Non-Targeted Screening and Quantitative Analysis of Pesticides and Veterinary Drug Residues in Brassica rapa chinensis Using an Improved Quechers Method Based on Magnetic Materials
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Instruments
2.2. Synthesis of Magnetic Multi-Walled Carbon Nanotubes (MG-MWCNTs)
2.3. Sample Preparation
2.4. Determination of Co-Extracts and ME
2.5. Instrument Conditions
2.6. Method Validation
3. Results and Discussion
3.1. Characterization of MG-MWCNTs
3.2. Optimization of Pre-Processing
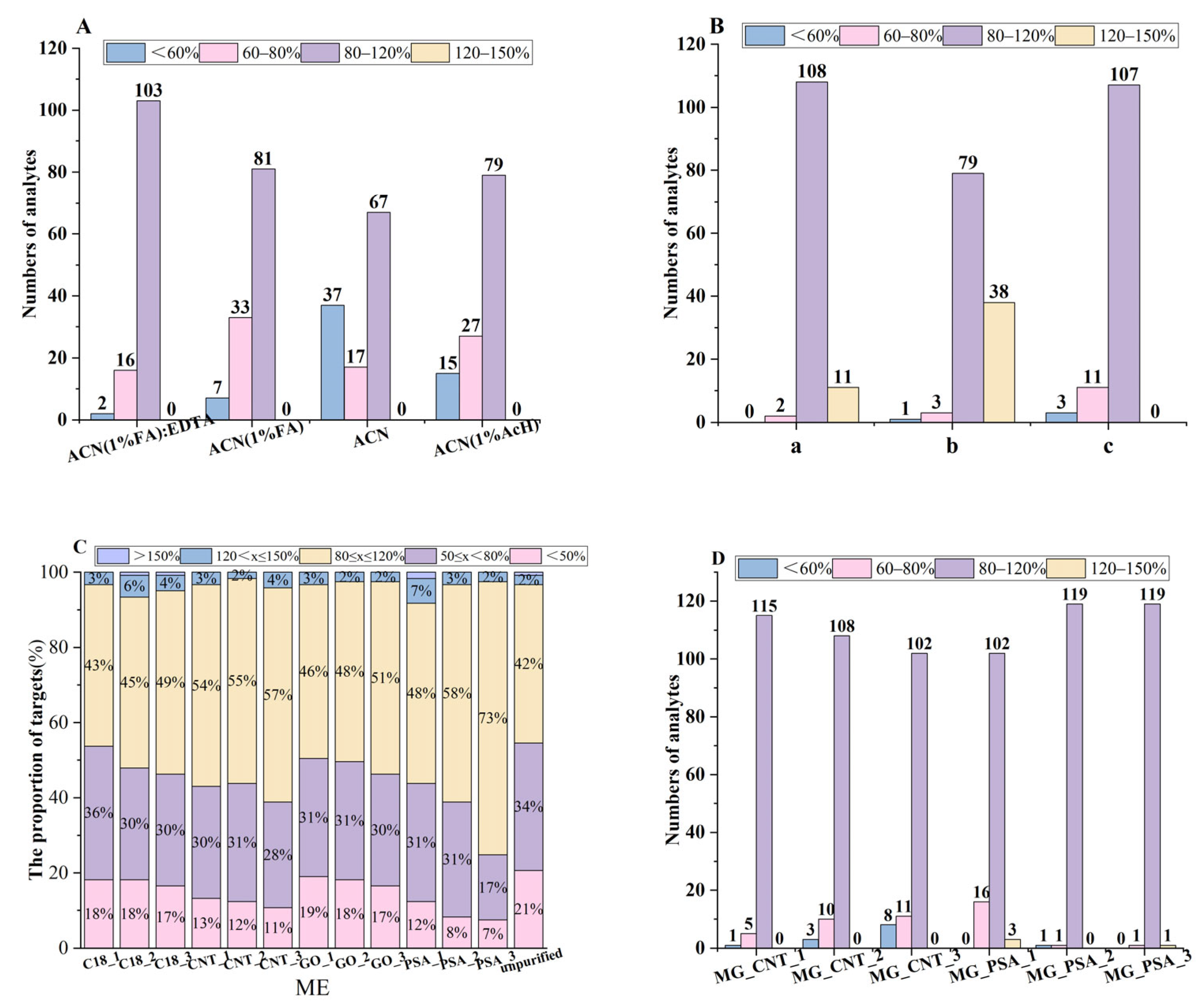
3.2.1. Optimization of Extraction Conditions
3.2.2. Optimization of Magnetic Nanomaterials
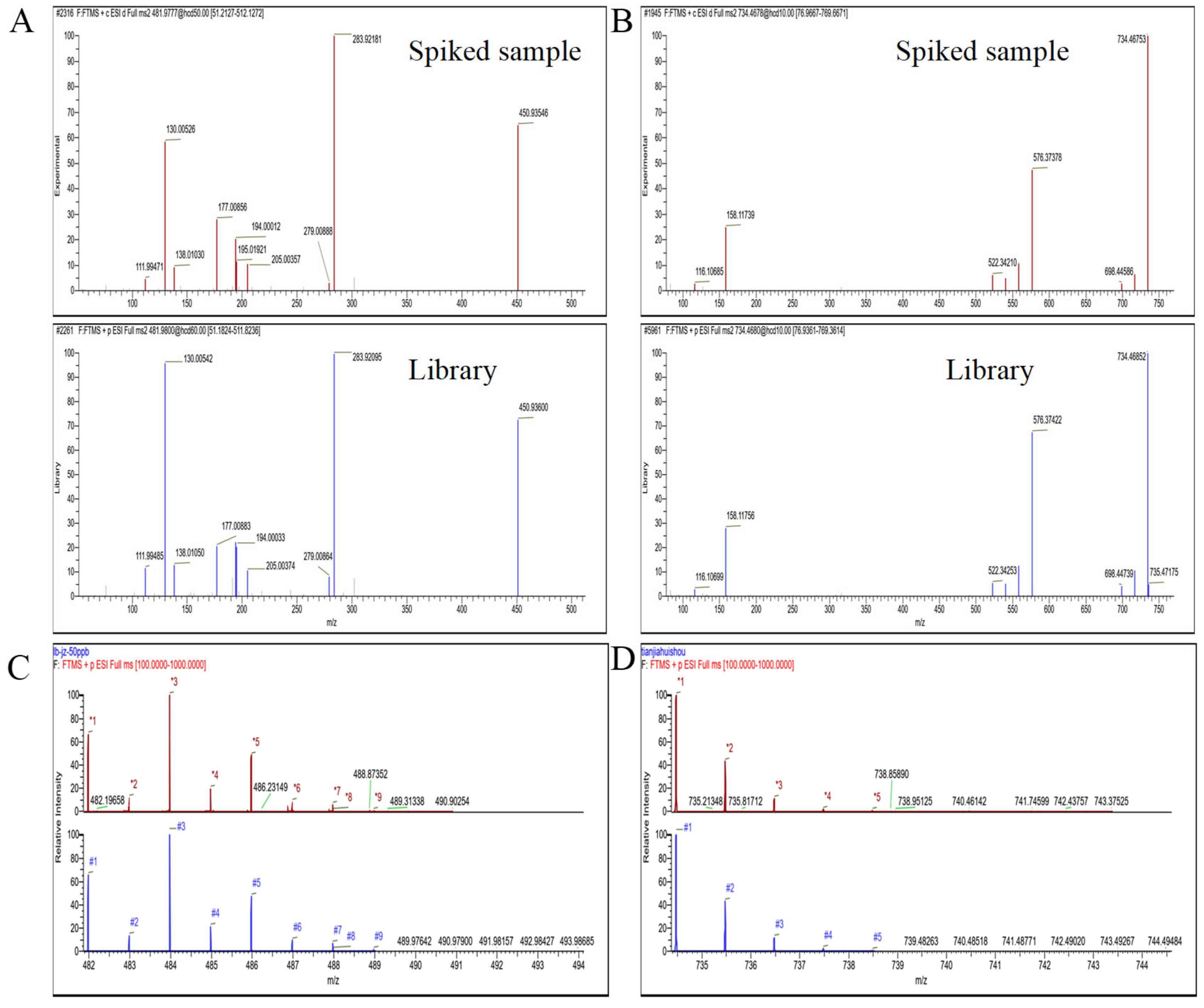
3.3. Qualitative Analysis
3.4. Quantitative Analysis
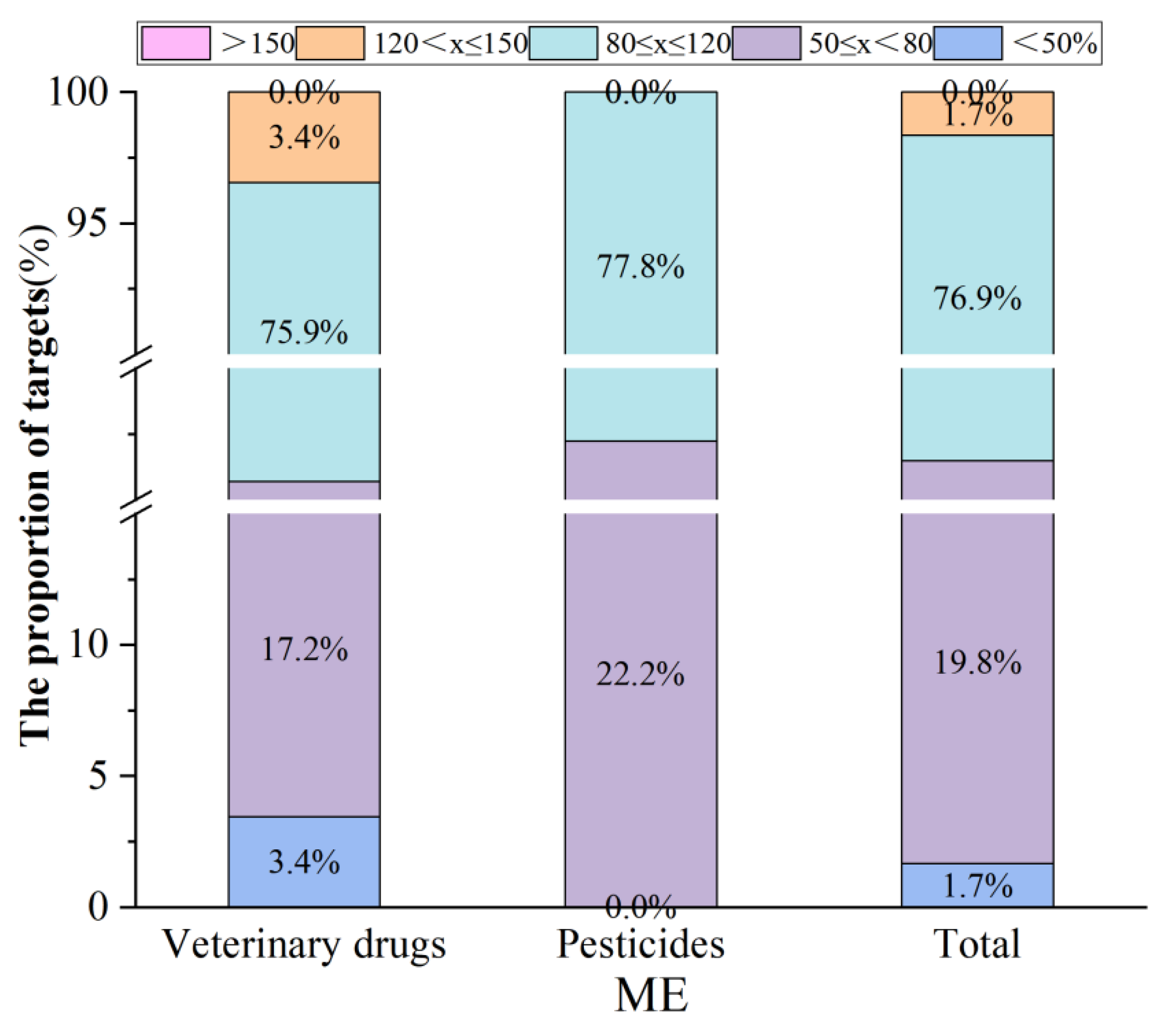
3.4.1. Matrix Effect (ME)
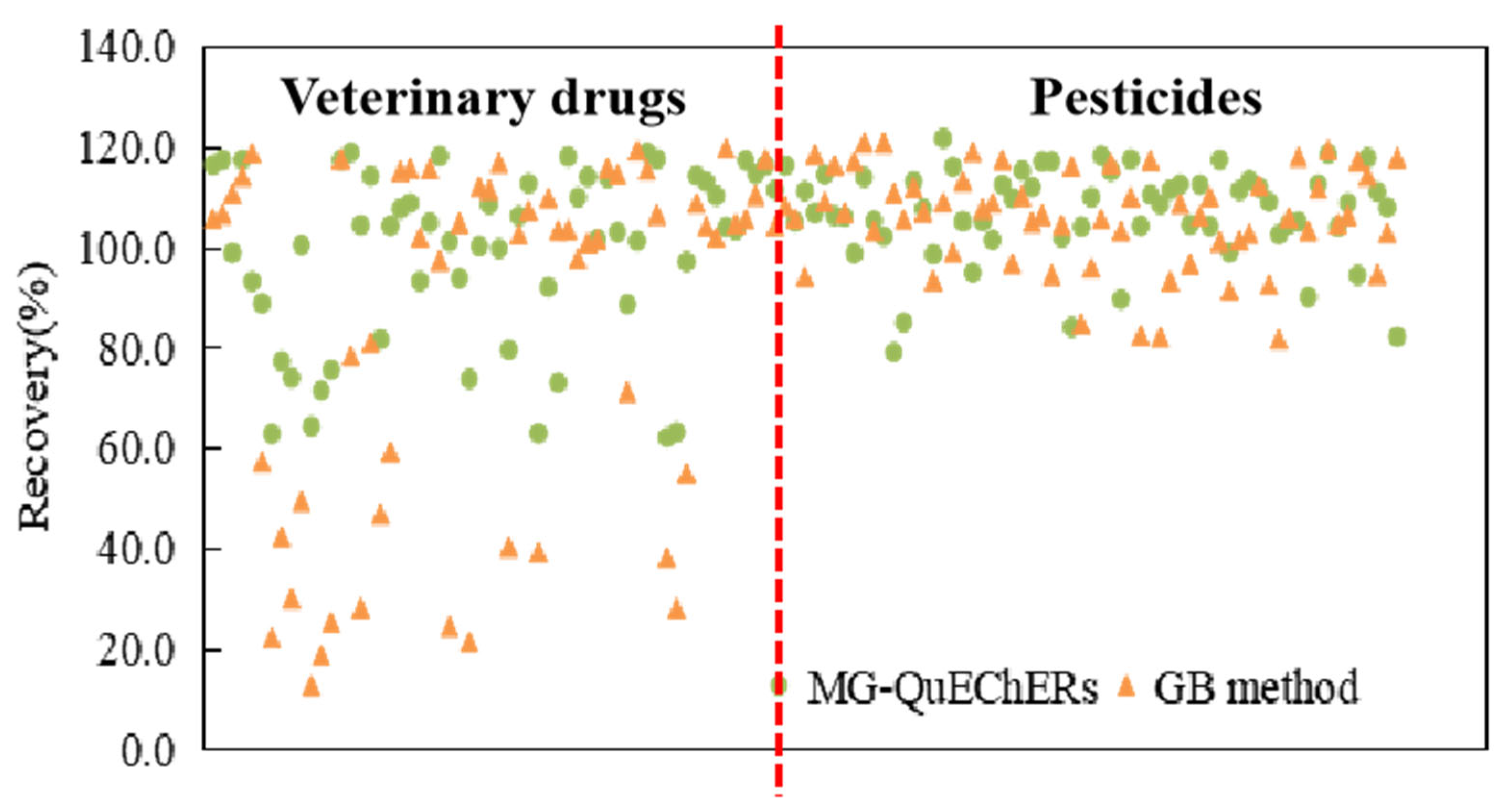
3.4.2. Linear Range, Quantification Limit, and Recovery Rates
3.5. The Detection of Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fang, H.; Han, Y.; Yin, Y.; Pan, X.; Yu, Y. Variations in dissipation rate, microbial function and antibiotic resistance due to repeated introductions of manure containing sulfadiazine and chlortetracycline to soil. Chemosphere 2014, 96, 51–56. [Google Scholar] [CrossRef]
- Fang, L.; Chen, C.; Zhang, F.; Ali, E.F.; Sarkar, B.; Rinklebe, J.; Shaheen, S.M.; Chen, X.; Xiao, R. Occurrence profiling and environmental risk assessment of veterinary antibiotics in vegetable soils at Chongqing region, China. Environ. Res. 2023, 227, 115799. [Google Scholar] [CrossRef]
- Khetagoudar, M.C.; Jinendra, U.; Kumar, A.P.; Bilehal, D.; Kollur, S.P. Multiresidue pesticide analysis in green chilli using GC–MS/MS using modified QuEChERS method with highly efficient Fe3O4@CFR@GO nanocomposite. Inorg. Chem. Commun. 2022, 137, 109195. [Google Scholar] [CrossRef]
- Yuan, H.; Li, B.; Wei, J.; Liu, X.; He, Z. Ultra-high performance liquid chromatography and gas chromatography coupled to tandem mass spectrometry for the analysis of 32 pyrethroid pesticides in fruits and vegetables: A comparative study. Food Chem. 2023, 412, 135578. [Google Scholar] [CrossRef]
- Na, T.W.; Seo, H.-J.; Jang, S.-N.; Kim, H.; Yun, H.; Kim, H.; Ahn, J.; Cho, H.; Hong, S.-H.; Kim, H.J.; et al. Multi-residue analytical method for detecting pesticides, veterinary drugs, and mycotoxins in feed using liquid- and gas chromatography coupled with mass spectrometry. J. Chromatogr. A 2022, 1676, 463257. [Google Scholar] [CrossRef]
- Ren, X.; Yao, Y.; Wu, X.; Wu, L.; Guo, H.; Yin, D.; Song, P.; Cui, J.; Jiang, N. Analysis of 16 pesticides in chive, cowpea and cabbage using a modified QuEChERS method with hydrophobic deep eutectic solvents as analyte protectants by GC-MS/MS. Microchem. J. 2025, 215, 114398. [Google Scholar] [CrossRef]
- Krauss, M.; Singer, H.; Hollender, J. Lc-high resolution MS in environmental analysis: From target screening to the identification of unknowns. Anal. Bioanal. Chem. 2010, 397, 943–951. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, Y.; Ge, N.; Liu, X.; Chang, Q.; Fan, C.; Pang, G.-F. Wide-scope screening of pesticides in fruits and vegetables using information-dependent acquisition employing UHPLC-QTOF-MS and automated MS/MS library searching. Anal. Bioanal. Chem. 2016, 408, 7795–7810. [Google Scholar] [CrossRef]
- Merlo, F.; Centenaro, D.; Maraschi, F.; Profumo, A.; Speltini, A. Green and efficient determination of fluoroquinolone residues in edible green fruits and leafy vegetables by ultrasound-assisted extraction followed by HPLC-MS/MS. Molecules 2022, 27, 6595. [Google Scholar] [CrossRef] [PubMed]
- Amelin, V.G.; Lavrukhina, O.I.; Tretyakov, A.V.; Batov, I.V.; Kish, L.K. Sample screening and determination of 214 veterinary drug residues in food using chromatography—high-resolution mass spectrometry. J. Anal. Chem. 2024, 79, 200–218. [Google Scholar] [CrossRef]
- Zhai, T.; Qiao, D.; Wang, J.; Li, C.-Y.; Yang, L.; Wang, J.; Wu, J.; Liu, Q.; Liu, J.-M.; Wang, S. Facile preparation of hollow covalent organic frameworks as superior and universal matrix clean-up micro-structures for high throughout determination of food hazards. Food Chem. 2024, 454, 139754. [Google Scholar] [CrossRef]
- Chen, W.L.; Yu, S.Y.; Liu, S.Y.; Lin, S.C.; Lee, T.H. Using HRMS fingerprinting to explore micropollutant contamination in soil and vegetables caused by swine wastewater irrigation. Sci. Total Environ. 2023, 862, 160830. [Google Scholar] [CrossRef]
- Anastassiades, M.; Lehotay, S.J.; Štajnbaher, D.; Schenck, F.J. Fast and easy multiresidue method employing acetonitrile extraction/partitioning and “dispersive solid-phase extraction” for the determination of pesticide residues in produce. J. AOAC Int. 2019, 86, 412–431. [Google Scholar] [CrossRef]
- Hercegova, A.; Domotorova, M.; Matisova, E. Sample preparation methods in the analysis of pesticide residues in baby food with subsequent chromatographic determination. J. Chromatogr. A 2007, 1153, 54–73. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-B.; Zhao, Q.; Mo, J.-Z.; Huang, Y.-Q.; Luo, Y.-B.; Yu, Q.-W.; Feng, Y.-Q. Quick, easy, cheap, effective, rugged and safe method with magnetic graphitized carbon black and primary secondary amine as adsorbent and its application in pesticide residue analysis. J. Chromatogr. A 2013, 1300, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Goon, A.; Khan, Z.; Oulkar, D.; Shinde, R.; Gaikwad, S.; Banerjee, K. simultaneous screening and quantitative method for the multiresidue analysis of pesticides in spices using ultra-high performance liquid chromatography-high resolution (orbitrap) mass spectrometry. J. Chromatogr. A 2018, 1532, 105–111. [Google Scholar] [CrossRef]
- Rajski, L.; Petromelidou, S.; Diaz-Galiano, F.J.; Ferrer, C.; Fernandez-Alba, A.R. Improving the simultaneous target and non-target analysis LC-amenable pesticide residues using high speed orbitrap mass spectrometry with combined multiple acquisition modes. Talanta 2021, 228, 122241. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Hao, D.; Chu, Q.; Wang, T.; Liu, S.; Lan, T.; Wang, F.; Pan, C. A one adsorbent QuEChERS method coupled with LC-MS/MS for simultaneous determination of 10 organophosphorus pesticide residues in tea. Food Chem. 2020, 321, 126657. [Google Scholar] [CrossRef]
- Li, A.; Zhu, A. Preparation of Fe3O4/PANI nanocomposite and its metal anticorrosive activity. Prog. Org. Coat. 2021, 161, 106477. [Google Scholar] [CrossRef]
- Rodríguez-Palazón, M.C.; Arroyo-Manzanares, N.; Viñas, P.; López-García, I.; Hernández-Córdoba, M.; Campillo, N. Dispersive magnetic solid-phase extraction for capsaicinoid compounds in human serum using LC-HRMS: Targeted and non-targeted approaches. Anal. Bioanal. Chem. 2023, 415, 2133–2145. [Google Scholar] [CrossRef]
- GB 23200.121-2021; National Food Safety Standards–Determination of 331 Pesticides and Metabolitesresidues in Foods of Plant Origin–Liquid Chromatography-Tandem Mass Spectrometry Method. Standards Press of China: Beijng, China, 2021. Available online: http://down.foodmate.net/standard/sort/3/97823.html (accessed on 10 April 2024).
- Geis-Asteggiante, L.; Lehotay, S.J.; Lightfield, A.R.; Dutko, T.; Ng, C.; Bluhm, L. Ruggedness testing and validation of a practical analytical method for >100 veterinary drug residues in bovine muscle by ultrahigh performance liquid chromatography—tandem mass spectrometry. J. Chromatogr. A 2012, 1258, 43–54. [Google Scholar] [CrossRef]
- Matuszewski, B.K.; Constanzer, M.L.; Chavez-Eng, C.M. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC−MS/MS. Anal. Chem. 2003, 75, 3019–3030. [Google Scholar] [CrossRef] [PubMed]
- Cantwell, H. (Ed.) Eurachem Guide: The Fitness for Purpose of Analytical Methods—A Laboratory Guide to Method Validation and Related Topics, 3rd ed.; 2025; Available online: http://www.eurachem.org (accessed on 25 August 2025).
- GB 5009.295-2023; National Food Safety Standards–General Rules for Validation of Chemical Analysis Methods. Standards Press of China: Beijng, China, 2021. Available online: http://down.foodmate.net/standard/sort/3/145846.html (accessed on 25 August 2025).
- Xu, X.; Xu, X.; Han, M.; Qiu, S.; Hou, X. Development of a modified QuEChERS method based on magnetic multiwalled carbon nanotubes for the simultaneous determination of veterinary drugs, pesticides and mycotoxins in eggs by UPLC-MS/MS. Food Chem. 2019, 276, 419–426. [Google Scholar] [CrossRef]
- Narenderan, S.T.; Meyyanathan, S.N.; Babu, B. Review of pesticide residue analysis in fruits and vegetables. Pre-treatment, extraction and detection techniques. Food Res. Int. 2020, 133, 109141. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, Z.; He, Y.; Huang, L.; Xu, X.; Pan, C.; Guo, F.; Yang, H.; Tang, S. Nontarget and high-throughput screening of pesticides and metabolites residues in tea using ultra-high-performance liquid chromatography and quadrupole-orbitrap high-resolution mass spectrometry. J. Chromatogr. B 2021, 1179, 122847. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lin, H.; Yao, J.; Chen, Y.-Q.; Shi, P.-Y.; Song, J.; Mao, R.; Xiao, Q.-W.; Dai, Q. Development of an enhanced quick, easy, cheap, effective, rugged, and safe method for determination of quinolone drug residues in quail eggs. Sep. Sci. Plus 2024, 7, e202400219. [Google Scholar] [CrossRef]
- Rejczak, T.; Tuzimski, T. A review of recent developments and trends in the QuEChERS sample preparation approach. Open Chem. 2015, 13, 980–1010. [Google Scholar] [CrossRef]
- Alcantara-Duran, J.; Moreno-Gonzalez, D.; Garcia-Reyes, J.F.; Molina-Diaz, A. Use of a modified QuEChERS method for the determination of mycotoxin residues in edible nuts by nano flow liquid chromatography high resolution mass spectrometry. Food Chem. 2019, 279, 144–149. [Google Scholar] [CrossRef]
- EN 15662: 2018; Foods of Plant Origin. Multimethod for the Determination of Pesticide Residues Using GC- and LC-Based Analysis Following Acetonitrile Extraction/Partitioning and Clean-Up by Dispersive SPE. Modular QuEChERS-Method. BSI standards publication: London, UK, 2018. Available online: https://www.cssn.net.cn/cssn/productDetail/939893d1fc10267cdf93c3213730ac6e (accessed on 10 April 2024).
- Chen, Z.; Han, R.; Zhong, C. Adsorption of fluoroquinolone by carbon nanotubes: A combined experimental and density functional theory study. Chem. Papers 2020, 74, 3847–3856. [Google Scholar] [CrossRef]
- GB 2763-2021; National Food Safety Standards–Maximum Residue Limits for Pesticides in Food. Standards Press of China: Beijng, China, 2021. Available online: http://down.foodmate.net/standard/sort/3/108497.html (accessed on 5 July 2024).
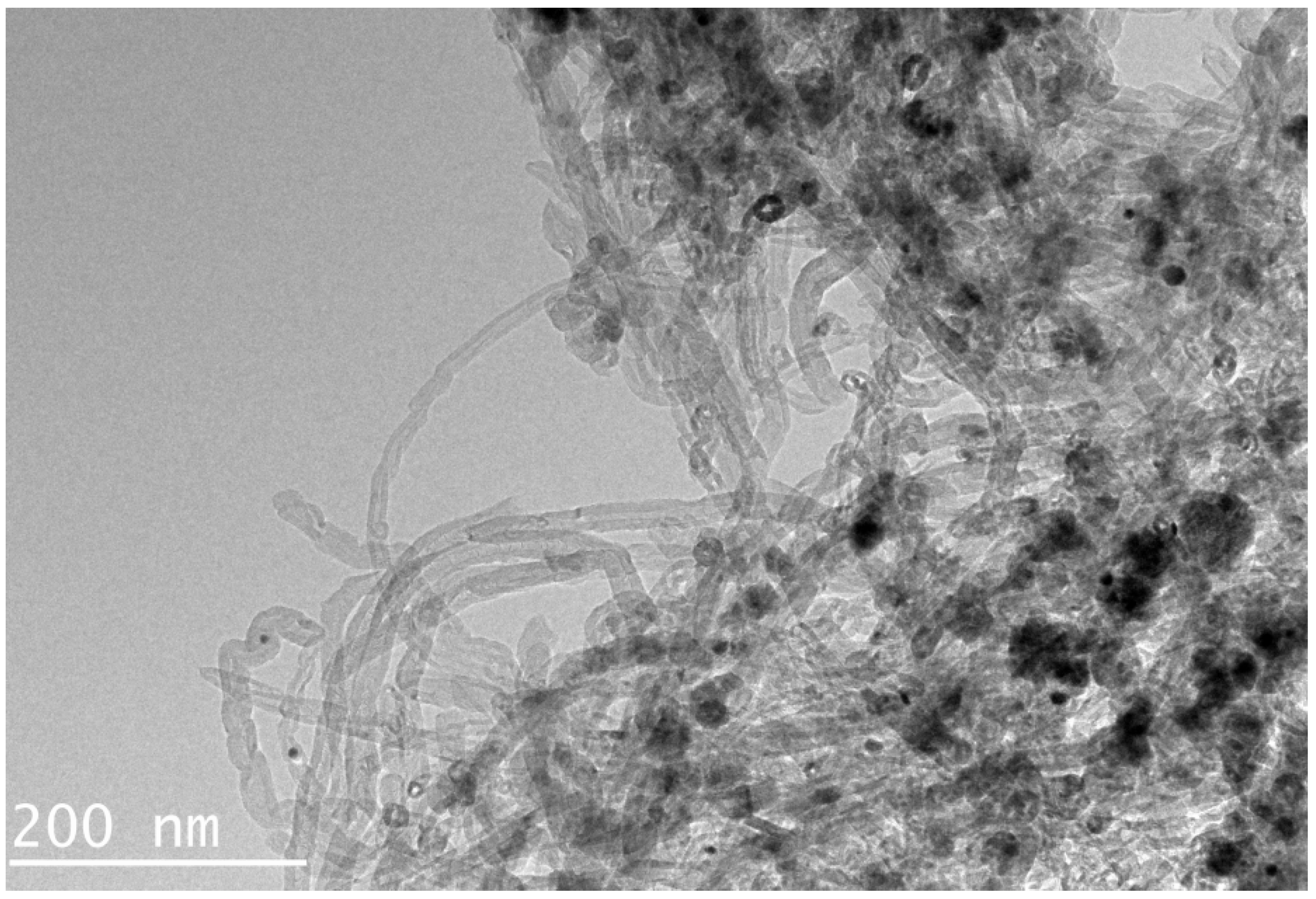
| Types of Magnetic Nanomaterials | MG-PSA | MG-C18 | MG-GO | MG-MWCNTs | Unpurified * | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 mg | 10 mg | 15 mg | 5 mg | 10 mg | 15 mg | 5 mg | 10 mg | 15 mg | 5 mg | 10 mg | 15 mg | ||
| Weight of co-extracts (mg) | 5.8 | 5.7 | 4.9 | 5.9 | 5.7 | 5.6 | 5.6 | 5.4 | 5.3 | 5.1 | 4.8 | 4.4 | 5.9 |
| Removal rate (%) | 1.7 | 3.4 | 16.9 | 0.0 | 3.4 | 5.1 | 5.1 | 8.5 | 10.2 | 13.6 | 18.6 | 25.4 | / |
| NO. | Compounds | Linear Range μg/kg | Calibration Curve | R2 | SDL μg/kg | LOQ μg/kg | Spiking Level (n = 3) | ||
|---|---|---|---|---|---|---|---|---|---|
| LOQ | 2 × LOQ | 20 × LOQ | |||||||
| 1 | Atrazine | 1–50 | y = 5.56 × 106x + 3.89 × 105 | 0.9999 | 1 | 2 | 117.6 (9.7) | 118.5 (4.4) | 114.4 (4.5) |
| 2 | Azoxystrobin | 1–50 | y = 6.39 × 106x − 4.13 × 106 | 0.9996 | 1 | 2 | 88.8 (10.2) | 95.9 (6.6) | 106.1 (5.9) |
| 3 | Betamethasone | 1–50 | y = 5.06 × 105x + 4.56 × 104 | 0.9998 | 1 | 2 | 87.8 (3.5) | 104.1 (9.6) | 108.4 (1.5) |
| 4 | Boscalid | 1–50 | y = 1.41 × 106x − 1.02 × 106 | 0.9999 | 1 | 2 | 119.1 (2.6) | 109.8 (10.9) | 98.7 (1.8) |
| 5 | Carbendazim | 1–50 | y = 4.31 × 106x − 1.64 × 106 | 0.9985 | 1 | 2 | 102.0 (3.6) | 105.4 (2.3) | 113.9 (5.0) |
| 6 | Carbofuran | 1–50 | y = 3.53 × 106x + 8.14 × 106 | 0.9993 | 1 | 2 | 72.2 (13.3) | 105.0 (5.0) | 105.2 (1.5) |
| 7 | Chlorantraniliprole | 1–50 | y = 5.62 × 105x − 5.81 × 105 | 0.9995 | 1 | 2 | 103.5 (19.5) | 98.2 (15.8) | 102.1 (2.5) |
| 8 | Chlorpyrifos | 1–50 | y = 1.53 × 106x − 4.25 × 105 | 0.9998 | 1 | 2 | 78.8 (7.9) | 94.5 (4.6) | 113.1 (3.5) |
| 9 | Clarithromycin | 1–50 | y = 2.84 × 105x − 4.30 × 104 | 1.0000 | 1 | 2 | 118.2 (3.2) | 109.3 (4.7) | 93.8 (2.1) |
| 10 | Dexamethasone | 1–50 | y = 5.06 × 105x + 4.57 × 104 | 0.9998 | 1 | 2 | 88.0 (1.3) | 101.9 (10.4) | 106.2 (1.4) |
| 11 | Dichlorvos | 1–50 | y = 1.27 × 106x − 2.69 × 105 | 1.0000 | 1 | 2 | 114.5 (19.6) | 117.1 (0.8) | 116.0 (4.5) |
| 12 | Difenoconazole | 1–50 | y = 1.47 × 106x − 8.57 × 105 | 0.9998 | 1 | 2 | 117.0 (16.7) | 115.3 (8.6) | 105.1 (4.1) |
| 13 | Diflubenzuron | 1–50 | y = 6.56 × 105x − 2.80 × 105 | 0.9993 | 1 | 2 | 97.2 (9.6) | 83.3 (6.7) | 94.8 (0.3) |
| 14 | Dimethomorph | 1–50 | y = 1.28 × 106x − 4.19 × 104 | 0.9992 | 1 | 2 | 109.6 (9.8) | 118.4 (0.9) | 116.3 (4.7) |
| 15 | Etofenprox | 1–50 | y = 2.84 × 106x − 3.18 × 106 | 0.9996 | 1 | 2 | 112.1 (9.5) | 108.1 (0.6) | 101.4 (4.2) |
| 16 | Fipronil | 1–50 | y = 1.57 × 106x + 1.05 × 105 | 0.9997 | 1 | 2 | 107.3 (6.2) | 97.7 (0.9) | 115.2 (2.3) |
| 17 | Fipronil desulfinyl | 1–50 | y = 1.78 × 106x + 1.17 × 104 | 0.9993 | 1 | 2 | 105.0 (3.6) | 112.7 (0.2) | 111.9 (1.7) |
| 18 | Fipronil sulfone | 1–50 | y = 4.12 × 106x + 2.51 × 105 | 0.9996 | 1 | 2 | 118.0 (10.1) | 108.7 (3.8) | 117.0 (0.9) |
| 19 | Fipronil sulfoxide | 1–50 | y = 1.93 × 106x + 1.75 × 105 | 0.9988 | 1 | 2 | 116.6 (9.9) | 95.9 (1.7) | 117.1 (3.4) |
| 20 | Flubendiamide | 1–50 | y = 1.06 × 106x − 1.82 × 105 | 0.9978 | 1 | 2 | 91.5 (8.2) | 96.3 (6.7) | 101.8 (1.9) |
| 21 | Forchlorfenuron | 1–50 | y = 1.85 × 106x − 4.60 × 105 | 0.9993 | 1 | 2 | 112.5 (4.4) | 115.0 (14.8) | 84.0 (2.1) |
| 22 | Imidacloprid | 1–50 | y = 1.40 × 106x + 5.42 × 104 | 0.9978 | 1 | 2 | 100.5 (8.2) | 108.4 (7.9) | 109.9 (6.6) |
| 23 | ISAZOFOS | 1–50 | y = 5.99 × 106x + 5.42 × 105 | 0.9997 | 1 | 2 | 105.8 (13.6) | 99.5 (5.3) | 118.2 (1.3) |
| 24 | Isocarbophos | 1–50 | y = 6.23 × 105x − 4.42 × 104 | 0.9988 | 1 | 2 | 106.1 (5.2) | 103.4 (10.2) | 115.1 (1.6) |
| 25 | Malathion | 1–50 | y = 1.46 × 106x − 3.01 × 104 | 0.9989 | 1 | 2 | 109.7 (5.5) | 116.0 (6.6) | 117.5 (2.8) |
| 26 | Metalaxyl | 1–50 | y = 5.59 × 106x + 8.35 × 105 | 0.9997 | 1 | 2 | 118.6 (3.4) | 117.9 (8.2) | 104.2 (0.4) |
| 27 | Metronidazole | 1–50 | y = 4.54 × 105x − 5.71 × 105 | 0.9995 | 1 | 2 | 115.1 (4.2) | 118.5 (12.1) | 113.8 (9.1) |
| 28 | Phosfolan | 1–50 | y = 5.81 × 106x + 3.86 × 105 | 0.9994 | 1 | 2 | 100.0 (0.3) | 117.7 (6.7) | 112.5 (1.3) |
| 29 | Phosmet | 1–50 | y = 1.26 × 106x + 1.78 × 105 | 0.9985 | 1 | 2 | 106.1 (3.3) | 89.4 (13.5) | 104.1 (0.9) |
| 30 | Phoxim | 1–50 | y = 4.70 × 106x + 3.46 × 106 | 0.9999 | 1 | 2 | 70.0 (14.1) | 110.9 (7.6) | 117.4 (2.4) |
| 31 | Prochloraz | 1–50 | y = 2.04 × 106x − 1.66 × 106 | 0.9998 | 1 | 2 | 106.2 (2.9) | 99.4 (1.3) | 98.9 (0.1) |
| 32 | Prochloraz-desimidazole-amino | 1–50 | y = 1.98 × 106x − 5.20 × 104 | 0.9996 | 1 | 2 | 114.2 (19.1) | 96.4 (6.5) | 111.2 (3.2) |
| 33 | Profenofos | 1–50 | y = 1.94 × 106x − 2.33 × 103 | 0.9998 | 1 | 2 | 99.9 (6.8) | 110.0 (17.6) | 113.4 (1.7) |
| 34 | Propamocarb free base | 1–50 | y = 3.60 × 106x − 8.19 × 104 | 0.9971 | 1 | 2 | 108.0 (6.3) | 115.2 (9.2) | 111.8 (1.2) |
| 35 | Propiconazole | 1–50 | y = 1.97 × 106x + 1.33 × 106 | 0.9999 | 1 | 2 | 102.9 (0.6) | 107.0 (6.9) | 109.0 (2.1) |
| 36 | Proponit | 1–50 | y = 5.04 × 106x + 4.19 × 105 | 0.9992 | 1 | 2 | 116.4 (6.3) | 110.5 (6.8) | 102.5 (0.2) |
| 37 | Pyraclostrobin | 1–50 | y = 5.02 × 106x + 1.19 × 105 | 0.9994 | 1 | 2 | 114.2 (0.7) | 86.5 (4.8) | 104.5 (1.2) |
| 38 | Pyridaben | 1–50 | y = 2.71 × 106x − 2.24 × 106 | 0.9997 | 1 | 2 | 91.8 (8.9) | 103.1 (6.3) | 105.0 (2.4) |
| 39 | Pyrimethanil | 1–50 | y = 5.86 × 106x + 3.70 × 105 | 0.9998 | 1 | 2 | 90.4 (14.4) | 72.0 (6.6) | 90.1 (1.3) |
| 40 | S-metolachlor | 1–50 | y = 5.04 × 106x + 4.19 × 105 | 0.9992 | 1 | 2 | 116.4 (6.3) | 110.5 (6.8) | 112.5 (0.2) |
| 41 | Spinetoram J | 1–50 | y = 2.49 × 106x − 4.40 × 104 | 0.9990 | 1 | 2 | 108.4 (14.9) | 97.0 (3.2) | 118.6 (1.8) |
| 42 | Sulfadimethoxine | 1–50 | y = 1.11 × 105x + 2.91 × 106 | 0.9889 | 1 | 2 | 105.6 (11.5) | 85.8 (6.9) | 93.2 (1.3) |
| 43 | Sulfadimidine | 1–50 | y = 2.75 × 105x + 5.0,8× 104 | 0.9992 | 1 | 2 | 81.4 (3.1) | 103.0 (2.7) | 117.4 (2.0) |
| 44 | Sulfadoxine | 1–50 | y = 3.09 × 106x − 2.03× 105 | 0.9982 | 1 | 2 | 102.9 (9.1) | 115.8 (1.2) | 110.2 (5.7) |
| 45 | Sulfamerazine | 1–50 | y = 1.73 × 106x − 4.06 × 103 | 0.9985 | 1 | 2 | 117.3 (3.6) | 115.7 (3.8) | 103.4 (4.0) |
| 46 | Sulfameter | 1–50 | y = 2.62 × 106x + 1.48 × 106 | 0.9998 | 1 | 2 | 86.8 (13.3) | 115.9 (1.3) | 114.0 (4.5) |
| 47 | Sulfamonomethoxine | 1–50 | y = 9.00 × 104x + 2.10 × 105 | 0.9999 | 1 | 2 | 97.0 (1.5) | 107.0 (3.3) | 98.8 (0.5) |
| 48 | Sulfapyridine | 1–50 | y = 1.54 × 106x + 1.90 × 106 | 0.9996 | 1 | 2 | 74.4 (4.9) | 112.8 (3.6) | 116.3 (0.8) |
| 49 | Sulfaquinoxaline | 1–50 | y = 1.32 × 105x + 1.29 × 104 | 0.9994 | 1 | 2 | 94.5 (2.7) | 102.3 (1.4) | 93.1 (2.0) |
| 50 | Sulfisomidine | 1–50 | y = 3.67 × 106x + 1.18 × 106 | 0.9980 | 1 | 2 | 91.8 (7.7) | 111.3 (4.6) | 111.4 (0.5) |
| 51 | Tebuconazole | 1–50 | y = 2.23 × 106x − 5.70 × 104 | 0.9997 | 1 | 2 | 90.4 (18.8) | 109.6 (6.7) | 108.8 (0.1) |
| 52 | Triadimefon | 1–50 | y = 2.18 × 106x − 3.30 × 105 | 0.9998 | 1 | 2 | 104.6 (0.8) | 119.1 (13.1) | 111.0 (0.5) |
| 53 | Triazophos | 1–50 | y = 7.79 × 106x − 1.96 × 106 | 0.9998 | 1 | 2 | 81.4 (1.7) | 113.9 (8.7) | 107.8 (1.2) |
| 54 | Tricyclazole | 1–50 | y = 7.44 × 106x − 3.43 × 106 | 0.9998 | 1 | 2 | 117.5 (1.9) | 80.6 (4.6) | 82.1 (3.9) |
| 55 | Trimethoprim | 1–50 | y = 1.02 × 105x + 1.46 × 105 | 0.9997 | 1 | 2 | 91.9 (1.6) | 114.7 (1.8) | 116.4 (1.5) |
| 56 | Acephate | 1–100 | y = 1.33 × 105x − 3.65 × 105 | 0.9965 | 1 | 5 | 117.0 (2.4) | 110.3 (18.5) | 105.0 (6.7) |
| 57 | Acetamiprid | 1–100 | y = 3.38 × 106x + 9.36 × 104 | 0.9982 | 1 | 5 | 89.1 (10.8) | 118.8 (3.0) | 111.1 (5.5) |
| 58 | Ciprofloxacin | 1–100 | y = 5.39 × 104x − 2.97 × 104 | 0.9999 | 1 | 5 | 88.4 (6.1) | 86.1 (2.5) | 62.8 (3.5) |
| 59 | Cortisol | 1–100 | y = 2.13 × 105x − 1.81 × 105 | 0.9997 | 1 | 5 | 117.3 (13.4) | 97.4 (1.5) | 99.7 (3.9) |
| 60 | Danofloxacin | 1–100 | y = 3.03 × 105x − 1.34 × 106 | 0.9908 | 1 | 5 | 86.3 (11.5) | 85.9 (9.8) | 79.5 (8.2) |
| 61 | Difloxacin | 1–100 | y = 1.46 × 105x + 5.75 × 105 | 0.9983 | 1 | 5 | 88.0 (6.0) | 97.0 (3.1) | 104.2 (6.0) |
| 62 | Dimethoate | 1–100 | y = 1.24 × 106x + 3.79 × 105 | 0.9998 | 1 | 5 | 75.0 (0.5) | 107.2 (12.8) | 105.1 (3.9) |
| 63 | Dimetridazole | 1–100 | y = 4.65 × 105x − 2.27 × 105 | 0.9978 | 1 | 5 | 114.3 (4.9) | 109.9 (8.3) | 112.7 (2.9) |
| 64 | Enoxacin | 1–100 | y = 8.54 × 104x − 1.33 × 106 | 0.9915 | 1 | 5 | 62.7 (10.5) | 68.4 (4.3) | 62.9 (1.6) |
| 65 | Erythromycin | 1–100 | y = 3.18 × 103x + 2.72 × 104 | 0.9919 | 1 | 5 | 81.2 (5.4) | 87.1 (4.6) | 104.3 (0.9) |
| 66 | Enrofloxacin | 1–100 | y = 1.44 × 105x + 1.62 × 105 | 0.9996 | 1 | 5 | 97.9 (3.1) | 70.2 (7.0) | 88.8 (0.6) |
| 67 | Fenthion | 1–100 | y = 4.82 × 105x + 2.06 × 105 | 0.9992 | 1 | 5 | 83.4 (6.4) | 91.6 (0.3) | 109.8 (0.7) |
| 68 | Fenpropathrin | 1–50 | y = 4.82 × 105x − 4.68 × 105 | 0.9996 | 1 | 5 | 91.2 (1.9) | 87.0 (3.9) | 112.5 (2.7) |
| 69 | Fleroxacin | 1–100 | y = 1.57 × 105x + 8.37 × 105 | 0.9982 | 1 | 5 | 75.9 (6.4) | 79.6 (0.2) | 81.5 (3.4) |
| 70 | Flumequine | 1–100 | y = 7.85 × 104x − 8.91 × 103 | 0.9997 | 1 | 5 | 79.0 (9.1) | 73.7 (3.6) | 73.0 (1.1) |
| 71 | Lomefloxacin | 1–100 | y = 1.11 × 105x − 1.28 × 105 | 0.9997 | 1 | 5 | 84.0 (7.6) | 89.1 (5.8) | 100.4 (1.6) |
| 72 | Methylprednisolone | 1–100 | y = 2.42 × 105x − 1.53 × 105 | 0.9989 | 1 | 5 | 115.2 (5.3) | 100.4 (5.2) | 101.5 (0.2) |
| 73 | Norfloxacin | 1–100 | y = 1.03 × 105x + 5.52 × 105 | 0.9980 | 1 | 5 | 64.9 (5.7) | 72.3 (3.8) | 73.9 (4.5) |
| 74 | Ofloxacin | 1–100 | y = 1.61 × 105x + 1.12 × 105 | 0.9997 | 1 | 5 | 105.0 (12.1) | 75.1 (5.4) | 77.2 (4.7) |
| 75 | Oleandomycin | 1–100 | y = 2.03 × 106x − 4.79 × 104 | 0.9991 | 1 | 5 | 108.5 (1.0) | 113.0 (0.1) | 103.0 (0.8) |
| 76 | Omethoate | 1–100 | y = 2.22 × 106x − 5.54 × 105 | 0.9974 | 1 | 5 | 118.4 (7.9) | 105.1 (15.0) | 108.4 (0.2) |
| 77 | Orbifloxacin | 1–100 | y = 2.00 × 106x − 1.71 × 103 | 0.9996 | 1 | 5 | 87.8 (3.9) | 89.4 (10.4) | 88.6 (6.7) |
| 78 | Pefloxacin | 1–100 | y = 2.50 × 105x − 2.95 × 106 | 0.9944 | 1 | 5 | 79.3 (6.8) | 65.3 (4.9) | 62.0 (2.2) |
| 79 | Prednisone | 1–100 | y = 1.01 × 105x − 5.44× 103 | 0.9997 | 1 | 5 | 72.6 (6.2) | 98.2 (9.2) | 118.8 (2.8) |
| 80 | Roxithromycin | 1–100 | y = 6.74 × 104x + 7.40 × 105 | 0.9971 | 1 | 5 | 89.5 (7.1) | 105.0 (5.7) | 117.4 (0.7) |
| 81 | Sarafloxacin | 1–100 | y = 8.34 × 104x + 3.36 × 105 | 0.9920 | 1 | 5 | 61.7 (9.3) | 68.4 (8.4) | 63.1 (2.0) |
| 82 | Sparfloxacin | 1–100 | y = 1.37 × 104x + 8.75 × 105 | 0.9920 | 1 | 5 | 81.0 (3.8) | 80.9 (4.2) | 97.1 (2.7) |
| 83 | Sulfamethoxazole | 1–100 | y = 9.82 × 104x + 2.59 × 105 | 0.9986 | 1 | 5 | 104.0 (6.8) | 115.9 (2.0) | 117.4 (1.4) |
| 84 | Sulfafurazole | 1–100 | y = 1.39 × 106x − 2.78 × 103 | 0.9963 | 1 | 5 | 97.8 (3.1) | 106.5 (2.5) | 103.9 (1.6) |
| 85 | Thiamethoxam | 1–100 | y = 8.55 × 105x + 1.83 × 105 | 0.9998 | 1 | 5 | 117.8 (8.6) | 118.7 (6.2) | 117.7 (3.5) |
| 86 | Tilmicosin | 1–100 | y = 3.79 × 104x − 8.01 × 102 | 0.9977 | 1 | 5 | 114.3 (0.6) | 104.9 (6.2) | 118.1 (2.8) |
| 87 | Tylosin | 1–100 | y = 5.40 × 104x + 4.29 × 105 | 0.9970 | 1 | 5 | 102.0 (4.2) | 94.2 (2.4) | 114.1 (3.4) |
| 88 | Aldicarb | 1–100 | y = 1.73 × 105x − 1.64 × 105 | 0.9998 | 2 | 5 | 118.8 (6.3) | 114.8 (11.4) | 106.8 (2.0) |
| 89 | Azithromycin | 1–100 | y = 6.29 × 104x + 7.50 × 105 | 0.9972 | 2 | 5 | 100.5 (19.0) | 100.6 (4.9) | 118.8 (2.2) |
| 90 | Chlorbenzuron | 1–100 | y = 8.13 × 105x − 2.62 × 105 | 0.9998 | 2 | 5 | 83.4 (1.9) | 78.7 (1.2) | 79.0 (3.2) |
| 91 | Clothianidin | 1–100 | y = 5.58 × 106x − 4.01 × 105 | 0.9991 | 2 | 5 | 104.8 (13.4) | 117.0 (0.2) | 107.6 (5.7) |
| 92 | Fludrocortisone acetate | 1–100 | y = 1.82 × 105x − 1.42 × 105 | 0.9993 | 2 | 5 | 64.0 (5.3) | 88.5 (8.2) | 91.9 (4.0) |
| 93 | Hexaconazole | 1–100 | y = 1.26 × 106x − 6.49 × 105 | 0.9994 | 2 | 5 | 106.5 (13.4) | 115.9 (13.5) | 104.0 (1.3) |
| 94 | Pendimethalin | 1–100 | y = 4.39 × 105x − 5.94 × 105 | 0.9989 | 2 | 5 | 77.4 (3.7) | 118.9 (8.4) | 112.5 (2.3) |
| 95 | Prednisolone | 1–100 | y = 2.29 × 105x − 1.62 × 105 | 0.9996 | 2 | 5 | 106.2 (3.9) | 96.6 (2.5) | 101.2 (2.2) |
| 96 | Spiromesifen | 1–100 | y = 3.56 × 105x − 2.29 × 105 | 0.9995 | 2 | 5 | 109.6 (7.8) | 104.4 (2.6) | 104.0 (5.3) |
| 97 | Sulfabenzamide | 1–100 | y = 7.85 × 105x + 5.97 × 105 | 0.9997 | 2 | 5 | 93.8 (12.8) | 116.3 (1.8) | 114.2 (6.8) |
| 98 | Sulfacetamide | 1–100 | y = 1.87 × 103x + 1.06 × 104 | 0.9998 | 2 | 5 | 91.6 (8.8) | 104.1 (17.7) | 108.7 (4.1) |
| 99 | Sulfadiazine | 1–100 | y = 1.26 × 105x − 4.62 × 104 | 0.9999 | 2 | 5 | 90.0 (6.3) | 101.6 (3.8) | 107.5 (0.2) |
| 100 | Sulfathiazole | 1–100 | y = 1.02 × 105x + 8.72 × 105 | 0.9983 | 2 | 5 | 95.5 (6.7) | 99.0 (3.5) | 104.8 (7.8) |
| 101 | Sulfamethizole | 1–100 | y = 4.88 × 105x + 1.81 × 104 | 0.9990 | 2 | 5 | 95.2 (13.9) | 115.0 (6.5) | 117.3 (1.5) |
| 102 | Tebufenozide | 1–100 | y = 2.25 × 105x − 8.14 × 105 | 0.9975 | 2 | 5 | 87.8 (0.5) | 101.3 (0.4) | 94.3 (0.4) |
| 103 | Tetracycline | 1–100 | y =1.17 × 105x – 9.01 × 104 | 0.9999 | 2 | 5 | 102.8 (3.5) | 67.5 (2.7) | 71.3 (1.6) |
| 104 | Avermectin B1a | 2–200 | y = 1.23 × 105x + 1.39 × 104 | 0.9958 | 2 | 10 | 86.0 (8.9) | 84.2 (6.4) | 106.2 (3.7) |
| 105 | Chlorfluazuron | 2–200 | y = 3.73 × 105x − 1.36 × 106 | 0.9953 | 2 | 10 | 81.2 (10.8) | 106.0 (0.6) | 84.9 (1.3) |
| 106 | Deltamethrin | 2–200 | y = 2.71 × 104x + 3.90 × 105 | 0.9848 | 2 | 10 | 75.4 (17.3) | 95.8 (16.3) | 98.5 (7.6) |
| 107 | Demeton | 2–200 | y = 3.88 × 104x − 7.48 × 103 | 0.9870 | 2 | 10 | 95.1 (8.9) | 110.3 (19.9) | 121.6 (15.4) |
| 108 | Methomyl | 2–200 | y = 2.55 × 105x − 5.70 × 105 | 0.9986 | 2 | 10 | 91.1 (15.0) | 116.8 (7.2) | 110.4 (6.6) |
| 109 | Beclometasone | 5–200 | y = 1.71 × 104x + 3.54 × 104 | 0.9959 | 5 | 10 | 95.1 (12.1) | 97.5 (8.7) | 100.2 (10.4) |
| 110 | Chlortetracycline | 5–200 | y = 2.27 × 104x + 2.91 × 104 | 0.9996 | 5 | 10 | 70.0 (10.5) | 77.1 (2.1) | 73.7 (6.5) |
| 111 | Dimetridazole-2-hydroxy | 5–200 | y = 1.32 × 105x + 5.25 × 104 | 0.9968 | 5 | 10 | 81.6 (4.5) | 87.3 (14.2) | 118.0 (2.2) |
| 112 | Doxycycline (anhydrous) | 5–200 | y = 1.45 × 105x − 2.36 × 105 | 0.9999 | 5 | 10 | 61.8 (11.8) | 79.4 (4.1) | 75.5 (1.9) |
| 113 | Hydroxymetronidazole | 5–200 | y = 2.00 × 105x − 1.24 × 104 | 0.9902 | 5 | 10 | 79.3 (9.5) | 105.3 (4.9) | 109.9 (7.8) |
| 114 | Lufenuron | 5–200 | y = 2.15 × 104x + 2.36 × 105 | 0.9951 | 5 | 10 | 86.3 (12.8) | 91.3 (1.9) | 89.6 (4.4) |
| 115 | Oxadixyl | 5–200 | y = 2.07 × 106x − 8.79 × 105 | 0.9999 | 5 | 10 | 118.0 (12.6) | 117.9 (11.2) | 111.3 (1.1) |
| 116 | Oxytetracycline | 5–200 | y = 7.42 × 104x + 6.40 × 104 | 0.9999 | 5 | 10 | 63.6 (13.6) | 70.8 (2.9) | 64.2 (5.1) |
| 117 | Spiramycin | 5–200 | y = 2.44 × 104x + 1.48 × 105 | 0.9969 | 5 | 10 | 116.7 (1.9) | 93.9 (5.8) | 101.0 (2.0) |
| 118 | Sulfachlorpyridazine | 5–200 | y = 4.95 × 105x + 2.18 × 105 | 0.9984 | 5 | 10 | 84.7 (5.2) | 99.6 (0.4) | 113.0 (0.9) |
| 119 | Sulfaphenazole | 5–200 | y = 1.12 × 106x − 1.43 × 105 | 0.9982 | 5 | 10 | 78.0 (14.9) | 113.6 (2.0) | 114.5 (1.0) |
| 120 | Permethrin | 5–200 | y = 1.32 × 104x + 4.75 × 105 | 0.9994 | 5 | 10 | 76.1 (17.2) | 98.8 (4.2) | 104.4 (4.3) |
| 121 | Ronidazole | 5–200 | y = 1.18 × 105x + 7.18 × 105 | 0.9928 | 5 | 10 | 81.2 (10.5) | 89.5 (6.4) | 117.5 (1.8) |
| Classification | Targets | Screening Numbers | Quantitative Numbers | Mean Value (µg/kg) | Range (µg/kg) | Sample ID |
|---|---|---|---|---|---|---|
| Pesticides | Dimethomorph | 21 | 14 | 211.37 | 2.13–1184 | sample 22, 24, 26–31, 34–49 |
| Pyraclostrobin | 13 | 12 | 8.90 | 2.01–24.4 | sample 22, 24, 26–28, 34–40, 44 | |
| Carbendazim | 12 | 12 | 11.7 | 2.24–31.7 | sample 21, 25–27, 29, 31, 36–40 | |
| Metalaxyl | 11 | 10 | 8.07 | 3.25–20.5 | sample 22–23, 32–40 | |
| Boscalid | 10 | 9 | 24.2 | 3.03–48.2 | sample 29, 31, 33, 35–40, 44 | |
| Chlorantraniliprole | 10 | 6 | 6.70 | 2.98–13.1 | sample 2–10, 21–25 | |
| Clothianidin | 8 | 5 | 133.34 | 15.5–501.6 | sample 22, 24–25, 27, 29, 37, 39–40 | |
| Propamocarb-free base | 7 | 6 | 184.6 | 2.49–299 | sample 29, 31, 34–36, 38, 44 | |
| Prochloraz-desimidazole-amino | 6 | 6 | 11.0 | 2.84–16.0 | sample 23, 25, 31, 37, 39–40 | |
| Thiamethoxam | 6 | 4 | 62.9 | 6.54–112.6 | sample 22, 25, 32, 37, 39–40 | |
| Difenoconazole | 5 | 2 | 2.55 | 2.07–3.03 | sample 34, 36–38, 40 | |
| Imidacloprid | 4 | 2 | 52.4 | 18.8–85.8 | sample 2, 15–16, 24 | |
| Pyridaben | 3 | 3 | 21.9 | 2.97–35.7 | sample 15, 21, 27 | |
| Chlorbenzuron | 2 | 1 | 9.95 | 9.95 | sample 23, 25 | |
| Dichlorvos | 2 | 1 | 15.7 | 15.7 | sample 39, 40 | |
| Tebuconazole | 2 | 2 | 35.5 | 18.2–52.7 | sample 37, 40 | |
| Azoxystrobin | 2 | 0 | — | — | sample 37, 39 | |
| Acetamiprid | 1 | 1 | 19.0 | 19.0 | sample 31 | |
| Chlorpyrifos | 1 | 1 | 157 | 157 | sample 31 | |
| Prochloraz | 1 | 1 | 12.6 | 12.6 | sample 40 | |
| Proponit | 1 | 0 | — | — | sample 28 | |
| Veterinary drugs | Doxycycline | 3 | 0 | — | — | sample 10, 26, 39 |
| Ofloxacin | 1 | 0 | — | — | sample 35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, M.; Ni, Y.; Zang, T.; Gao, W.; Song, J.; Zou, J.; Xu, D. Non-Targeted Screening and Quantitative Analysis of Pesticides and Veterinary Drug Residues in Brassica rapa chinensis Using an Improved Quechers Method Based on Magnetic Materials. Foods 2025, 14, 3288. https://doi.org/10.3390/foods14193288
Tang M, Ni Y, Zang T, Gao W, Song J, Zou J, Xu D. Non-Targeted Screening and Quantitative Analysis of Pesticides and Veterinary Drug Residues in Brassica rapa chinensis Using an Improved Quechers Method Based on Magnetic Materials. Foods. 2025; 14(19):3288. https://doi.org/10.3390/foods14193288
Chicago/Turabian StyleTang, Minmin, Yongbiao Ni, Tianli Zang, Wei Gao, Jinzhu Song, Jie Zou, and Danke Xu. 2025. "Non-Targeted Screening and Quantitative Analysis of Pesticides and Veterinary Drug Residues in Brassica rapa chinensis Using an Improved Quechers Method Based on Magnetic Materials" Foods 14, no. 19: 3288. https://doi.org/10.3390/foods14193288
APA StyleTang, M., Ni, Y., Zang, T., Gao, W., Song, J., Zou, J., & Xu, D. (2025). Non-Targeted Screening and Quantitative Analysis of Pesticides and Veterinary Drug Residues in Brassica rapa chinensis Using an Improved Quechers Method Based on Magnetic Materials. Foods, 14(19), 3288. https://doi.org/10.3390/foods14193288






