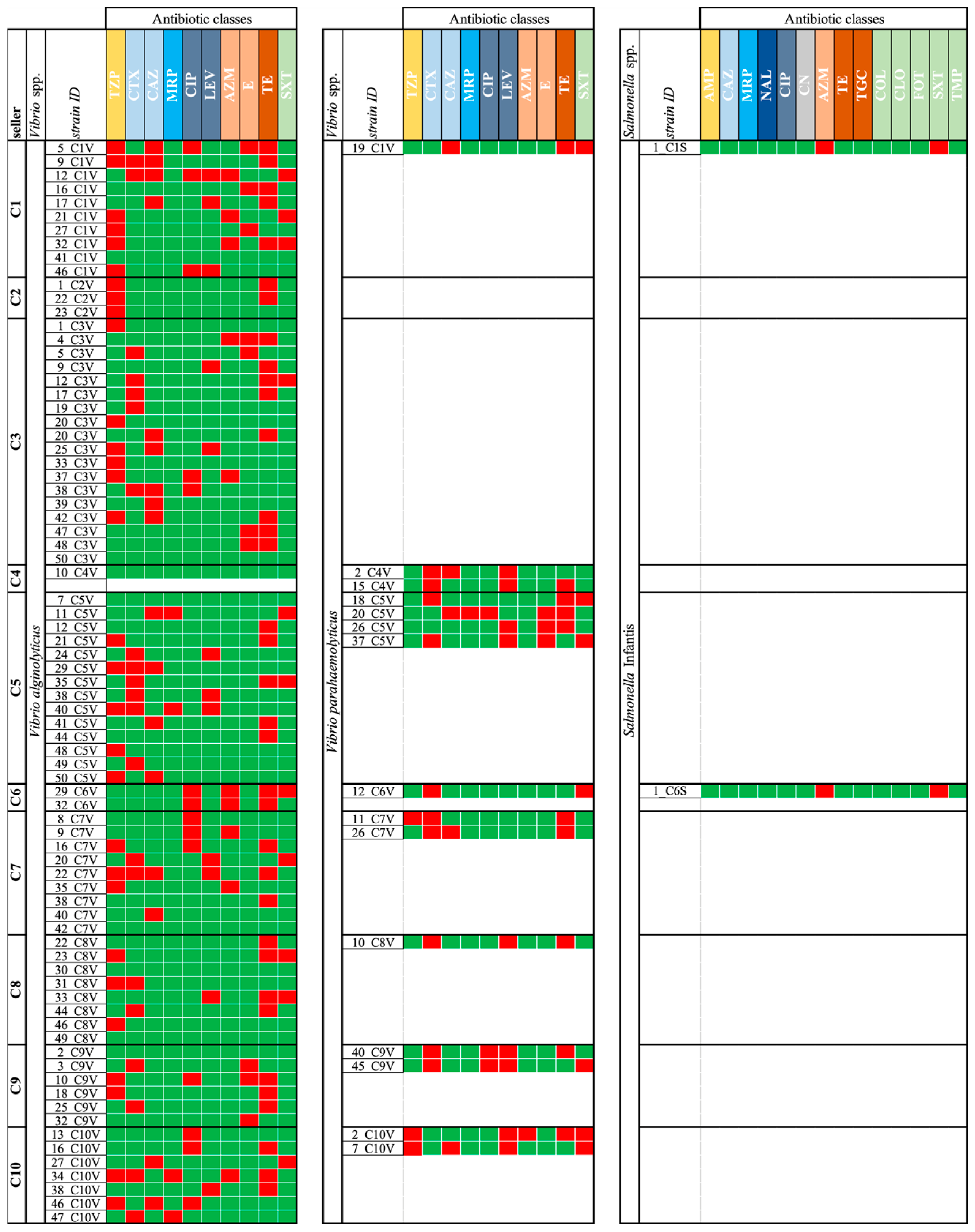
3.1. Detection and Quantification of Target Bacteria in Compliance with Food Safety Criteria
The sale of mussels in conditions that do not comply with current European regulations is a practice that is unfortunately rooted in Campania, although it is increasingly rare. It involves the sale of molluscs operated by unlicensed sellers and/or molluscs without traceability and not in mesh bags (
Table S1). Consumers are not aware of the hazards that may be hidden in these products, especially biological ones. When we refer to bivalve molluscs on the market for their retail sale,
E. coli and
Salmonella spp. must be studied to validate the compliance of the products with the European Regulation (EC) 2073/2005 [
7]. In addition, emerging hazards, such as bacterial species or antimicrobial genes, must be paid attention to.
E. coli is a well-known indicator of fecal contamination, and its presence is notably significant when isolated in both food and environmental contexts.
E. coli is widely recognized as a key indicator of fecal contamination and is used to assess the microbiological quality of seafood, particularly mussels [
11]. The levels of fecal contamination in bivalve molluscs are measured using the MPN method; the analysis involves the entire body of the animal and the intravalvar liquid. Any excess above the limits established by the European legislation is considered a violation of food safety criteria, although
E. coli is not a strictly pathogenic bacterium, and bivalve molluscs should be eaten well cooked. However, its presence in high quantities can represent a health problem, especially if related to the consumption of the product in unsafe ways such as undercooked or raw; furthermore, some
E. coli species are responsible for foodborne disease (e.g., STEC) and the presence of
E. coli is often correlated to the presence of other pathogens, both bacterial and viral.
In this context, the origin of the mussels from illegal stalls is not negligible, especially in a cultural context in which it is customary to consume this product undercooked. Although these products are not sold in compliance with the law and regulations, to correctly interpret the results it is necessary to classify them as “products placed on the market during their shelf life”. Based on these assumptions, it can be said that in all the samples analyzed, the levels of
E. coli exceeded the limit established by Regulation (EC) 2073/2005 [
7] (
Table S2).
Salmonella spp. poses a notable risk considering the food safety criteria in Regulation (EC) 2073/2005 [
7], which requires its absence in live molluscs sold on the market. According to the European Food Safety Authority (EFSA), in 2023, 10.59% of outbreaks linked to fish and fishery products consumption were attributed to
Salmonella spp., including multidrug-resistant (MDR) strains, escalating its threat [
21]. The EFSA report, related to the years 2020/2021, reveals that resistance to ampicillin, sulfonamides, and tetracyclines was noted at elevated levels in
Salmonella spp. isolated from humans in 2021, with a range from moderate to very high in isolates from food-producing animals and poultry carcasses. Multidrug resistance was generally high among
Salmonella spp. reported in human cases in the EU (22.5%). Similarly, MDR has been observed at moderate-to-very high levels in
Salmonella spp. recovered from turkey and broiler carcasses (19.1% and 53.6%, respectively) and at high levels in fattening pigs (39.1%), calves (30.4%), fattening turkeys (41.7%), and broilers for fattening (44%). Resistance to multiple antibiotics has been observed in
S. Infantis and other serotypes, bolstering their epidemiological prominence [
22].
Our analyses aimed at detecting
Salmonella spp. indeed unveiled the presence of
S. Infantis in two samples. Additionally, the isolated strains exhibited notable resistance to two antibiotics: trimethoprim-sulfamethoxazole and azithromycin (
Figure 2). While in the European Union territory
S. Enteritidis is the most common serovars responsible for non-typhoidal salmonellosis,
S. Infantis is the most isolated in foods in Italy (approximately 47.55% in 2022 [
23], and the first serovar isolated from the “broiler” source (96.1%) in 2023 [
21].
3.2. Vibrio spp., Emerging Pathogenic Species
The dynamism of EU food legislation is driven by the emergence of new global public health challenges. Analyzing new risks is necessary to identify the critical concentration limits above which food containing a hazard could be harmful to the consumer. New hazards and challenges are reported by both scientific communities, through their publications, and by Food Business Operators and Competent Authorities, through self-control and official control activities. In this scenario, some
Vibrio species are gaining ground among emerging bacterial pathogens due to their role in causing foodborne disease worldwide. The main pathogen vehicle is seafood, involved in 32 cases of foodborne outbreaks (FBOs) with strong and weak evidence, especially per the presence of
V. parahaemolyticus (30/32). This bacterium was responsible for 221 human cases and 57 hospitalizations. The list of virulence and pathogenicity genes is also evolving; until a few years ago, the genes encoding the hemolysins TDH and TRH (thermostable direct hemolysin, TDH, or TDH-related hemolysin, TRH) were the only ones considered. However, a small portion of
V. parahaemolyticus isolated from those human cases (~10%) did not show positivity for these genes, highlighting the plausible presence of other virulence factors.
V. alginolyticus may also occasionally be involved in human cases of disease, especially in the most susceptible target individuals (immunocompromised) [
24].
The challenges related to this bacterial genus are more than those mentioned above; for instance, depuration, the standard method for depurating bivalve molluscs from coliforms (
E. coli, in particular; Regulation UE 2019/627 [
13]), is not properly effective in removing and cleaning filter-feeding organisms from
Vibrio [
25]. Food business operators should control and monitor all the variables that can influence the depuration of organisms from these bacteria, such as water temperature, salinity, and pH, and the dwell time of molluscs in water. European legislation does not set the hours or days necessary for depuration, and food business operators refer to scientific data that confirm the validity of their applied methods. However, until the legislators consider
Vibrio, the methods will be evaluated on microorganisms such as
E. coli, which are more sensitive to depuration processes. Bivalve molluscs could be vehicles of these bacteria and, therefore, of the antibiotic-resistant genes (ARGs) commonly present in their genome [
26].
In this study, particular attention was paid to the
Vibrio genus, investigating their presence in 10 batches of mussels. In each mussel batch, characteristic colonies of
Vibrio were isolated; identification with MALDI-TOF confirmed our suspicions and highlighted the presence of potentially dangerous enteropathogenic species. In more detail, out of the total of 300 colonies subjected to identification (30 characteristic colonies per mussel batch), 177 colonies were identified at the species level (58%). The most frequently isolated bacterial species were
Vibrio alginolyticus (44.07%),
Shewanella putrefaciens (24.29%), and
Proteus mirabilis (19.77%) (
Figure S1). It is not uncommon to identify genera other than
Vibrio that are capable of growing on TCBS agar [
27].
As shown in
Figure S1, the main bacterial species identified was
V. alginolyticus. In humans, this bacterium is usually a vehicle of extraintestinal infections, mainly skin infections resulting from contact with seawater. However, the gastrointestinal pathogenic potential of
V. alginolyticus in humans has also been recognized, even capable of causing mortality in immunocompromised patients [
28]. The other
Vibrio species isolated was
V. parahaemolyticus, a ubiquitous bacterium, with a global distribution, primarily inhabiting coastal marine ecosystems. Its prevalence poses a significant food safety concern, particularly in marine-derived products from temperate or tropical waters, including bivalve molluscs, crustaceans, and fish. The ingestion of raw or contaminated seafood harboring
V. parahaemolyticus can lead to acute gastroenteritis, manifested by symptoms such as diarrhea, vomiting, and abdominal cramps. Consequently,
V. parahaemolyticus stands as the predominant causative agent of seafood-associated gastroenteritis worldwide.
Despite the implementation of diverse preservation methodologies,
V. parahaemolyticus exhibits resilience within seafood matrices, contributing to its sustained prevalence. This persistence is notably accentuated by the emergence of specific serotypes, notably those carrying the
tdh gene [
29]. However, it is not only virulence and pathogenicity genes that are cause of concern, but also those related to antibiotic resistance. When a strain is equipped with them, it not only acquires resistance to a certain antibiotic, but becomes a carrier and donor of the same genes to other bacteria.
Aware of the importance and need to contain the spread of ARGs and, therefore, of MDR bacteria, a phenotypic evaluation of the antibiotic resistance profile of
Vibrio strains isolated from the mussels was carried out. In particular, a total of 93
Vibrio strains were subjected to antimicrobial susceptibility testing toward ten antibiotics. After incubation, the inhibition zones were recorded for each strain and data were analyzed according to EUCAST guidelines [
14]. The specific antibiotic resistance profile of each
Vibrio isolated in this study is shown in
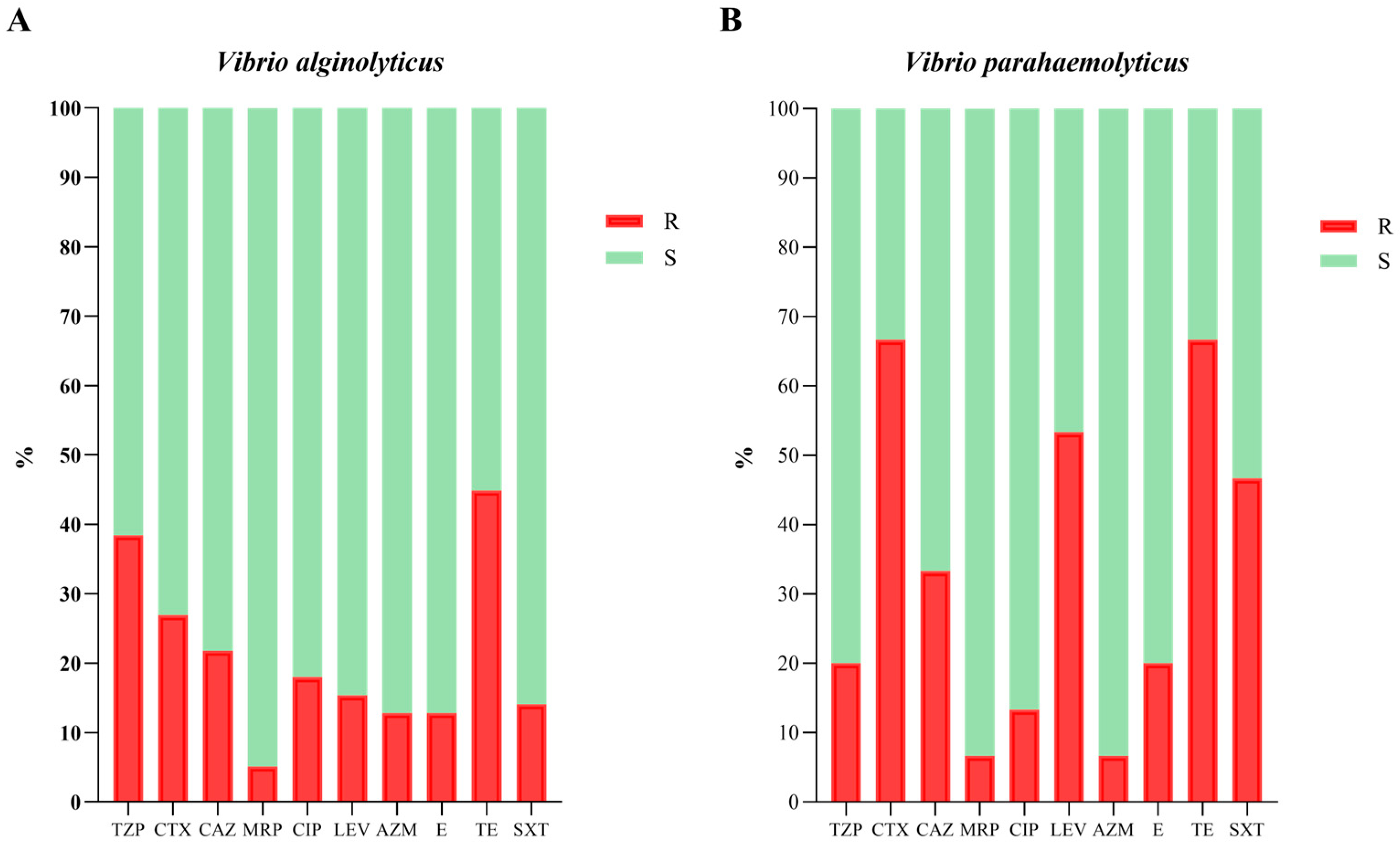
Figure 2. No strain showed resistance to more than six antibiotics. In more detail, 1.08% (1/93 strains), 4.30% (4/93), and 10.75% (10/93) showed resistance to six, five, and four antibiotics, respectively; 27.96% (26/93), 29.03% (27/93) and 18.28% (17/93) were resistant to three, two, and one antibiotic, respectively; and finally, only 8.60% (8/93) were sensitive to all tested antibiotics (10/10). Overall, the highest percentage of resistance was detected against TE (48.39%, 45/93 strains), followed by resistance to TZP (35.48%, 33/93), CTX (33.33%, 31/93), CAZ (23.66%, 22/93), and LEV (22.58%, 21/93). On the other hand, most of the
Vibrio strains were found to be susceptible to MRP (94.62%, 88/93), AZM (88.17%, 82/93) and E (86.02%, 80/93). Analyzing the data by species, the results were slightly different (
Figure 3). Higher resistance rates have been described for
V. parahaemolyticus strains. However, it is worth noting that fewer strains belonging to the
parahaemolyticus species were isolated in the present study than those belonging to the
alginolyticus species. For these reasons, the percentages were calculated on different numbers of strains (78 strains of
V. a. and 15 strains of
V. p.) and this may influence the calculation of the percentages that are difficult to compare with each other. In any case, for both species the highest resistance rate was against TE (66.67% and 44.87% for
V. p. and
V. a., respectively), while the lowest was against MRP (6.67% and 5.13% for
V. p. and
V. a., respectively). Meanwhile, the principal differences were in the resistance to CTX, LEV, and SXT, with
V. parahaemolyticus strains showing higher resistance (
Figure 3B).
In summary, the analyses performed revealed a widespread and varied presence of antibiotic resistance which made the strains resistant to multiple classes of antibiotics (
Figure S2). In particular, 37.63% (35/93
Vibrio strains) of the strains examined were found to be multidrug-resistant (
Figure 4), meaning resistant to at least three different classes of antibiotics. It is worth underlining that almost all
V. parahaemolyticus strains were multidrug-resistant (80%, 12/15 strains); lower percentages were obtained for
V. alginolyticus (29.49%, 23/78 strains).
Multidrug-resistant strains showed resistance to several classes of antibiotics, with the highest rates observed for tetracyclines and cephalosporins, both affecting 68.57% of Vibrio isolates. High levels of resistance were also recorded for fluoroquinolones (62.86%), followed by penicillins (48.57%), miscellaneous agents (45.71%), and macrolides (37.14%). Particularly concerning is the resistance to carbapenems, identified in 11.43% of the resistant strains. Although relatively low, this finding is significant given the critical role of carbapenems as last-resort antibiotics. Their reduced efficacy, even in a small proportion of isolates, raises serious concerns about treatment options and the potential for further dissemination of resistance.
Recent studies suggest that multidrug resistance in vibrios and other enteric pathogenic bacteria is mainly attributable to horizontal gene transfer. This process involves the movement of mobile genetic elements, such as plasmids, transposons, and integrons, between different bacteria, regardless of their phylogenetic relationship. These genetic elements are highly dynamic and facilitate the rapid transfer of resistance genes, contributing to the spread of antibiotic resistance [
30]. These horizontal transfer mechanisms could explain the isolation of several MDR strains from the same sample, as well as the simultaneous presence of resistance to trimethoprim-sulfamethoxazole and azithromycin in
Salmonella and
Vibrio strains isolated from samples C1 and C6.
A more in-depth analysis of antibiotic resistance patterns further highlights the critical situation regarding multidrug resistance among
Vibrio isolates. The Multiple Antibiotic Resistance (MAR) index for
V. parahaemolyticus was particularly high (0.34 ± 0.07), exceeding the critical threshold of 0.2 across all isolates, suggesting that these strains likely originate from environments with intense antibiotic pressure.
V. alginolyticus showed greater variability (mean = 0.21 ± 0.13; min = 0; max = 0.6), indicating a mixed population of resistant and susceptible strains. This variability is due both to the greater number of
V. alginolyticus strains isolated in this study and to the variability among the seized mussel samples. Indeed, referring to
Figure S3 it is possible to note the different placement of the 10 samples with respect to the impact of antibiotic use and persistence in the environment. Overall, in fact, the MAR value was higher for the strains isolated from samples C1, C6, and C10 (0.32 ± 0.05, 0.30 ± 0.06, and 0.29 ± 0.05, respectively; mean ± error standard), followed by C7, C9, and C5 (0.22 ± 0.07, 0.23 ± 0.05, and 0.24 ± 0.08, respectively; mean ± error standard). Aggregating data from all
Vibrio isolates, the mean MAR index was also above the critical threshold (0.23 ± 0.13), again highlighting the importance of the observed antibiotic resistance profiles and raising concerns about the ability to spread these types of resistance.
Furthermore, the antibiotic resistance pattern (ARPA) index for the 93 Vibrio strains was 2.31. Since this index is calculated as the ratio between the total resistances found and the total bacteria subjected to the antibiotic sensitivity study, it is possible to state that each Vibrio strain was resistant, on average, to more than two antibiotics. More specifically, the index was 2.10 for V. alginolyticus strains and 3.40 for V. parahaemolyticus strains.
The values of the MAR and ARPA indices underscore the severity of antibiotic resistance in these Vibrio strains, highlighting the likelihood that the mussels were exposed to environments with high antibiotic pressure.
3.3. Mussels’ Traceability
It is well known that NIRS is a widely used technique in several fields, like food analysis [
31]. Using the infrared part of the spectrum (about 800–2500 nm), it examines how electromagnetic waves interact with a sample, exploring its physical–chemical properties. The underlying principle of NIRS involves the interaction of infrared radiation with molecular vibrations. When molecules absorb energy at specific wavelengths within the near-infrared spectrum, they undergo a transition to an excited vibrational state. Subsequent relaxation to the ground state results in the emission of infrared light, which is quantitatively measured by a specialized detector. This process yields a unique absorption profile characteristic of the analyzed sample’s distinct physicochemical properties. Physically, the NIR radiation primarily influences combination bands and overtones, which are mostly linked to C-C, O-H, and N-H bonds. The resulting signal is generally weak, but highly characteristic. Compared to traditional chemical analysis methods, NIRS stands out for its speed and simplicity. Measurements typically take only 5–10 s and deliver a high accuracy and reproducibility. Crucially, NIRS requires no preliminary sample preparation or specialized technical training for the operator, making it a highly accessible tool. As reported by [
32], NIRS provides valuable information on the chemical composition and physical properties of samples, as demonstrated in studies on mussel tissues and shells, including the bioaccumulation of inorganic elements. These characteristics are significantly influenced by factors such as the mussels’ geographical origin, size, and spatial distribution. Consequently, analyzing trace element levels in samples through NIRS can help differentiate between various geographical origins.
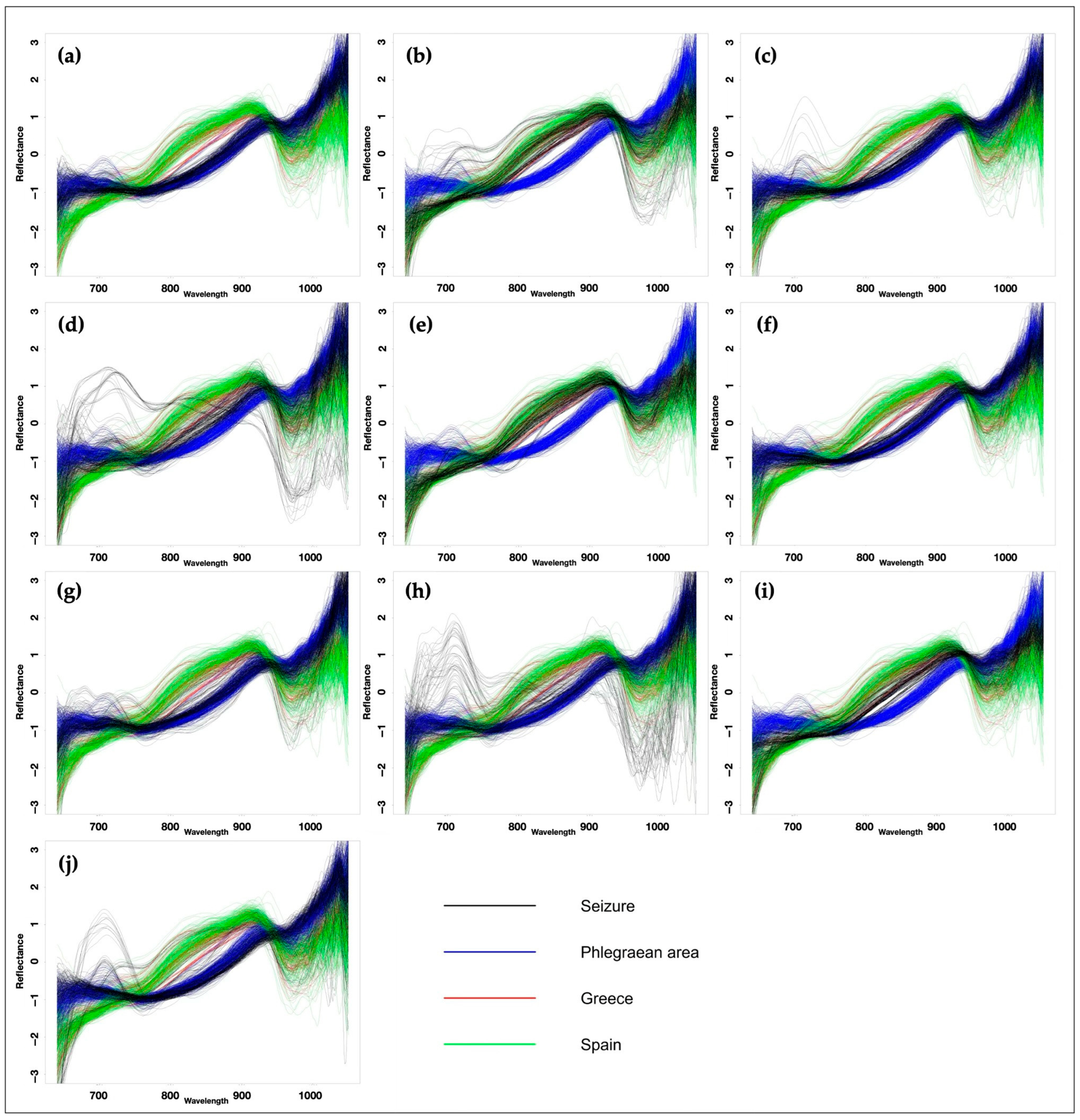
In this study, for each seized sample the spectra were overlaid and compared with the reference standards, i.e., with calibration spectra of mussels of Phlegraean origin, Greek origin, and Spanish origin (
Figure 5). The hypothesized match was finally attributed to the origin that showed the highest percentage among the three loaded into the dataset (detailed information is reported as
supplementary data, in Table S3).
The random forest model classified ten seized samples based on their correspondence to standard references. Therefore, by superimposing the spectra of the samples with those of the reference standards, it was possible to state that seven samples (seized samples 1, 3, 4, 6, 7, 8 and 10) should have Phlegraean origin, due to the highest percentage of overlap with the respective reference spectra (
Figure 5); in contrast, in three samples the highest percentage of overlap with the reference spectra of Greek mussels suggests a Greek origin (seized samples 2, 5 and 9). Percentages of overlap seem to suggest that no sample has a Spanish origin. Regarding the percentage of correspondence, in four cases (seized samples 1, 3, 6 and 7) a very strict correspondence (>90%) with the reference spectra of Phlegraean origin was reported; in particular, only seized sample 1 (C1) showed 100% correspondence. On the other hand, seized samples 2 and 5 showed the weakest correspondence with the standard references, with a 59% and 61.5% match, respectively (
Table S3). The
Random Forest model employed in this study classifies seized samples based on the three geographical origins established in the training dataset. The percentage assignment to each class is derived from the affinity of the sampled Near-Infrared (NIR) spectra. It was observed that certain groups, specifically C2, C4, and C5, exhibited relatively low affinity percentages, approximating 60%. This comparatively modest value may stem from several contributing factors: potential sampling errors—given the inherent difficulty of obtaining consistent measurements from the curved shells of molluscs—or the poorly defined boundaries among the three designated origin groups. Future implementations will focus on two key improvements: refining the sampling methodologies and substantially increasing the number of spectra utilized in the construction of the training dataset.
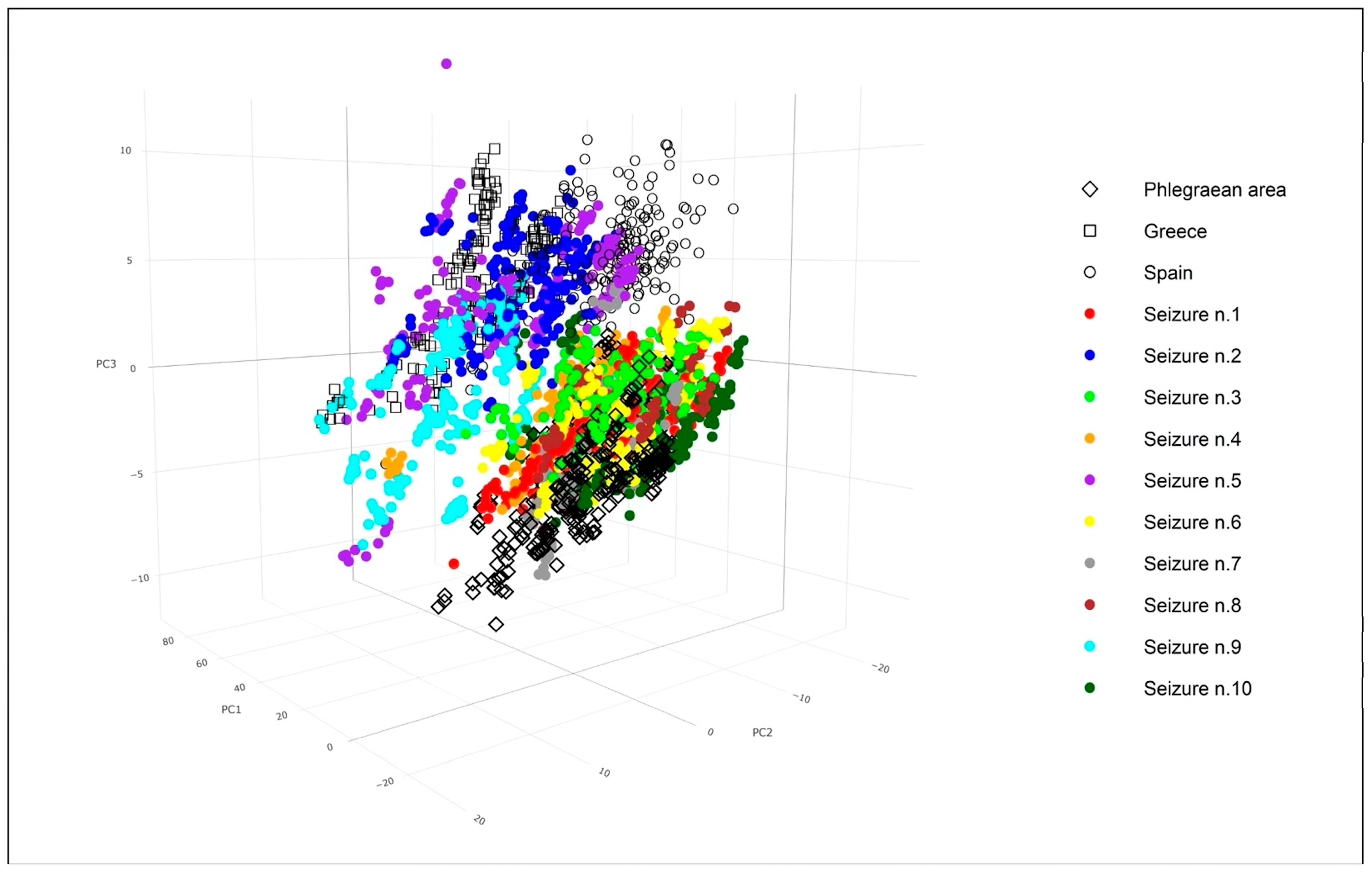
A Principal Component Analysis (PCA) was performed to evaluate the spectral overlap between seized samples and established reference standards.
Figure 6, which visualizes the PCA scores for the first three components, reveals that PC1, PC2, and PC3 account for 72.7%, 14.4%, and 4.5% of the total explained variance, respectively.
As evident from
Figure 6, the standard reference samples tended to group into three distinct clusters, corresponding to their three different geographical origins. Notably, the spectra of the seized samples exhibited varying degrees of overlap within the same three-dimensional space as these standard references. The degree of overlap between the different clusters is a critical indicator of how effectively the PCA embedding distinguishes them. Significant overlap signifies similarity, while clear separation implies distinct underlying characteristics. Looking at the standard references, the “Phlegraean area” (black diamonds) and “Greece” (black squares) data points form relatively distinct clusters, showing good separation from each other and from most of the seized sample categories. In contrast, the “Spain” points (black circles) are notably more scattered, indicating greater variability within this reference group.
Among the seized samples, there is a discernible attempt to differentiate between various types. Some seized samples form relatively compact clusters, suggesting internal homogeneity. For instance, samples 1, 3, 6, 7, 8, and 10 visually cluster together in the lower-right quadrant of the plot.
There was notable overlap between the “Phlegraean area” reference samples and several seized samples (specifically C1, C3, and C6). This overlap could suggest shared underlying characteristics or a less definitive distinction between these particular seized samples and the Phlegraean origin when based solely on these principal components. Conversely, “Phlegraean area” and “Greece” showed better separation, as do some pairwise comparisons between seized samples (e.g., C1 versus C9). This improved separation suggests that these groups possess more distinctive features captured by the principal components.
A more detailed examination reveals that seized samples 1, 3, 4, 6, 7, 8, and 10 exhibited overlap within the three-dimensional space corresponding to the Phlegraean standard references. Conversely, seized sample 9 demonstrated an overlap with the Greek standard references. Lastly, seized samples 2 and 5 were observed to share common three-dimensional space with both the Greek and Spanish standard references.
The 3D PCA plot in
Figure 6 effectively visualizes the complex relationships and distinctions among the various data points, immediately highlighting both well-separated clusters and significant areas of overlap based on their underlying spectral characteristics. This figure also provides direct insights into the geographical origins of the seized samples. Consequently, these findings lead to the hypothesis that most of the seized mussels are derived from local aquaculture operations.
The application of NIRS technology, in conjunction with robust decision-making models and blockchain technology [
33], emerges as a reliable methodology for ensuring the quality and traceability of mussels, thus preserving the interests of both consumers and producers. Beyond regulatory applications, this technique could also be adopted by food industry operators seeking to authenticate and certify product origin, thus valorizing their production. Concurrently, blockchain technology would ensure unassailable transparency and legality, thereby empowering consumers with informed and confident choices. The BluDev
® technology, employed in this investigation, was specifically engineered to fulfill these needs, providing not only expeditious support for origin identification but also the capacity to render information immutable and transparent for end-users.