Decoding the Molecular Mechanisms of Menthol Isomer Perception Based on Computational Simulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Sensory Analysis
2.2.1. Recruitment and Training of the Panelists
2.2.2. Sensory Quantitative Descriptive Analysis
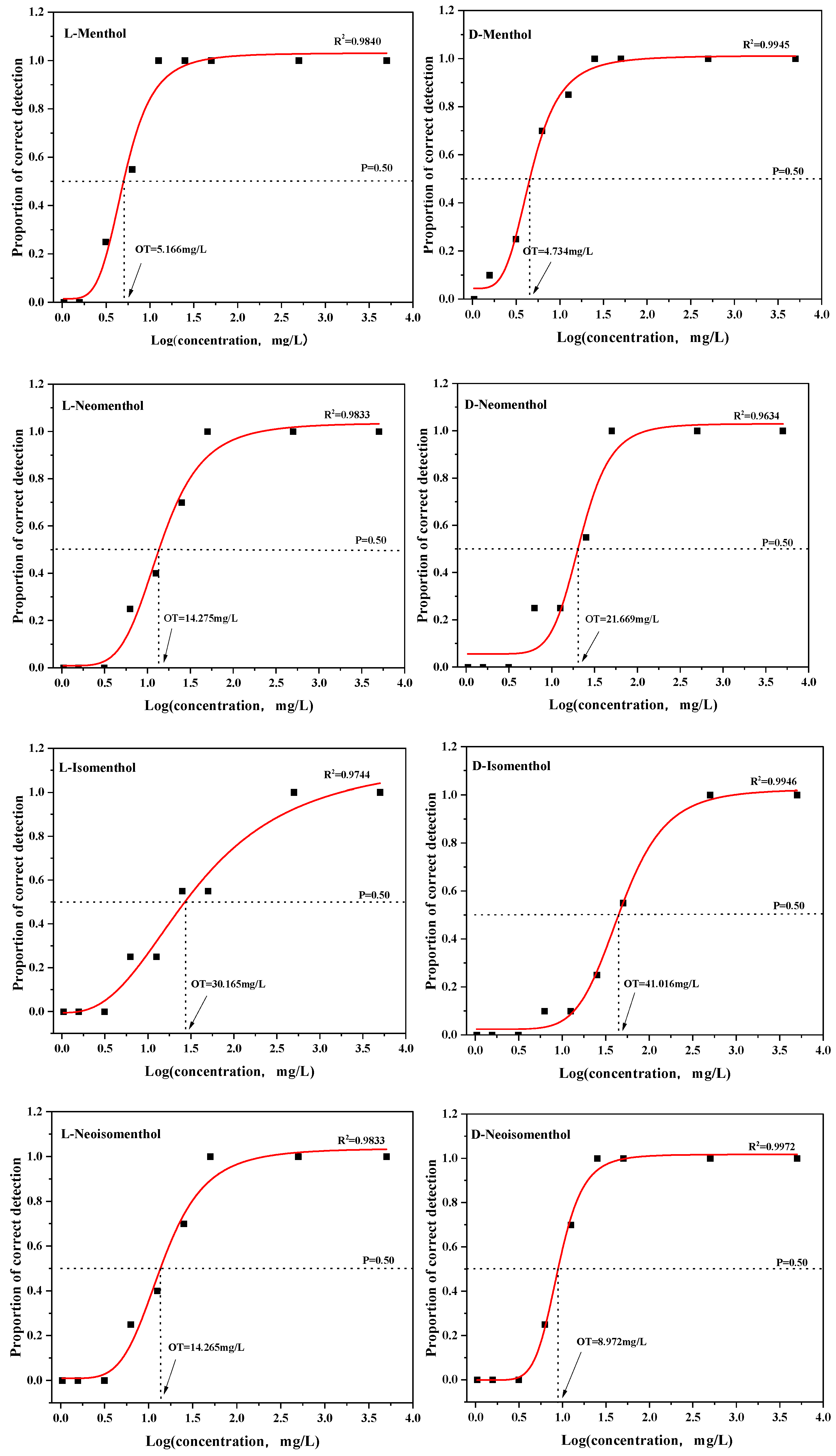
2.2.3. Determination of Odor Detection Thresholds
2.3. Molecular Docking
2.3.1. Preparatory Work
2.3.2. Model Construction and Optimization
2.3.3. Molecular Docking
3. Results and Discussion
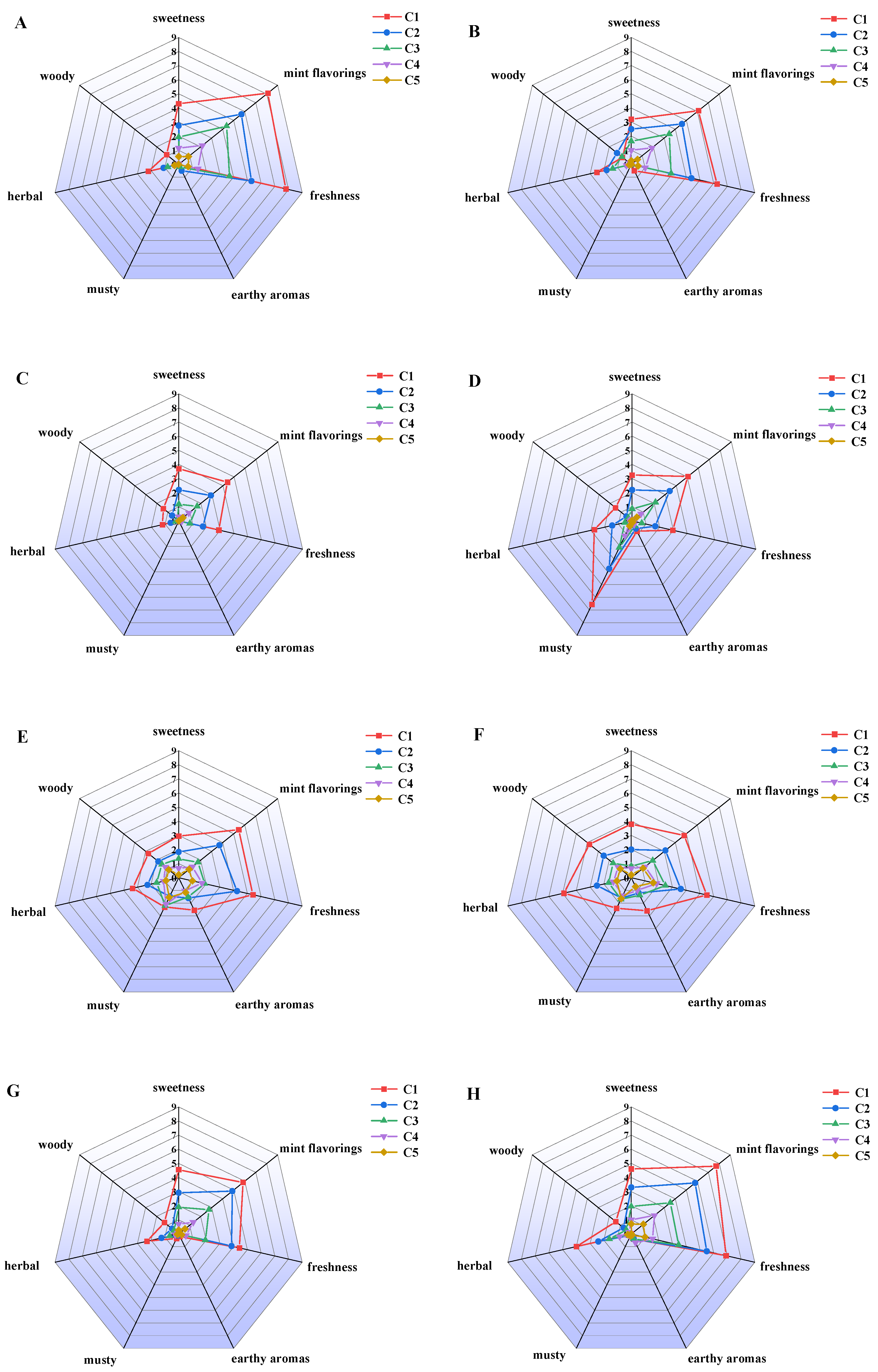
3.1. Aroma Profiles of Eight Menthol Isomers
3.2. Threshold Analysis of Eight Menthol Isomers
3.3. Analysis of Interactions Between ORs and Menthol Isomers by Molecular Docking
3.3.1. Binding Regions Between ORs and Menthol Isomers
3.3.2. Comparison of Binding Energy Between Different ORs and Menthol Isomers
3.3.3. Analyzing the Binding Forces and Sites Between ORs and Menthol Isomers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kamatou, G.P.P.; Vermaak, I.; Viljoen, A.M.; Lawrence, B.M. Menthol: A simple monoterpene with remarkable biological properties. Phytochemistry 2013, 96, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Merckel, C.; Pragst, F.; Ratzinger, A.; Aebi, B.; Bernhard, W.; Sporkert, F. Application of headspace solid phase microextraction to qualitative and quantitative analysis of tobacco additives in cigarettes. J. Chromatogr. A. 2006, 1116, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Casares, N.; Alfaro, M.; Cuadrado-Tejedor, M.; Lasarte-Cia, A.; Navarro, F.; Vivas, I.; Lasarte, J.J. Improvement of cognitive function in wild-type and Alzheimer´s disease mouse models by the immunomodulatory properties of menthol inhalation or by depletion of T regulatory cells. Front. Immunol. 2023, 14, 1130044. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Han, Y.; Chen, X.; Aierken, A.; Wen, H.; Zheng, W.; Wang, H.; Lu, X.; Zhao, Z.; Ma, C.; et al. Molecular mechanisms underlying menthol binding and activation of TRPM8 ion channel. Nat. Commun. 2020, 11, 3790. [Google Scholar] [CrossRef] [PubMed]
- Takai, Y.; Touhara, K. Enantioselective recognition of menthol by mouse odorant receptors. Biosci. Biotechnol. Biochem. 2015, 79, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, M.; Li, Y.; Wan, X.; Yang, X. Characterization of the orchid-like aroma contributors in selected premium tea leaves. Food Res. Int. 2019, 129, 108841. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, X.; Lin, Y.; Ke, Q.; Niu, Y.; Zhang, J.; Xiao, Z. Unraveling the characteristic chestnut aroma compounds in MeiTanCuiYa green tea and their interaction mechanisms with broad-spectrum olfactory receptors using molecular docking. Lebensm. Wiss. Technol. 2024, 194, 115785. [Google Scholar] [CrossRef]
- Zhou, Y.; He, Y.; Zhu, Z. Understanding of formation and change of chiral aroma compounds from tea leaf to tea cup provides essential information for tea quality improvement. Food Res. Int. 2023, 167, 112703. [Google Scholar] [CrossRef] [PubMed]
- Eccles, R. Menthol and related cooling compounds. J. Pharm. Pharmacol. 1994, 46, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Gusain, P.; Ohki, S.; Hoshino, K.; Tsujino, Y.; Shimokawa, N.; Takagi, M. Chirality-Dependent Interaction of d- and l-Menthol with Biomembrane Models. Membranes 2017, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Zhang, F.; Liu, Z.; Chen, H.; He, P.; Liu, C.; Zhu, R. Separation of Eight Optical Isomers of Menthol by Tandem Capillary Chiral Columns. Food Sci. 2021, 42, 339–345. [Google Scholar]
- Bear, D.M.; Lassance, J.M.; Hoekstra, H.E.; Datta, S.R. The Evolving Neural and Genetic Architecture of Vertebrate Olfaction. Curr. Biol. 2016, 26, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.-L.; Wu, W.; Tang, C.-Y.; Ren, J.-L.; Jiang, D.; Li, J.-T. Transcriptome Analysis Reveals Olfactory System Expression Characteristics of Aquatic Snakes. Front. Genet. 2022, 13, 825974. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Koo, J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012, 45, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Kalra, S.; Mittal, A.; Gupta, K.; Singhal, V.; Gupta, A.; Mishra, T.; Naidu, S.; Sengupta, D.; Ahuja, G. Analysis of single-cell transcriptomes links enrichment of olfactory receptors with cancer cell differentiation status and prognosis. Commun. Biol. 2020, 3, 506. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Cho, H.J.; Lee, C.; Koo, J. Odorant receptors in cancer. BMB Rep. 2022, 55, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.S.; Ye, J.S.; Shim, J. Exploring the influence of olfactory receptors in metabolic diseases and cancers: Beyond sensory functions. Kosin. Med. J. 2025, 40, 15–20. [Google Scholar] [CrossRef]
- Jiang, E. Response Pattern Analysis of Human Olfactory Receptors to Indole. Ph.D. Dissertation, Seoul National University, Seoul, Republic of Korea, 2020. [Google Scholar]
- Florentinus-Mefailoski, A.; Bowden, P.; Scheltens, P.; Killestein, J.; Teunissen, C.; Marshall, J.G. The plasma peptides of Alzheimer’s disease. Clin. Proteom. 2021, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Ben Khemis, I.; Sagaama, A.; Issaoui, N.; Ben Lamine, A. Steric and energetic characterizations of mouse and human musk receptors activated by nitro musk smelling compounds at molecular level: Statistical physics treatment and molecular docking analysis. Int. J. Biol. Macromol. 2021, 188, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Ben Khemis, I.; Knani, S.; Aouaini, F.; Graba, B.; Ben Lamine, A. New insights into the docking mechanism of 4-methylphenol adsorption on cow and human olfactory receptors: Molecular docking simulation statistical physics modeling. Mater. Chem. Phys. 2025, 341, 130929. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, X.; Sun, Z.; Shen, T.; Kou, X.; Niu, Y.; Xiao, Z. Unraveling the interaction mechanism between enantiomers of lactone compounds (γ-octalactone and γ-undecalactone) in Longjing tea and OR1A1 olfactory receptor using molecular docking and molecular dynamics simulation. Food Biosci. 2025, 66, 106282. [Google Scholar] [CrossRef]
- Jia, X.; Gao, Y.; Xi, H.; Cui, C.; Yang, X.; He, B.; Li, T. A flavor imitation method for Osmanthus aroma based on molecular docking screening and odor activity value analysis. LWT—Food Sci. Technol. 2025, 223, 117697. [Google Scholar] [CrossRef]
- Xiao, Z.; Gao, J.; Niu, Y.; Wang, Z.; Zhou, R.; Zhang, J.; Zhu, J. Elucidation of the sweetening mechanism of sweet orange fruit aroma compounds on sucrose solution using sensory evaluation, electronic tongue, molecular docking, and molecular dynamics simulation. LWT—Food Sci. Technol. 2024, 205, 116555. [Google Scholar] [CrossRef]
- Xiao, Z.; Qu, H.; Mao, C.; Niu, Y. Study on the sweetening mechanism of aroma compounds in yangshan peach using sensory analysis, molecular docking, and molecular dynamics simulation techniques. LWT—Food Sci. Technol. 2023, 191, 115562. [Google Scholar] [CrossRef]
- Jiang, J.; Ji, S.; Pan, G.; Tao, X.; An, F.; Liu, Q.; Wu, R. Machine learning combined with sensory evaluation and multi-sensor technology to evaluate the overall quality of commercial soybean paste in China. J. Future Foods 2025, in press. [Google Scholar] [CrossRef]
- GB/T 22366-2022/ISO 13301:2018; Sensory Analysis–Methodology–General Guidance for Measuring Odour, Flavour and Taste Detection Thresholds by a Three-Alternative Forced-Choice (3-AFC) Procedur. ISO: Geneva, Switzerland, 2022.
- Wang, J.; Zhang, C.; Qian, J.; Wang, S.; Fan, W.; Shi, Q.; Mao, J.; Xie, J.; Zhang, Q.; Chai, G. Structure-threshold relationship in food aroma molecules: Insights from S-curve method, molecular docking, and dynamics simulations. Curr. Res. Food Sci. 2025, 10, 101073. [Google Scholar] [CrossRef] [PubMed]
- Sharmeen, J.; Mahomoodally, F.; Zengin, G.; Maggi, F. Essential Oils as Natural Sources of Fragrance Compounds for Cosmetics and Cosmeceuticals. Molecules 2021, 26, 666. [Google Scholar] [CrossRef] [PubMed]
- Sonstrom, R.E.; Cannon, D.M.; Neill, J.L. Chiral Analysis of Linalool, an Important Natural Fragrance and Flavor Compound, by Molecular Rotational Resonance Spectroscopy. Symmetry 2022, 14, 917. [Google Scholar] [CrossRef]
- Cerutti-Delasalle, C.; Mehiri, M.; Cagliero, C.; Rubiolo, P.; Bicchi, C.; Meierhenrich, U.J.; Baldovini, N. The (+)-cis- and (+)-trans-Olibanic Acids: Key Odorants of Frankincense. Angew. Chem. Int. Ed. 2016, 55, 13719–13723. [Google Scholar] [CrossRef] [PubMed]
- Masashi, K.; Saki, K.; Takuya, I.; Hiroyuki, M.; Yasuo, T.; Naoki, T. Synthesis and odor properties of Phantolide analogues. Tetrahedron 2017, 73, 2089–2099. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, Z.; Liu, Z.; Zhu, R.; Zhang, F.; Liu, Z.; Si, X. Botanical discrimination and classification of Mentha plants applying two-chiral column tandem GC-MS analysis of eight menthol enantiomers. Food Res. Int. 2022, 162, 112035. [Google Scholar] [CrossRef] [PubMed]
- Gondal, H.Y.; Choudhary, M.I.; Khan, A.A. Microbial Transformations of (+)-Isomenthol by Fusarium lini and Rhizopus stolonifer. Chem. Pharm. Bull. 2011, 59, 874–875. [Google Scholar] [CrossRef] [PubMed]
- Dilshod, A.M.; Alisher Kh, K.; Khamid Kh, K.; Khamza, S.T.; Enrico, B. Relationship between structural properties and biological activity of (-)-menthol and some menthyl esters. Comput. Biol. Chem. 2025, 115, 108357. [Google Scholar] [CrossRef] [PubMed]
- Haifang, M.; Yucheng, Z.; Zhengyang, X.; Yun, Z. Enhanced enantioselective separation of racemic menthol via reverse-phase high-performance liquid chromatography: Method development and computational insights for pre-screening. Talanta 2024, 282, 127062. [Google Scholar]
- Tian, H.; Yu, B.; Yu, H.; Chen, C. Evaluation of the synergistic olfactory effects of diacetyl, acetaldehyde, and acetoin in a yogurt matrix using odor threshold, aroma intensity, and electronic nose analyses. J. Dairy Sci. 2020, 103, 7957–7967. [Google Scholar] [CrossRef] [PubMed]
- Oliveira da Silva, A.L.; Lempert, L.K.; Glantz, S.A. More than a “characterizing flavor”: Menthol at subliminal levels in tobacco products. Drug Alcohol Depend. 2024, 261, 111346. [Google Scholar] [CrossRef] [PubMed]
- Rosenblatt, M.R.; Olmstead, R.E.; Iwamoto-Schaap, P.N.; Jarvik, M.E. Olfactory thresholds for nicotine and menthol in smokers (abstinent and nonabstinent) and nonsmokers. Physiol. Behav. 1999, 65, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Krüsemann, E.J.Z.; Cremers, J.W.J.M.; Visser, W.F.; Punter, P.H.; Talhout, R. The Sensory Difference Threshold of Menthol Odor in Flavored Tobacco Determined by Combining Sensory and Chemical Analysis. Chem. Senses 2017, 42, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Dylong, D.; Hausoul, P.J.C.; Palkovits, R.; Eisenacher, M. Synthesis of (−)-menthol: Industrial synthesis routes and recent development. Flavour Fragr. J. 2022, 37, 195–209. [Google Scholar] [CrossRef]
- Tandon, K.S.; Baldwin, E.A.; Shewfelt, R.L. Aroma perception of individual volatile compounds in fresh tomatoes (Lycopersicon esculentum, Mill.) as affected by the medium of evaluation. Postharvest Biol. Technol. 2000, 20, 261–268. [Google Scholar] [CrossRef]
- Plotto, A.; Margaría, C.A.; Goodner, K.L.; Baldwin, E.A. Odour and flavour thresholds for key aroma components in an orange juice matrix: Esters and miscellaneous compounds. Flavour Fragr. J. 2008, 23, 398–406. [Google Scholar] [CrossRef]
- Schmiedeberg, K.; Shirokova, E.; Weber, H.-P.; Schilling, B.; Meyerhof, W.; Krautwurst, D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 2007, 159, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Nicoli, A.; Haag, F.; Marcinek, P.; He, R.; Kreißl, J.; Stein, J.; Di Pizio, A. Modeling the Orthosteric Binding Site of the G Protein-Coupled Odorant Receptor OR5K1. J. Chem. Inf. Model. 2023, 63, 2014–2029. [Google Scholar] [CrossRef] [PubMed]
- Man, O.; Gilad, Y.; Lancet, D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. A Publ. Protein Sci. 2004, 13, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Pils, B.; Copley, R.R.; Schultz, J. Variation in structural location and AA conservation of functional sites in protein domain families. BMC Bioinf. 2005, 6, 210. [Google Scholar] [CrossRef] [PubMed]
- Floriano, W.B.; Vaidehi, N.; Goddard, W.A. Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptors. Chem. Senses 2004, 29, 269–290. [Google Scholar] [CrossRef] [PubMed]
- De March, C.A.; Yu, Y.; Ni, M.J.; Adipietro, K.A.; Matsunami, H.; Ma, M.; Golebiowski, J. Conserved Residues Control Activation of Mammalian G Protein-Coupled Odorant Receptors. J. Am. Chem. Soc. 2015, 137, 8611–8616. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Ding, J.; Chen, X. Identification of differential volatile and non-volatile compounds in coffee leaves prepared from different tea processing steps using HS-SPME/GC–MS and HPLC-Orbitrap-MS/MS and investigation of the binding mechanism of key phytochemicals with olfactory and taste receptors using molecular docking. Food Res. Int. 2023, 168, 112760. [Google Scholar] [PubMed]
- Corey, E.A.; Ukhanov, K.; Bobkov, Y.V.; McIntyre, J.C.; Martens, J.R.; Ache, B.W. Inhibitory signaling in mammalian olfactory transduction potentially mediated by Gαo. Mol. Cell. Neurosci. 2021, 110, 103585. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, R.; Aier, I.; Semwal, R.; Tyagi, P.; Varadwaj, P. Sense of Smell: Structural, Functional, Mechanistic Advancements and Challenges in Human Olfactory Research. Curr. Neuropharmacol. 2019, 17, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Li, R.C.; Molday, L.L.; Lin, C.C.; Ren, X.; Fleischmann, A.; Molday, R.S.; Yau, K.W. Low signaling efficiency from receptor to effector in olfactory transduction: A quantified ligand-triggered GPCR pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2121225119. [Google Scholar] [CrossRef] [PubMed]
- Fukutani, Y.; Abe, M.; Saito, H.; Eguchi, R.; Tazawa, T.; de March, C.A.; Matsunami, H. Antagonistic interactions between odorants alter human odor perception. Curr. Biol. 2023, 33, 2235–2245. [Google Scholar] [CrossRef] [PubMed]
- Wachowiak, M.; Shipley, M.T. Coding and synaptic processing of sensory information in the glomerular layer of the olfactory bulb. Semin. Cell Dev. Biol. 2006, 17, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Zhang, L.; Li, P.; Pu, D.; Fu, Y.; Zheng, R.; Xi, H.; Qiao, K.; Wang, D.; Sun, B.; et al. Molecular mechanisms of caramel-like odorant-olfactory receptor interactions based on a computational chemistry approach. Food Res. Int. 2023, 171, 113063. [Google Scholar] [CrossRef] [PubMed]
- Katada, S.; Hirokawa, T.; Oka, Y.; Suwa, M.; Touhara, K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: Mapping the odorant-binding site. J. Neurosci. 2005, 25, 1806–1815. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Inoue, N.; Ito, Y.; Kubota, K.; Sugimoto, A.; Kakuda, T.; Fushiki, T. Sedative effects of the jasmine tea odor and (R)-(-)-linalool, one of its major odor components, on autonomic nerve activity and mood states. Eur. J. Appl. Physiol. 2005, 95, 107–114. [Google Scholar] [CrossRef] [PubMed]







| Ligands | Binding Energy (kcal/mol) | ||
|---|---|---|---|
| Olfr874 | OR8B8 | OR8B12 | |
| L-menthol | −5.1 | −5.7 | −5.5 |
| D-menthol | −5.5 | −7.3 | −5.4 |
| L-Neomenthol | −5.3 | −5.6 | −5.2 |
| D-Neomenthol | −5.3 | −5.2 | −5.2 |
| L-Isomenthol | −5.7 | −5.8 | −5.6 |
| D-Isomenthol | −5.6 | −5.4 | −5.5 |
| L-Neoisomenthol | −6.3 | −6.7 | −5.6 |
| D-Neoisomenthol | −6.4 | −7.1 | −5.6 |
| Receptor | Ligand | Hydrogen Bonding Amino Acid Residues | Hydrophobic Amino Acid Residues |
|---|---|---|---|
| Olfr874 | L-Menthol | - | Ile-48, His-55, Phe-60, Phe-63 |
| D-Menthol | - | Ile-48, His-55, Phe-60, Phe-63, Tyr-140 | |
| L-Neomenthol | - | Ile-48, His-55, Phe-60, Phe-63, Tyr-140 | |
| D-Neomenthol | - | Ile-48, His-55, Phe-60, Phe-63, Tyr-140 | |
| L-Isomenthol | His-55 Thr-56 | Ile-48, His-55, Phe-60, Phe-63, Tyr-140 | |
| D-Isomenthol | - | Ile-48, His-55, Phe-60, Phe-63, Tyr-140 | |
| L-Neoisomenthol | - | Val-107, Val-202, Ile-206, Phe-250, Phe-251, Tyr-258 | |
| D-Neoisomenthol | - | Val-107, Val-108, Val-202, Ile-206, Phe-250, Phe-251, Tyr-277 | |
| OR8B8 | L-Menthol | Thr-57 | His-56, Phe-61, Tyr-64 |
| D-Menthol | - | Leu-105, Val-108, Val-109, Val-203, Ile-207, Phe-251, Phe-252, Tyr-259, Tyr-278 | |
| L-Neomenthol | Tyr-94 | Phe-8, Val-9, Phe-168 | |
| D-Neomenthol | - | Phe-8, Val-9, Tyr-94, Phe-168 | |
| L-Isomenthol | Leu-55 | Ile-49, Arg-50, His-56, Phe-61, Tyr-64 | |
| D-Isomenthol | Thr-57 | Ile-49, His-56, Phe-61, Tyr-64 | |
| L-Neoisomenthol | - | Leu-105, Val-108, Val-109, His-159, Val-203, Ile-207, Phe-251, Phe-252, Tyr-259 | |
| D-Neoisomenthol | - | Ile-49, Arg-50, His-56, Phe-61, Tyr-64 | |
| OR8B12 | L-Menthol | - | Ile-210, Val-213, Tyr-217, Ile-244, Val-247, Ser-248, Phe-251 |
| D-Menthol | - | Ile-210, Val-213, Tyr-217, Ile-244, Val-247, Phe-251 | |
| L-Neomenthol | - | Ile-48, His-55, Phe-60, Phe-63 | |
| D-Neomenthol | - | Ile-210, Val-213, Tyr-217, Ile-244, Val-247, Phe-251 | |
| L-Isomenthol | Ser-248 | Ile-210, Val-213, Tyr-217, Ile-244, Val-247, Phe-251 | |
| D-Isomenthol | - | Ile-210, Val-213, Tyr-217, Ile-244, Val-247, Ser-248, Phe-251 | |
| L-Neoisomenthol | - | Ile-210, Val-213, Phe-214, Tyr-217, Ile-244, Val-247, Phe-251 | |
| D-Neoisomenthol | Phe-199 | Leu-181, Val-198, Val-202, Tyr-258, Leu-259, Leu-262 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wen, F.; Zhang, L.; Sun, B.; Xie, J.; Sun, S.; Zhang, Y. Decoding the Molecular Mechanisms of Menthol Isomer Perception Based on Computational Simulations. Foods 2025, 14, 2494. https://doi.org/10.3390/foods14142494
Wang M, Wen F, Zhang L, Sun B, Xie J, Sun S, Zhang Y. Decoding the Molecular Mechanisms of Menthol Isomer Perception Based on Computational Simulations. Foods. 2025; 14(14):2494. https://doi.org/10.3390/foods14142494
Chicago/Turabian StyleWang, Mengxue, Fengge Wen, Lili Zhang, Baoguo Sun, Jianping Xie, Shihao Sun, and Yuyu Zhang. 2025. "Decoding the Molecular Mechanisms of Menthol Isomer Perception Based on Computational Simulations" Foods 14, no. 14: 2494. https://doi.org/10.3390/foods14142494
APA StyleWang, M., Wen, F., Zhang, L., Sun, B., Xie, J., Sun, S., & Zhang, Y. (2025). Decoding the Molecular Mechanisms of Menthol Isomer Perception Based on Computational Simulations. Foods, 14(14), 2494. https://doi.org/10.3390/foods14142494







