A Simple, Rapid, and Contamination-Free Ultra-Sensitive Cronobacter sakazakii Visual Diagnostic Platform Based on RPA Combined with CRISPR/Cas12a
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Bacterial Strain Cultivation and Genomic DNA Extraction
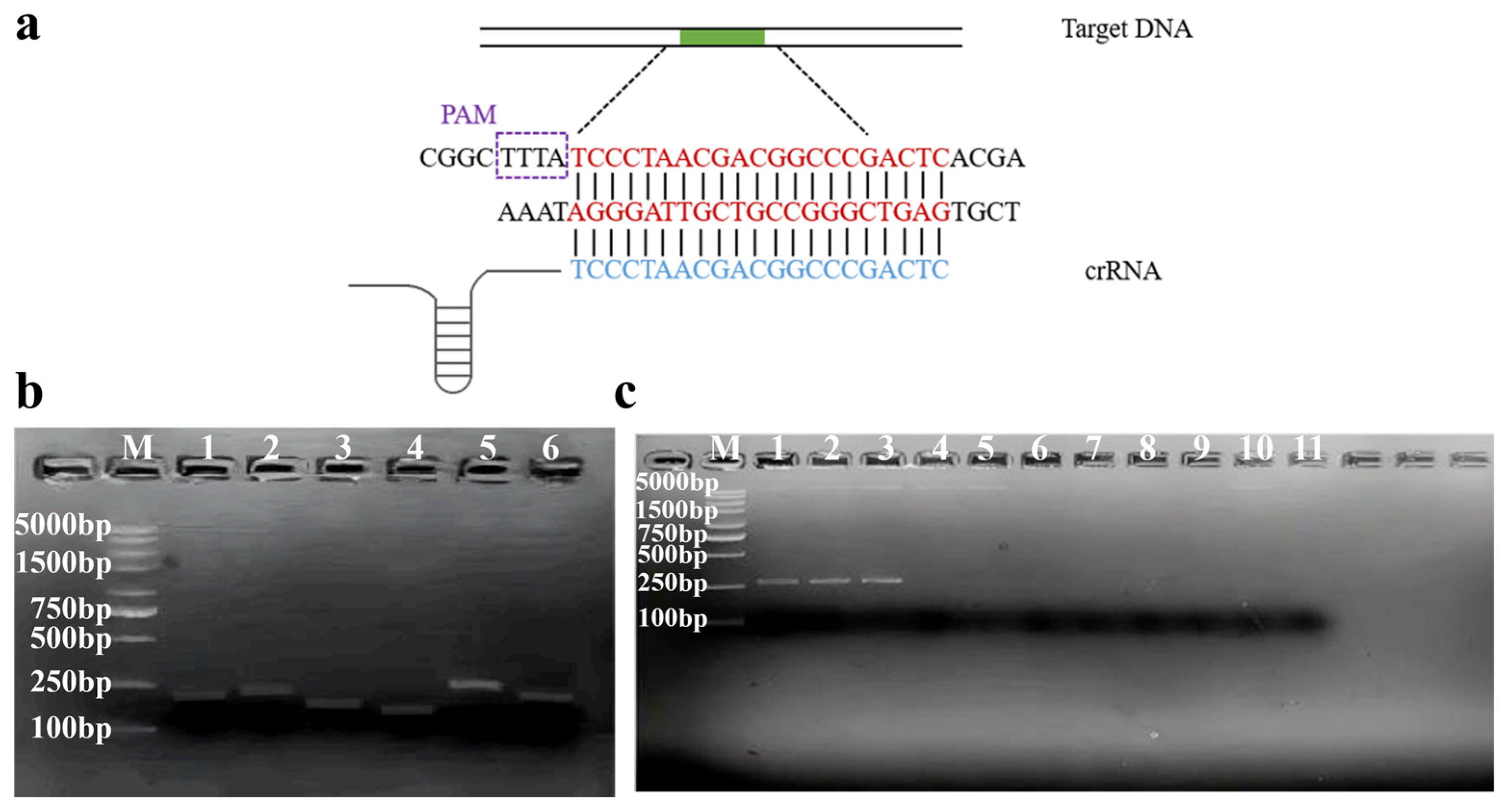
2.3. Design and Screening for RPA Primers and crRNA
2.4. Two-Step RPA-CRISPR/Cas12a System Construction
2.5. Preliminary Test of the PTIT (Pipette Tip-in-Tube) Method
2.6. The PTIT Method Procedure
2.7. Establishment of the PTIT System
2.8. Evaluation of Specificity and Sensitivity
2.9. Sample Detection
2.10. Method Validation
3. Results
3.1. Screening of RPA Primers and crRNA
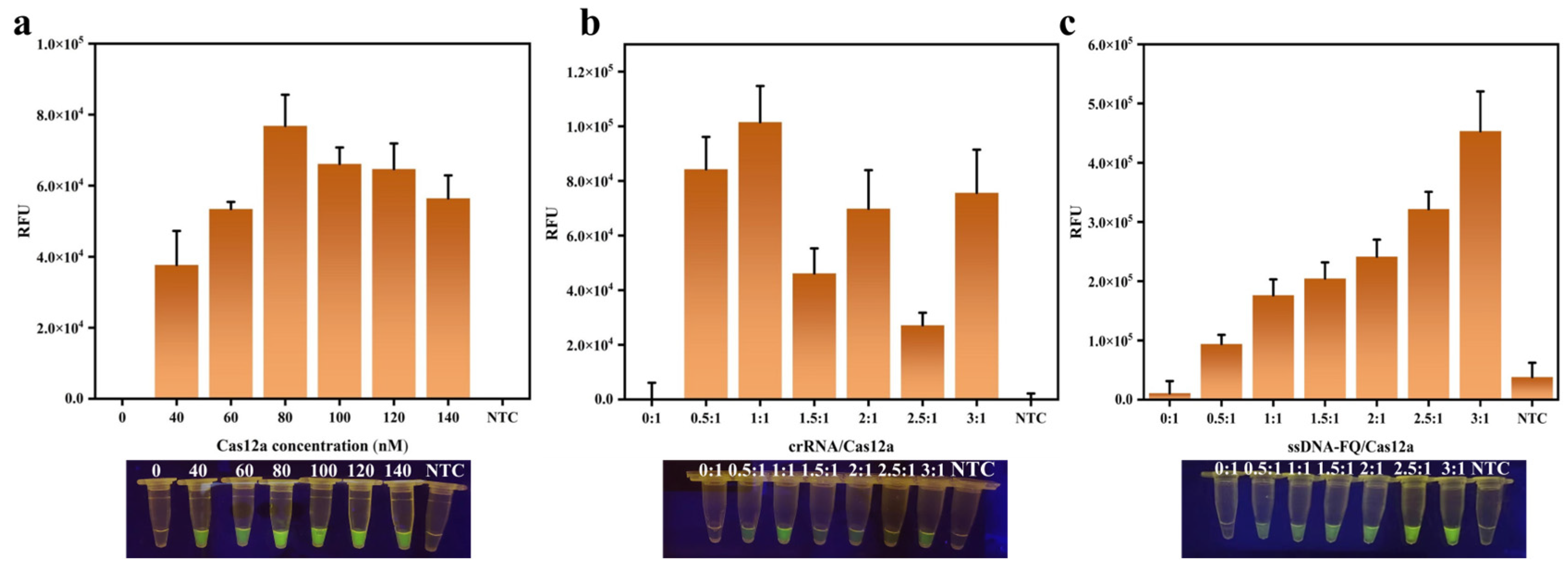
3.2. Optimization of RPA Reaction
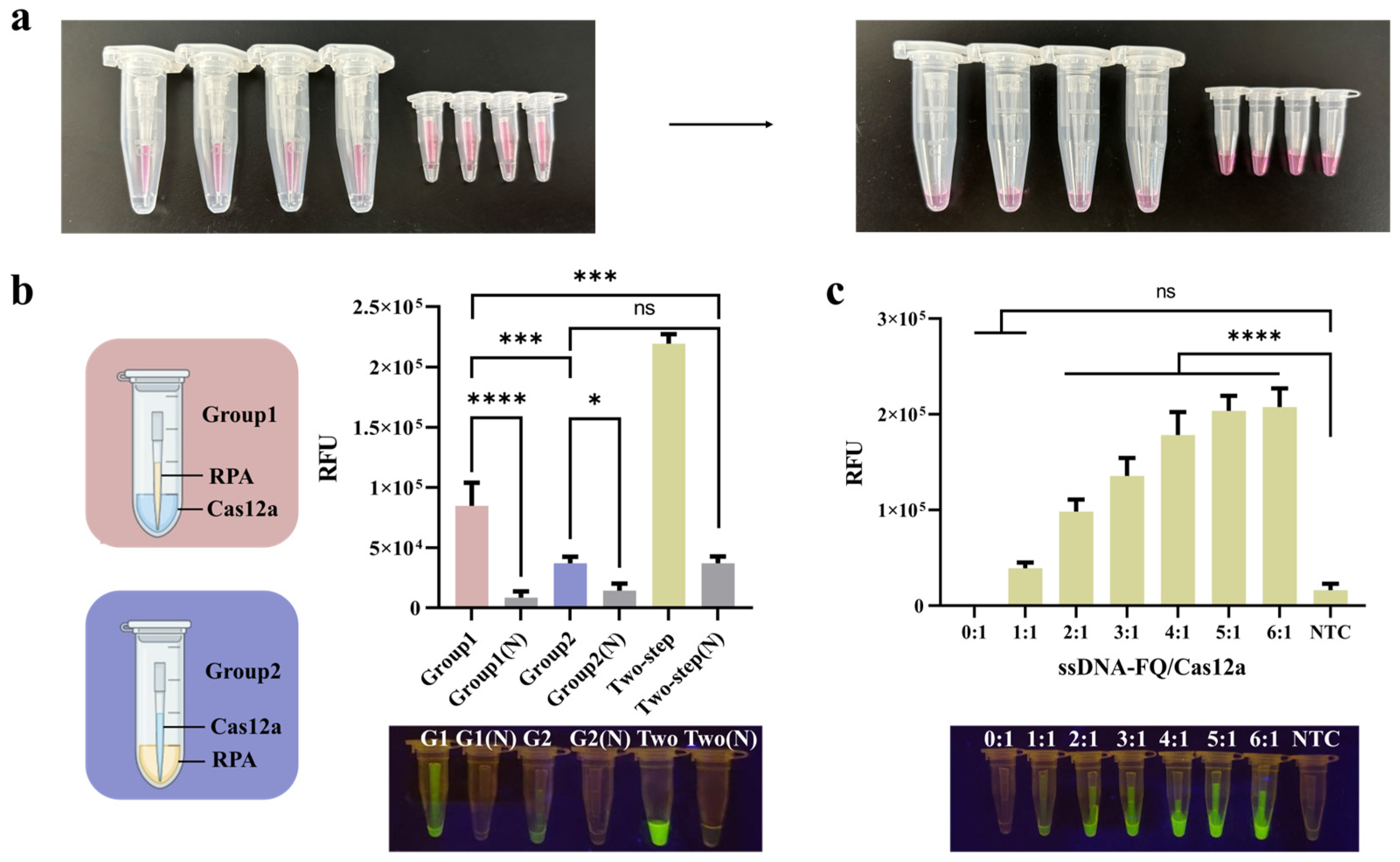
3.3. Establishment of the Two-Step RPA-CRISPR/Cas12a Detection Platform
3.4. The Development of PTIT Method
3.5. The Optimization of the PTIT System
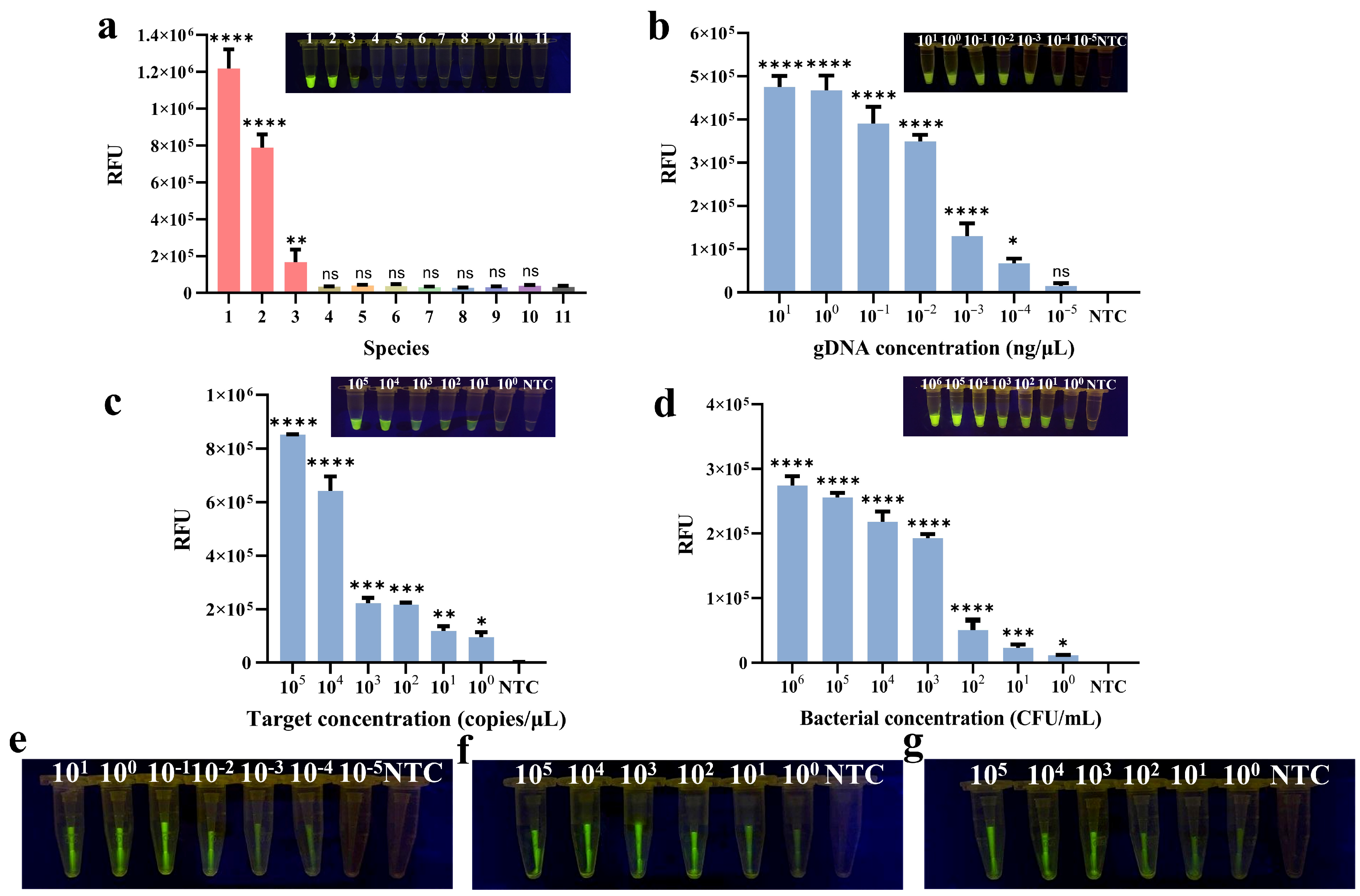
3.6. Evaluation of the Assays’ Specificity and Sensitivity
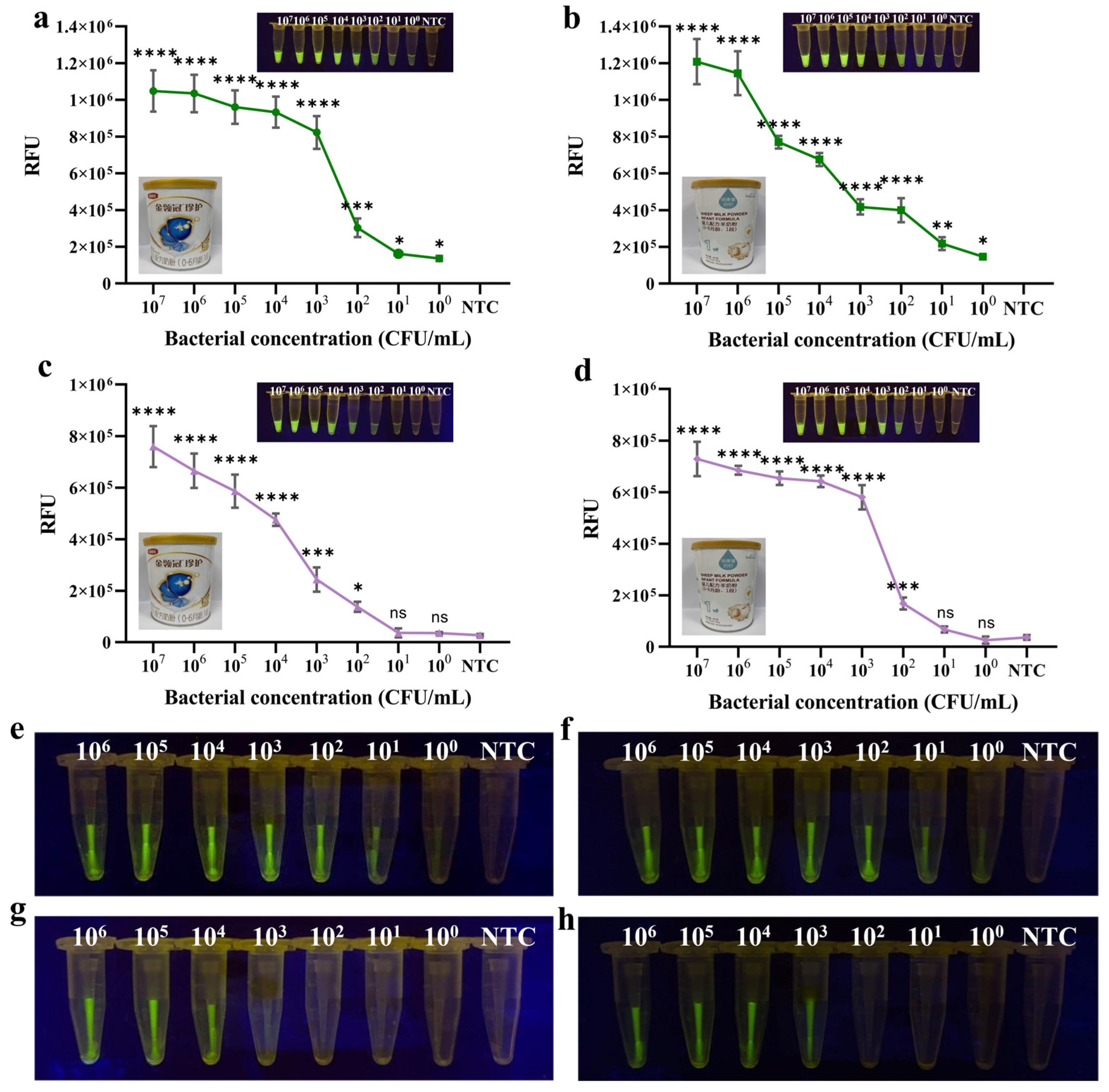
3.7. Application of Methods in Food Samples
3.8. Method Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, P.-R.; Wang, Z.-X.; Xu, Z.-K.; Wang, J.; Li, B.; Shen, X.; Xu, Z.-L. An RPA-Assisted CRISPR/Cas12a Assay Combining Fluorescence and Lateral Flow Strips for the Rapid Detection of Enterotoxigenic Bacillus cereus. J. Agric. Food Chem. 2024, 72, 14000–14010. [Google Scholar] [CrossRef]
- Li, L.; Shen, G.; Wu, M.; Jiang, J.; Xia, Q.; Lin, P. CRISPR-Cas-mediated diagnostics. Trends Biotechnol. 2022, 40, 1326–1345. [Google Scholar] [CrossRef]
- Song, R.; Chen, Z.; Xiao, H.; Wang, H. The CRISPR-Cas system in molecular diagnostics. Clin. Chim. Acta 2024, 561, 119820. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C. Making the cut(s): How Cas12a cleaves target and non-target DNA. Biochem. Soc. Trans. 2019, 47, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Lin, M.; Yue, H.; Tian, T.; Xiong, E.; Zhu, D.; Jiang, Y.; Zhou, X. Glycerol Additive Boosts 100-fold Sensitivity Enhancement for One-Pot RPA-CRISPR/Cas12a Assay. Anal. Chem. 2022, 94, 8277–8284. [Google Scholar] [CrossRef]
- Chen, L.; Hu, M.; Zhou, X. Trends in developing one-pot CRISPR diagnostics strategies. Trends Biotechnol. 2024, 43, 98–110. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley Ann, E.; Segel, M.; Barretto Robert, P.J.; Ranu, A.; Macrae Rhiannon, K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Liu, J.-K.; Nie, X.-Q.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Lin, K.; Guo, J.; Guo, X.; Li, Q.; Li, X.; Sun, Z.; Zhao, Z.; Weng, J.; Wu, J.; Zhang, R.; et al. Fast and visual detection of nucleic acids using a one-step RPA-CRISPR detection (ORCD) system unrestricted by the PAM. Anal. Chim. Acta 2023, 1248, 340938. [Google Scholar] [CrossRef]
- Lu, S.; Tong, X.; Han, Y.; Zhang, K.; Zhang, Y.; Chen, Q.; Duan, J.; Lei, X.; Huang, M.; Qiu, Y.; et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 2022, 6, 286–297. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, X.; Wang, J.; Zhang, Y.; Zeng, W.; Liu, Y.; Zhang, X. Photoactivatable CRISPR/Cas12a Strategy for One-Pot DETECTR Molecular Diagnosis. Anal. Chem. 2022, 94, 9724–9731. [Google Scholar] [CrossRef]
- Hu, M.; Liu, R.; Qiu, Z.; Cao, F.; Tian, T.; Lu, Y.; Jiang, Y.; Zhou, X. Light-Start CRISPR-Cas12a Reaction with Caged crRNA Enables Rapid and Sensitive Nucleic Acid Detection. Angew. Chem. Int. Ed. 2023, 62, e202300663. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Lin, Y.; Zhuo, X.; Zeng, J.; Chen, B.; Zou, Z.; Liu, G.; Xiong, E.; Yang, R. Universal crRNA Acylation Strategy for Robust Photo-Initiated One-Pot CRISPR–Cas12a Nucleic Acid Diagnostics. Angew. Chem. Int. Ed. 2024, 63, e202401486. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Liu, Y.; Zhao, S.; Zhang, Z.; Li, X.; Peng, N.; Jiang, Z. A one-pot CRISPR/Cas13a-based contamination-free biosensor for low-cost and rapid nucleic acid diagnostics. Biosens. Bioelectron. 2022, 202, 113994. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xiu, L.; Guo, X.; Hu, Q.; Yin, K. Spatiotemporal dual-dimensional separation enables ultra-simple one-pot CRISPR-based molecular diagnostics. Chem. Eng. J. 2024, 482, 148872. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, T.; Liu, C.; Xu, Q.; Fang, S.; Wu, Y.; Wu, M.; Liu, Q. An ultrasensitive and contamination-free on-site nucleic acid detection platform for Listeria monocytogenes based on the CRISPR-Cas12a system combined with recombinase polymerase amplification. LWT 2021, 152, 112166. [Google Scholar] [CrossRef]
- Uno, N.; Li, Z.; Avery, L.; Sfeir, M.M.; Liu, C. CRISPR gel: A one-pot biosensing platform for rapid and sensitive detection of HIV viral RNA. Anal. Chim. Acta 2023, 1262, 341258. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef] [PubMed]
- Kalyantanda, G.; Shumyak, L.; Archibald, L.K. Cronobacter Species Contamination of Powdered Infant Formula and the Implications for Neonatal Health. Front. Pediatr. 2015, 3, 56. [Google Scholar] [CrossRef]
- Hayman, M.M.; Edelson-Mammel, S.G.; Carter, P.J.; Chen, Y.; Metz, M.; Sheehan, J.F.; Tall, B.D.; Thompson, C.J.; Smoot, L.A. Prevalence of Cronobacter spp. and Salmonella in Milk Powder Manufacturing Facilities in the United States. J. Food Prot. 2020, 83, 1685–1692. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xie, G.; Xiong, Q.; Mu, D.; Xu, H. A simple and sensitive aptasensor with rolling circle amplification for viable Cronobacter sakazakii detection in powdered infant formula. J. Dairy Sci. 2021, 104, 12365–12374. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.J. Updates on the Cronobacter Genus. Annu. Rev. Food Sci. Technol. 2018, 9, 23–44. [Google Scholar] [CrossRef]
- Kang, Q.; Zhang, S.-Q.; Lin, T.; Li, J.-Z.; Ma, C.-J.; Jiao, J.-B.; Li, C.; Du, X.-J.; Wang, S. Ultrasensitive detection assay for Cronobacter sakazakii based on nucleic acid-driven aggregation-induced emission of gold nanoclusters and cascaded signal amplification. Sens. Actuators B Chem. 2024, 408, 135565. [Google Scholar] [CrossRef]
- Barron, J.C.; Forsythe, S.J. Dry Stress and Survival Time of Enterobacter sakazakii and Other Enterobacteriaceae in Dehydrated Powdered Infant Formula. J. Food Prot. 2007, 70, 2111–2117. [Google Scholar] [CrossRef]
- Fakruddin, M.; Rahaman, M.; Ahmed, M.M.; Hoque, M.M. Stress tolerant virulent strains of Cronobacter sakazakii from food. Biol. Res. 2014, 47, 63. [Google Scholar] [CrossRef]
- Kim, D.; Kim, M. Sensitive detection of viable Cronobacter sakazakii by bioluminescent reporter phage emitting stable signals with truncated holin. Food Res. Int. 2023, 174, 113665. [Google Scholar] [CrossRef]
- Townsend, S.M.; Hurrell, E.; Gonzalez-Gomez, I.; Lowe, J.; Frye, J.G.; Forsythe, S.; Badger, J.L. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 2007, 153, 3538–3547. [Google Scholar] [CrossRef]
- Song, X.; Teng, H.; Chen, L.; Kim, M. Cronobacter Species in Powdered Infant Formula and Their Detection Methods. Korean J. Food Sci. Anim. Resour. 2018, 38, 376–390. [Google Scholar]
- Li, Y.; Wu, L.; Wang, Z.; Tu, K.; Pan, L.; Chen, Y. A magnetic relaxation DNA biosensor for rapid detection of Listeria monocytogenes using phosphatase-mediated Mn(VII)/Mn(II) conversion. Food Control 2021, 125, 107959. [Google Scholar] [CrossRef]
- Huang, C.; Zhao, J.; Lu, R.; Wang, J.; Nugen, S.R.; Chen, Y.; Wang, X. A phage-based magnetic relaxation switching biosensor using bioorthogonal reaction signal amplification for Salmonella detection in foods. Food Chem. 2023, 400, 134035. [Google Scholar] [CrossRef]
- Tamarapu, S.; McKillip, J.L.; Drake, M. Development of a Multiplex Polymerase Chain Reaction Assay for Detection and Differentiation of Staphylococcus aureus in Dairy Products†. J. Food Prot. 2001, 64, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, G.; Li, X.; Xu, Z.; Lei, H.; Shen, X. Development of rapid and easy detection of Salmonella in food matrics using RPA-CRISPR/Cas12a method. LWT 2022, 162, 113443. [Google Scholar] [CrossRef]
- GB 4789.40-2016; National Food Safety Standard—Food Microbiological Examination—Examination of Cronobacter spp. (Enterobacter sakazakii). Standards Press of China: Beijing, China, 2016.
- SN/T 1632.3-2013; Determination of Enterobacter sakazakii (Cronobacter spp.) from Dehydrated Powdered Milk for Export-Part 3: Real-Time PCR Method. Standards Press of China: Beijing, China, 2013.
- Xie, Y.; Liu, L.; Cao, X.-M.; Gai, Z.-Q.; Zhang, X.; Lei, H.-T.; Xu, Z.-L.; Shen, X. A super convenient and specific CRISPR/Cas12a diagnostic platform for toxigenic Burkholderia gladioli based on virulence genes. Food Control 2025, 168, 110904. [Google Scholar] [CrossRef]
- Jianwei, L.; Jobichen, C.; Machida, S.; Meng, S.; Read, R.J.; Hongying, C.; Jian, S.; Yuan, Y.A.; Sivaraman, J. Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage. PLoS Biol. 2023, 21, e3002023. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; van der Oost, J.; Jinek, M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 2017, 66, 221–233.e4. [Google Scholar] [CrossRef]
- Yi, M.; He, P.; Li, J.; Zhang, J.; Lin, L.; Wang, L.; Zhao, L. A portable toolbox based on time-resolved fluoroimmunoassay and immunomagnetic separation for Cronobacter sakazakii on-site detection in dairy. Int. Dairy J. 2022, 133, 105425. [Google Scholar] [CrossRef]
- Gao, S.; Sun, C.; Hong, H.; Gooneratne, R.; Mutukumira, A.; Wu, X. Rapid detection of viable Cronobacter sakazakii in powdered infant formula using improved propidium monoazide (PMAxx) and quantitative recombinase polymerase amplification (qRPA) assay. Food Control 2021, 124, 107899. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xie, Y.; Wang, Z.; Gai, Z.; Zhang, X.; Chen, J.; Lei, H.; Xu, Z.; Shen, X. A Simple, Rapid, and Contamination-Free Ultra-Sensitive Cronobacter sakazakii Visual Diagnostic Platform Based on RPA Combined with CRISPR/Cas12a. Foods 2025, 14, 3120. https://doi.org/10.3390/foods14173120
Liu Y, Xie Y, Wang Z, Gai Z, Zhang X, Chen J, Lei H, Xu Z, Shen X. A Simple, Rapid, and Contamination-Free Ultra-Sensitive Cronobacter sakazakii Visual Diagnostic Platform Based on RPA Combined with CRISPR/Cas12a. Foods. 2025; 14(17):3120. https://doi.org/10.3390/foods14173120
Chicago/Turabian StyleLiu, Yan, Yu Xie, Zhangli Wang, Zuoqi Gai, Xu Zhang, Jiahong Chen, Hongtao Lei, Zhenlin Xu, and Xing Shen. 2025. "A Simple, Rapid, and Contamination-Free Ultra-Sensitive Cronobacter sakazakii Visual Diagnostic Platform Based on RPA Combined with CRISPR/Cas12a" Foods 14, no. 17: 3120. https://doi.org/10.3390/foods14173120
APA StyleLiu, Y., Xie, Y., Wang, Z., Gai, Z., Zhang, X., Chen, J., Lei, H., Xu, Z., & Shen, X. (2025). A Simple, Rapid, and Contamination-Free Ultra-Sensitive Cronobacter sakazakii Visual Diagnostic Platform Based on RPA Combined with CRISPR/Cas12a. Foods, 14(17), 3120. https://doi.org/10.3390/foods14173120






