Microbial and Biochemical Analyses of High-Quality, Long-Ripened, Blue-Veined Cabrales Cheese
Abstract
1. Introduction
2. Materials and Methods
2.1. Cheese Sampling and Microbial Analyses
2.2. Metataxonomic Analysis
2.2.1. Isolation of Total Microbial DNA from Cheese Microbes
2.2.2. Amplification and Sequencing of Ribosomal Sequences
2.3. Metabolic Analysis of Cheeses
2.3.1. Organic Acids and Sugars
2.3.2. Amino Acids and Biogenic Amines
2.3.3. Volatile Compounds (VOCs)
2.4. Statistical Analysis
3. Results
3.1. Microbial Counts
3.2. Molecular Microbiology
3.3. Analysis of Cheese Metabolites
3.4. Correlations Between Microorganisms and Metabolites in Cabrales Cheese
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Carlino, N.; Blanco-Míguez, A.; Punčochář, M.; Mengoni, C.; Pinto, F.; Tatti, A.; Manghi, P.; Armanini, F.; Avagliano, M.; Barcenilla, C.; et al. Unexplored microbial diversity from 2,500 food metagenomes and links with the human microbiome. Cell 2024, 16, 5775–5795. [Google Scholar] [CrossRef]
- Ceugniez, A.; Taminiau, B.; Coucheney, F.; Jacques, P.; Delcenserie, V.; Daube, G.; Drider, D. Use of a metagenetic approach to monitor the bacterial microbiota of Tomme d′Orchies cheese during the ripening process. Int. J. Food Microbiol. 2017, 247, 65–69. [Google Scholar] [CrossRef]
- Alessandria, V.; Ferrocino, I.; De Filippis, F.; Fontana, M.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Microbiota of an Italian Grana-like cheese during manufacture and ripening, unraveled by 16S rRNA-based approaches. Appl. Environ. Microbiol. 2016, 82, 3988–3995. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 16–27. [Google Scholar] [CrossRef]
- Monnet, C.; Dugat-Bony, E.; Swennen, D.; Beckerich, J.M.; Irlinger, F.; Fraud, S.; Bonnarme, P. Investigation of the activity of the microorganisms in a Reblochon-style cheese by metatranscriptomic analysis. Front. Microbiol. 2016, 7, 536. [Google Scholar] [CrossRef]
- Rampanti, G.; Ferrocino, I.; Harasym, J.; Foligni, R.; Cardinali, F.; Orkusz, A.; Milanović, V.; Franciosa, I.; Garofalo, C.; Mannozzi, C.; et al. Queijo Serra da Estrela PDO cheese: Investigation into its morpho-textural traits, microbiota, and volatilome. Foods 2023, 12, 169. [Google Scholar] [CrossRef]
- Unno, R.; Suzuki, T.; Matsutani, M.; Ishikawa, M. Evaluation of the relationships between microbiota and metabolites in soft-type ripened cheese using an integrated omics approach. Front. Microbiol. 2021, 12, 681185. [Google Scholar] [CrossRef]
- Walsh, A.M.; Macori, G.; Kilcawley, K.N.; Cotter, P.D. Meta-analysis of cheese microbiotas highlights contributions to multiple aspects of quality. Nat. Foods 2020, 1, 500–510. [Google Scholar] [CrossRef]
- Lee, J.; Seo, Y.; Ha, J.; Kim, S.; Choi, Y.; Oh, H.; Lee, Y.; Kim, Y.; Kang, J.; Park, E.; et al. Influence of milk microbiota on Listeria monocytogenes survival during cheese ripening. Food Sci. Nutr. 2020, 8, 5071–5076. [Google Scholar] [CrossRef]
- Nam, J.H.; Cho, Y.S.; Rackerby, B.; Goddik, L.; Park, S.H. Shifts of microbiota during cheese production: Impact on production and quality. Appl. Microbiol. Biotechnol. 2021, 105, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Mo, L.; Pan, L.; Hou, Q.; Li, C.; Darima, I.; Yu, J. Using PacBio sequencing to investigate the bacterial microbiota of traditional Buryatian cottage cheese and comparison with Italian and Kazakhstan artisanal cheeses. J. Dairy Sci. 2018, 101, 6885–6896. [Google Scholar] [CrossRef] [PubMed]
- Ritschard, J.S.; Schuppler, M. The Microbial diversity on the surface of smear-ripened cheeses and its impact on cheese quality and safety. Foods 2024, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Núñez, M. Microflora of Cabrales cheese: Changes during maturation. J. Dairy Res. 1978, 45, 501–508. [Google Scholar] [CrossRef]
- Flórez, A.B.; López-Díaz, T.M.; Álvarez-Martín, P.; Mayo, B. Microbial characterisation of the traditional Spanish blue-veined Cabrales cheese: Identification of dominant lactic acid bacteria. Eur. Food Res. Technol. 2006, 223, 503–508. [Google Scholar] [CrossRef]
- Alexa, E.A.; Cobo-Díaz, J.F.; Renes, E.; OʼCallaghan, T.F.; Kilcawley, K.; Mannion, D.; Skibinska, I.; Ruiz, L.; Margolles, A.; Fernández-Gómez, P.; et al. The detailed analysis of the microbiome and resistome of artisanal blue-veined cheeses provides evidence on sources and patterns of succession linked with quality and safety traits. Microbiome 2024, 12, 78. [Google Scholar] [CrossRef]
- Rodríguez, J.; González-Guerra, A.; Vázquez, L.; Fernández-López, R.; Flórez, A.B.; de la Cruz, F.; Mayo, B. Isolation and phenotypic and genomic characterization of Tetragenococcus spp. from two Spanish traditional blue-veined cheeses made of raw milk. Int. J. Food Microbiol. 2022, 371, 109670. [Google Scholar] [CrossRef]
- International Standard FIL/IDF 50 B; Methods of Sampling Milk and Milk Products. FIL/IDF (Fédération Internationale de Laiterie/International Dairy Federation): Brussels, Belgium, 1985.
- Kisand, V.; Cuadros, R.; Wikner, J. Phylogeny of culturable estuarine bacteria catabolizing riverine organic matter in the northern Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 379–388. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols. A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Meola, M.; Rifa, E.; Shani, N.; Delbès, C.; Berthoud, H.; Chassard, C. DAIRYdb: A manually curated reference database for improved taxonomy annotation of 16S rRNA gene sequences from dairy products. BMC Genom. 2019, 20, 560. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Alegría, A.; González, P.; Delgado, S.; Flórez, A.B.; Hernández-Barranco, A.; Rodríguez, A.; Mayo, B. Characterisation of the technological behaviour of mixtures of mesophilic lactic acid bacteria isolated from traditional cheeses made of raw milk without added starters. Int. J. Dairy Technol. 2016, 69, 507–519. [Google Scholar] [CrossRef]
- Redruello, B.; Ladero, V.; Cuesta, I.; Álvarez-Buylla, J.R.; Martín, M.C.; Fernández, M.; Alvarez, M.A. A fast, reliable, ultra high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013, 139, 1029–1035. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electronica 2001, 4, 1. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Schwertman, N.; Owens, M.; Adnan, R. A simple more general boxplot method for identifying outliers. Comput. Stat. Data Anal. 2004, 47, 165–174. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Ferroukhi, I.; Bord, C.; Alvarez, S.; Fayolle, K.; Theil, S.; Lavigne, R.; Chassard, C.; Mardon, J. Functional changes in Bleu d’Auvergne cheese during ripening. Food Chem. 2022, 397, 133850. [Google Scholar] [CrossRef]
- Diezhandino, I.; Fernández, D.; Gonzáalez, L.; McSweeney, P.L.H.; Fresno, J.M. Microbiological, physicochemical and proteolytic changes in a Spanish blue cheese during ripening (Valdeón cheese). Food Chem. 2015, 168, 134–141. [Google Scholar] [CrossRef]
- Cantor, M.D.; van den Tempel, T.; Hansen, T.K.; Ardö, Y. Blue cheese. In Cheese, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: New York, NY, USA, 2017; pp. 929–954. [Google Scholar] [CrossRef]
- Decadt, H.; Vermote, L.; Díaz-Muñoz, C.; Weckx, S.; de Vuyst, L. Decarboxylase activity of the non-starter lactic acid bacterium Loigolactobacillus rennini gives crack defects in Gouda cheese through the production of gamma-aminobutyric acid. Appl. Environ. Microbiol. 2024, 90, e0165523. [Google Scholar] [CrossRef]
- Haastrup, M.K.; Johansen, P.; Malskær, A.H.; Castro-Mejía, J.L.; Kot, W.; Krych, L.; Arneborg, N.; Jespersen, L. Cheese brines from Danish dairies reveal a complex microbiota comprising several halotolerant bacteria and yeasts. Int. J. Food Microbiol. 2018, 285, 173–187. [Google Scholar] [CrossRef]
- Place, R.B.; Hiestand, D.; Gallmann, H.R.; Teuber, M. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 2003, 26, 30–37. [Google Scholar] [CrossRef]
- Kastman, E.K.; Kamelamela, N.; Norville, J.W.; Cosetta, C.M.; Dutton, R.J.; Wolfe, B.E. Biotic interactions shape the ecological distributions of Staphylococcus species. Mbio 2016, 7, e01157-16. [Google Scholar] [CrossRef]
- Pangallo, D.; Saková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, L.; Kuchta, T. Microbial diversity and dynamics during the production of May Bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef]
- Biolcati, F.; Ferrocino, I.; Bottero, M.T.; Dalmasso, A. The bacterial and fungal microbiota of “Robiola di Roccaverano” protected designation of origin raw milk cheese. Front. Microbiol. 2022, 12, 776862. [Google Scholar] [CrossRef]
- Rocha, R.; Vaz Vello, M.; Santos, J.; Fernandes, P. Serra da Estrela PDO cheese microbiome as revealed by next generation sequencing. Microorganisms 2021, 9, 2007. [Google Scholar] [CrossRef]
- WHO (World Health Organization). WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Sallami, L.; Kheadr, E.E.; Fliss, I.; Vuillemard, J.C. Impact of autolytic, proteolytic, and nisin-producing adjunct cultures on biochemical and textural properties of cheddar cheese. J. Dairy Sci. 2004, 87, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef]
- Masotti, F.; Hogenboom, J.A.; Rosi, V.; De Noni, I.; Pellegrino, L. Proteolysis indices related to cheese ripening and typicalness in PDO Grana Padano cheese. Int. Dairy J. 2010, 20, 352–359. [Google Scholar] [CrossRef]
- Linares, D.M.; Martín, M.C.; Ladero, V.; Álvarez, M.A.; Fernández, M. Biogenic amines in dairy products. Crit. Rev. Food Sci. Nutr. 2011, 51, 691–703. [Google Scholar] [CrossRef]
- Ardö, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef]
- Fernández, M.; Linares, D.M.; del Río, B.; Ladero, V.; Álvarez, M.A. HPLC quantification of biogenic amines in cheeses: Correlation with PCR-detection of tyramine-producing microorganisms. J. Dairy Res. 2007, 74, 276–282. [Google Scholar] [CrossRef]
- Han, J.; Zhao, X.; Zhao, X.; Wang, Q.; Li, P.; Gu, Q. Microbial-derived gamma-aminobutyric acid: Synthesis, purification, physiological function, and applications. J. Agric. Food Chem. 2023, 71, 14931–14946. [Google Scholar] [CrossRef]
- Valenzuela, J.A.; Vázquez, L.; Rodríguez, J.; Flórez, A.B.; Vasek, O.M.; Mayo, B. Phenotypic, technological, safety, and genomic profiles of gamma-aminobutyric acid-producing Lactococcus lactis and Streptococcus thermophilus strains isolated from cow’s milk. Int. J. Mol. Sci. 2024, 25, 2328. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.J.; Wang, Y.D.; Cao, Y.P.; Wang, B.; Liu, Y. The key aroma compounds and sensory characteristics of commercial Cheddar cheeses. J. Dairy Sci. 2021, 104, 7555–7571. [Google Scholar] [CrossRef]
- Capozzi, V.; Lonzarich, V.; Khomenko, I.; Cappellin, L.; Navarini, L.; Biasioli, F. Unveiling the molecular basis of Mascarpone cheese aroma: VOCs analysis by SPME-GC/MS and PTR-ToF-MS. Molecules 2020, 25, 1242. [Google Scholar] [CrossRef]
- Masotti, F.; Cattaneo, S.; Stuknytė, M.; Battelli, G.; Vallone, L.; De Noni, I. Composition, proteolysis, and volatile profile of Strachítunt cheese. J. Dairy Sci. 2017, 100, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
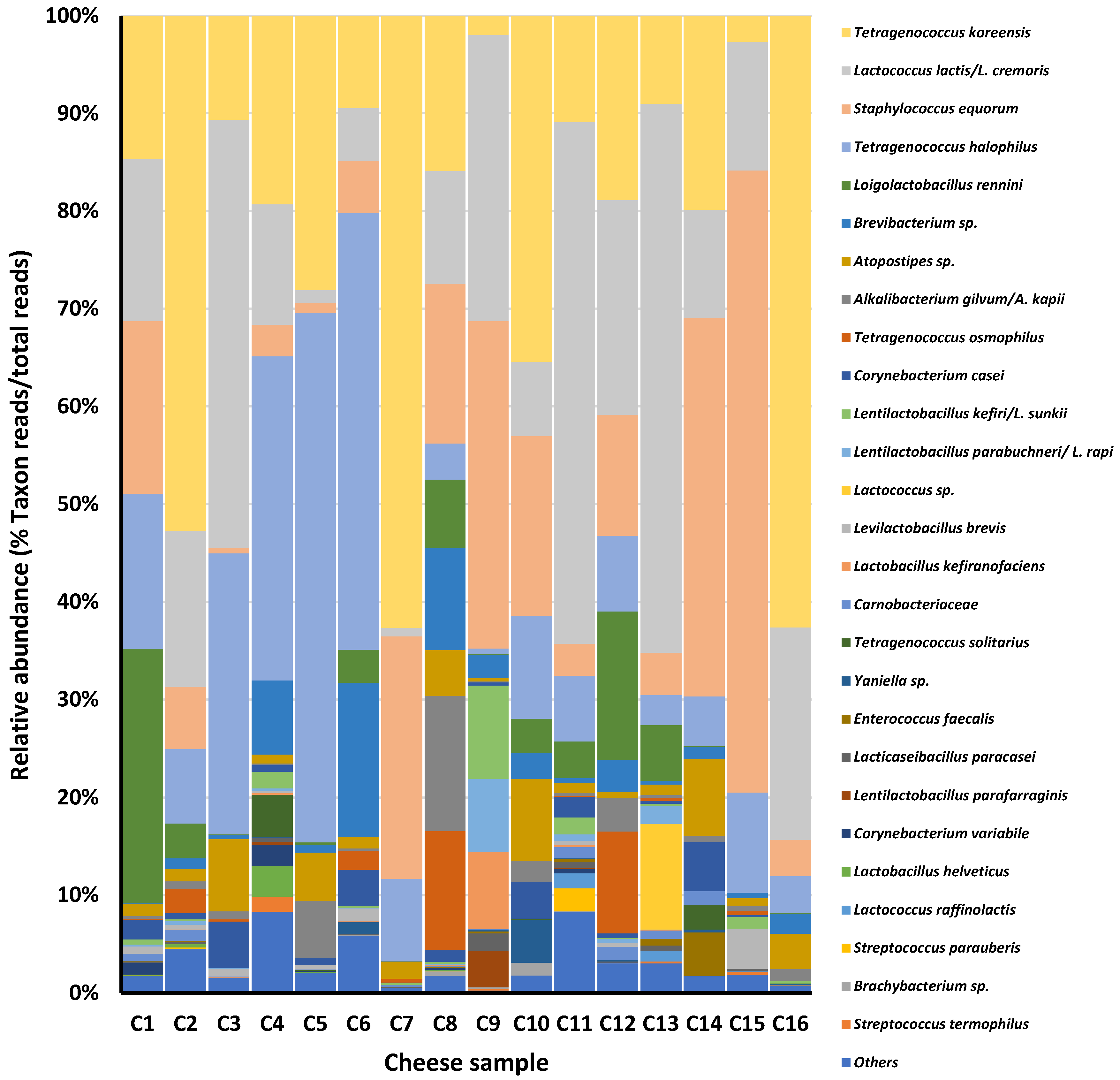
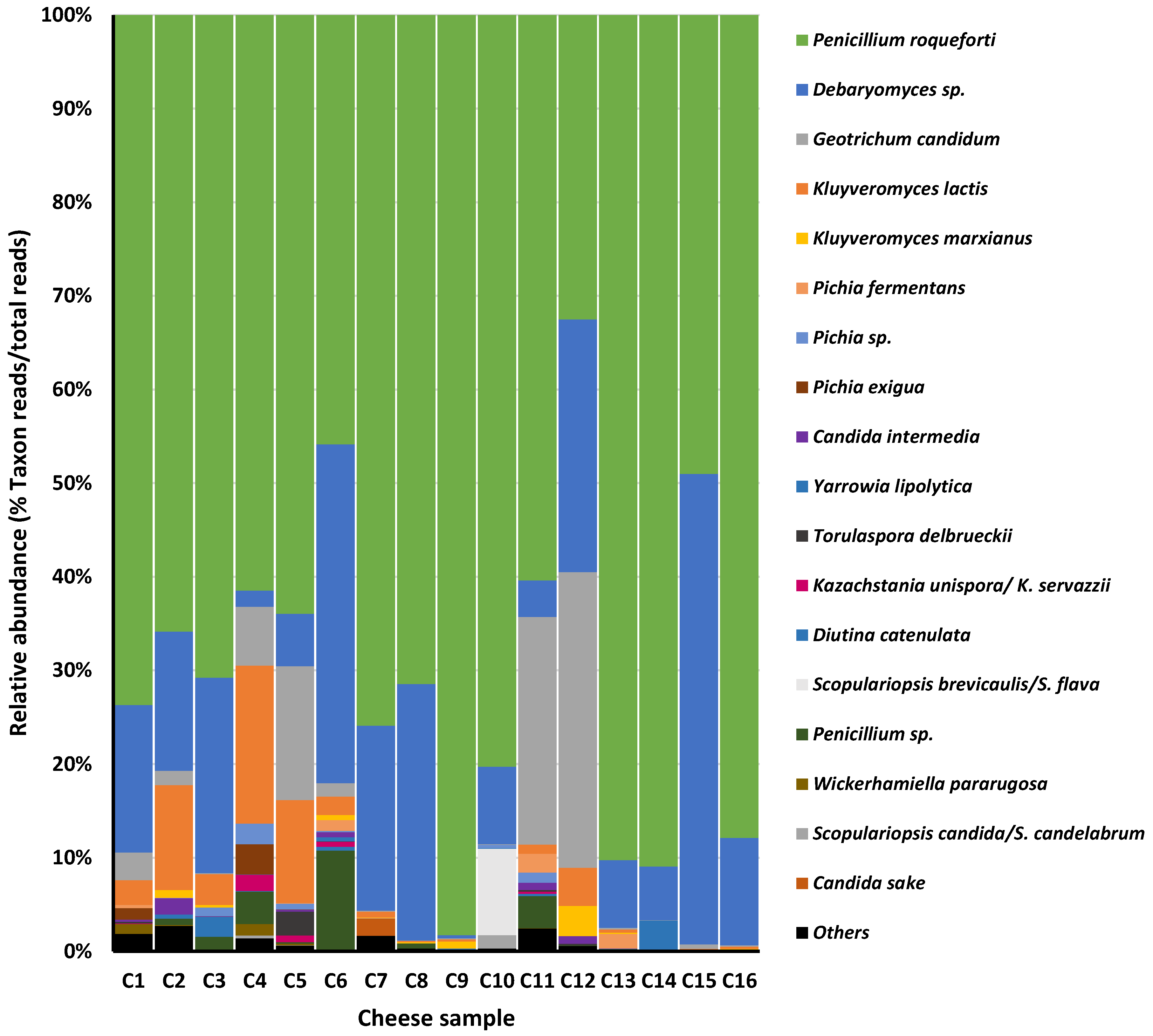
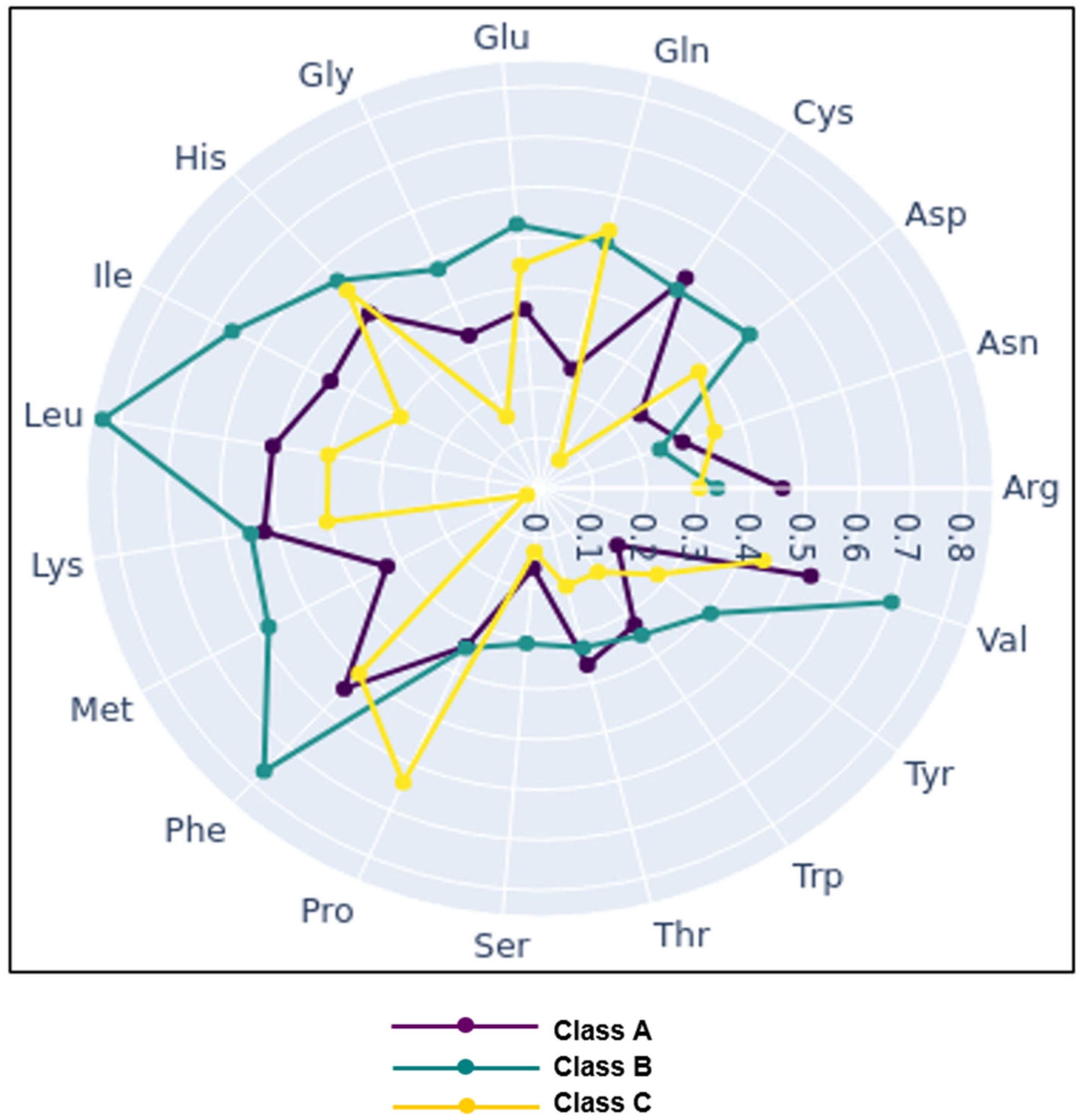



| Microbial Group (Counting Medium) | Cheese Sample | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 | C13 | C14 | C15 | C16 | |
| Total viable mesophilic counts (PCAM) | 5.97 a | 5.56 | 5.00 | 4.51 | 2.00 | 5.42 | 6.95 | 3.56 | 7.39 | 5.53 | 6.47 | 5.46 | 5.38 | 7.65 | 7.14 | 4.94 |
| Lactococci/streptococci (GM17A) | 4.63 | 4.67 | 3.55 | 4.34 | 2.60 | 5.26 | 4.65 | 2.30 | 5.49 | 3.60 | 5.27 | 3.04 | 4.06 | 7.16 | 4.89 | 3.00 |
| Lactobacillaceae (MRSA) | 4.70 | 4.58 | 2.78 | 3.96 | <2 b | 4.43 | <2 | 2.60 | 5.31 | <2 | 5.11 | 2.48 | 3.27 | 6.70 | 4.69 | 2.30 |
| Enterococci (S-BA) | 3.68 | 4.71 | <2 | 4.17 | <2 | 4.57 | 5.30 | 2.00 | 3.31 | 4.23 | 4.86 | 2.90 | 4.11 | 4.46 | 3.65 | 2.00 |
| Micrococci/staphylococci (B-PA) | <2 | 4.54 | 3.72 | 4.37 | <2 | 3.18 | 4.31 | <2 | 5.68 | 4.41 | 4.71 | 2.85 | 3.30 | 6.18 | 3.45 | <2 |
| Enterobacteriaceae (VRBGA) | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 |
| Coliforms (VRBLA) | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 |
| Yeasts and moulds (YGCA) | 6.79 | 3.85 | 6.35 | 4.94 | 2.60 | 6.52 | 6.42 | 5.11 | 4.61 | 6.47 | 6.08 | 6.15 | 4.26 | 7.31 | 7.12 | 4.00 |
| Cheese Sample | Organic Acid/Compound | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetic a | Butyric | Formic | Lactic | Orotic | Pyruvic | Uric | Lactose | Glucose | Galactose | Ethanol | Ammonia | |
| C1 | 199 | 95 | - | 397 | 3 | 16 b | 10 | 714 | 24 | - | - | 4590 |
| C2 | 201 | 209 | - | 125 | 1 | 8 | 13 | 566 | 22 | - | 36 | 4016 |
| C3 | 385 | 326 | - | 247 | 1 | 5 | 4 | 562 | 26 | 17 b | 101 | 1505 b |
| C4 | 322 | 430 | 14 b | 422 | 1 | 7 | 18 | 707 | 4 | - | 105 | 3989 |
| C5 | 381 | 373 | 21 b | 351 | 1 | 9 | 12 | 536 | 6 | - | - | 4569 |
| C6 | 227 | 454 | - | 405 | 2 | 11 | 24 | 598 | 11 | 7 b | - | 4036 |
| C7 | 91 | 66 | - | 187 | 2 | 17 b | 21 | 446 | 13 | - | - | 5104 |
| C8 | 360 | 431 | - | 234 | 1 | 11 | 19 | 588 | 9 | 7 b | 22 | 4009 |
| C9 | 390 | 345 | - | 190 | 3 | 1 | 13 | 583 | 4 | - | 148 | 4753 |
| C10 | 352 | 199 | - | 140 | - | 9 | 14 | 421 | 4 | - | 120 | 3802 |
| C11 | 220 | 206 | - | 70 | 3 | 6 | 24 | 517 | 5 | - | - | 3448 |
| C12 | 96 | 136 | - | 199 | 1 | 7 | 21 | 540 | 13 | 5 b | - | 3662 |
| C13 | 349 | 308 | - | 165 | 1 | 4 | 8 | 597 | 2 | - | 100 | 3507 |
| C14 | 178 | 105 | - | 14 | 1 | 3 | 21 | 422 | 10 | - | 81 | 3696 |
| C15 | 23 | 142 | - | 99 | 1 | 7 | 11 | 473 | - | - | 82 | 2997 |
| C16 | 388 | 125 | - | 169 | 1 | 7 | 12 | 451 | - | - | 120 | 4248 |
| Cheese Sample | Compound | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Agmatine | Cadaverine | Ethylamine | Histamine | Ornithine | Putrescine | Tyramine | Tryptamine | β-Phenylethylamine | GABA | |
| C1 | - | 58 | - | 40 | 237 | 8 | 279 | 82 | 7 | 1337 a |
| C2 | - | 73 | - | 7 | 161 | 2 | 172 | 65 | 9 | 235 |
| C3 | - | 37 | - | 14 | 197 | 13 | 245 | - | 5 | 9 |
| C4 | - | 39 | - | 85 | 268 | 12 | 290 | 25 | 5 | 313 |
| C5 | - | 118 | 2 a | - | 317 | 1 | 513 | 144 | 14 | 16 |
| C6 | - | 152 | 2 a | 74 | 211 | 22 | 508 | 139 | 18 | 392 |
| C7 | 11 a | 247 | - | - | 186 | 10 | 145 | 174 | 30 | 15 |
| C8 | 9 | 169 | - | - | 224 | 3 | 130 | 64 | 20 | 74 |
| C9 | 3 | 31 | - | 60 | 423 | 1 | 457 | - | 4 | 5 |
| C10 | - | 9 | - | - | 322 | - | 186 | 61 | 1 | 354 |
| C11 | 4 | 251 | - | 27 | 161 | 68 a | 412 | 110 | 30 | 213 |
| C12 | 4 | 121 | - | 13 | 132 | 7 | 220 | 107 | 15 | 129 |
| C13 | 4 | 59 | - | 70 | 298 | 2 | 302 | 29 | 7 | 637 |
| C14 | - | 6 | - | - | 244 | - | 225 | 11 | 1 | 5 |
| C15 | - | 173 | - | - | 174 | - | 145 | 3 | 21 | 9 |
| C16 | - | 341 | - | - | 158 | 11 | 224 | 116 | 41 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, J.; Suárez, P.R.; Das, S.; Vázquez, L.; Lama, S.; Flórez, A.B.; Tamang, J.P.; Mayo, B. Microbial and Biochemical Analyses of High-Quality, Long-Ripened, Blue-Veined Cabrales Cheese. Foods 2025, 14, 2366. https://doi.org/10.3390/foods14132366
Rodríguez J, Suárez PR, Das S, Vázquez L, Lama S, Flórez AB, Tamang JP, Mayo B. Microbial and Biochemical Analyses of High-Quality, Long-Ripened, Blue-Veined Cabrales Cheese. Foods. 2025; 14(13):2366. https://doi.org/10.3390/foods14132366
Chicago/Turabian StyleRodríguez, Javier, Paula Rosa Suárez, Souvik Das, Lucía Vázquez, Sonam Lama, Ana Belén Flórez, Jyoti Prakash Tamang, and Baltasar Mayo. 2025. "Microbial and Biochemical Analyses of High-Quality, Long-Ripened, Blue-Veined Cabrales Cheese" Foods 14, no. 13: 2366. https://doi.org/10.3390/foods14132366
APA StyleRodríguez, J., Suárez, P. R., Das, S., Vázquez, L., Lama, S., Flórez, A. B., Tamang, J. P., & Mayo, B. (2025). Microbial and Biochemical Analyses of High-Quality, Long-Ripened, Blue-Veined Cabrales Cheese. Foods, 14(13), 2366. https://doi.org/10.3390/foods14132366










