [89] | |
| Osmotically activated L-carnitine/choline ABC transporter, permease protein | OpuCD | [88] | |
| L-proline glycine betaine binding ABC transporter protein | ProX (TC 3.A.1.12.1) | [87] | |
| Channel that opens in response to stretch forces in the membrane lipid bilayer. May participate in the regulation of osmotic pressure changes within the cell | mscL | [84] | |
| Bile salt | Linear amide C-N hydrolases, choloylglycine hydrolase family | - | [90] |
| Linear amide C-N hydrolases, choloylglycine hydrolase family | - | [91] | |
| Sodium bile acid symporter family | mleP | [92] | |
| Catalyzes the addition of an amino acid to the nucleotide precursor UDP-N-acetylmuramoyl-L-alanyl-D-glutamate (UMAG) in the biosynthesis of bacterial cell-wall peptidoglycan | murE | [51] | |
| Malic enzyme | mleS | [92] | |
| Adhesion | Biosynthesis protein | epsB | [93,94] |
| Glycosyltransferase like family 2 | epsG | [95] | |
| Fibronectin-binding protein | FbpA | [14,96] | |
| MucBP domain | inlJ | [97,98] | |
| CHAP domain | p40 | [99,100] | |
| NlpC P60 family protein | p75 | [99,101] | |
| Transketolase, C-terminal domain protein | pdhB | [102] | |
| hydrolase, family 65, central catalytic | mapA | [103] | |
| pH | Produces ATP from ADP in the presence of a proton gradient across the membrane | atpC | [104,105,106] |
| ATP synthase (with catalytic β subunits like AtpD in bacteria) produces ATP from ADP using energy from a proton gradient | atpD | [105,106,107] | |
| ATP synthase produces ATP from ADP using a proton gradient, with the γ subunit regulating enzyme activity and proton flow through CF0 | atpG | [106,108] | |
| ATP synthase produces ATP from ADP using a proton gradient, with its alpha subunit serving a regulatory role | atpA | [105,106] | |
| F1F0 ATP synthase generates ATP from ADP using a proton/sodium gradient, coupling catalytic F1 rotation with F0 proton translocation | atpH | [105,106] | |
| Component of the F(0) channel; it forms part of the peripheral stalk, linking F(1) to F(0) | atpF | [105,106] | |
| F1F0 ATP synthase harnesses proton/sodium gradients to drive ATP synthesis via rotational coupling between F1 catalytic core and F0 proton channel | atpE | [105,106] | |
| atpB plays a direct role in the translocation of protons across the membrane | atpB | [105,106,109] |
| Protein ID | Gene Name | Identity | Coverage | e-Value | Function | Gene ID |
|---|---|---|---|---|---|---|
| S4E4Q5 | WalR | 83.40 | 99.57 | 1.24 × 10−115 | Transcriptional regulatory protein | EPH95667 |
| Q82ZX2 | CspR | 89.39 | 98.51 | 6.13 × 10−37 | Cold shock protein | AAO82613 |
| Sequence | Spacer/Gene | CRISPR_Id | CRISPR_Start | CRISPR_End | CRISPR_Length | Direction | Consensus_Repeat | Evidence Level |
|---|---|---|---|---|---|---|---|---|
| NODE_20_length_63718_cov_19.819975 | 2 | NODE_20_length_63718_cov_19.819975_crispr_1 | 41,374 | 41,641 | 267 | ND | TATGTGGAGGTTTCTGCGACTGTGAGCGCGTTTCCGAGCGAAGCGTGGC | 1 |
| NODE_27_length_38928_cov_17.618979 | 31 | NODE_27_length_38928_cov_17.618979_1 | 30,678 | 32,759 | 2081 | ND | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abitayeva, G.; Kurmangali, D.; Baikonys, T.; Bekshin, Z. Lacticaseibacillus paracasei subsp. paracasei 2LB: Identification of Genes to Assess the Safety and Probiotic Potential of the Strain. Foods 2025, 14, 3449. https://doi.org/10.3390/foods14193449
Abitayeva G, Kurmangali D, Baikonys T, Bekshin Z. Lacticaseibacillus paracasei subsp. paracasei 2LB: Identification of Genes to Assess the Safety and Probiotic Potential of the Strain. Foods. 2025; 14(19):3449. https://doi.org/10.3390/foods14193449
Chicago/Turabian StyleAbitayeva, Gulyaim, Diana Kurmangali, Temirlan Baikonys, and Zhandarbek Bekshin. 2025. "Lacticaseibacillus paracasei subsp. paracasei 2LB: Identification of Genes to Assess the Safety and Probiotic Potential of the Strain" Foods 14, no. 19: 3449. https://doi.org/10.3390/foods14193449
APA StyleAbitayeva, G., Kurmangali, D., Baikonys, T., & Bekshin, Z. (2025). Lacticaseibacillus paracasei subsp. paracasei 2LB: Identification of Genes to Assess the Safety and Probiotic Potential of the Strain. Foods, 14(19), 3449. https://doi.org/10.3390/foods14193449





