Antioxidant Activity In Vitro and Protective Effects Against Lipopolysaccharide-Induced Oxidative Stress and Inflammation in RAW264.7 Cells of Ulva prolifera-Derived Bioactive Peptides Identified by Virtual Screening, Molecular Docking, and Dynamics Simulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Computer-Aided Virtual Screening
2.3. Molecular Docking
2.4. MD Simulations
2.5. Synthesis of Peptides
2.6. In Vitro Antioxidant Activity Assay
2.7. Cell Culture of RAW264.7
2.8. MTT Assay
2.9. NO Production Assay
2.10. Detection of SOD and CAT Activities and MDA Level
2.11. RNA Extraction and qRT-PCR Analysis
2.12. Western Blot Assay
2.13. Statistical Analysis
3. Results
3.1. Virtual Screening of Ulva Prolifera Proteins
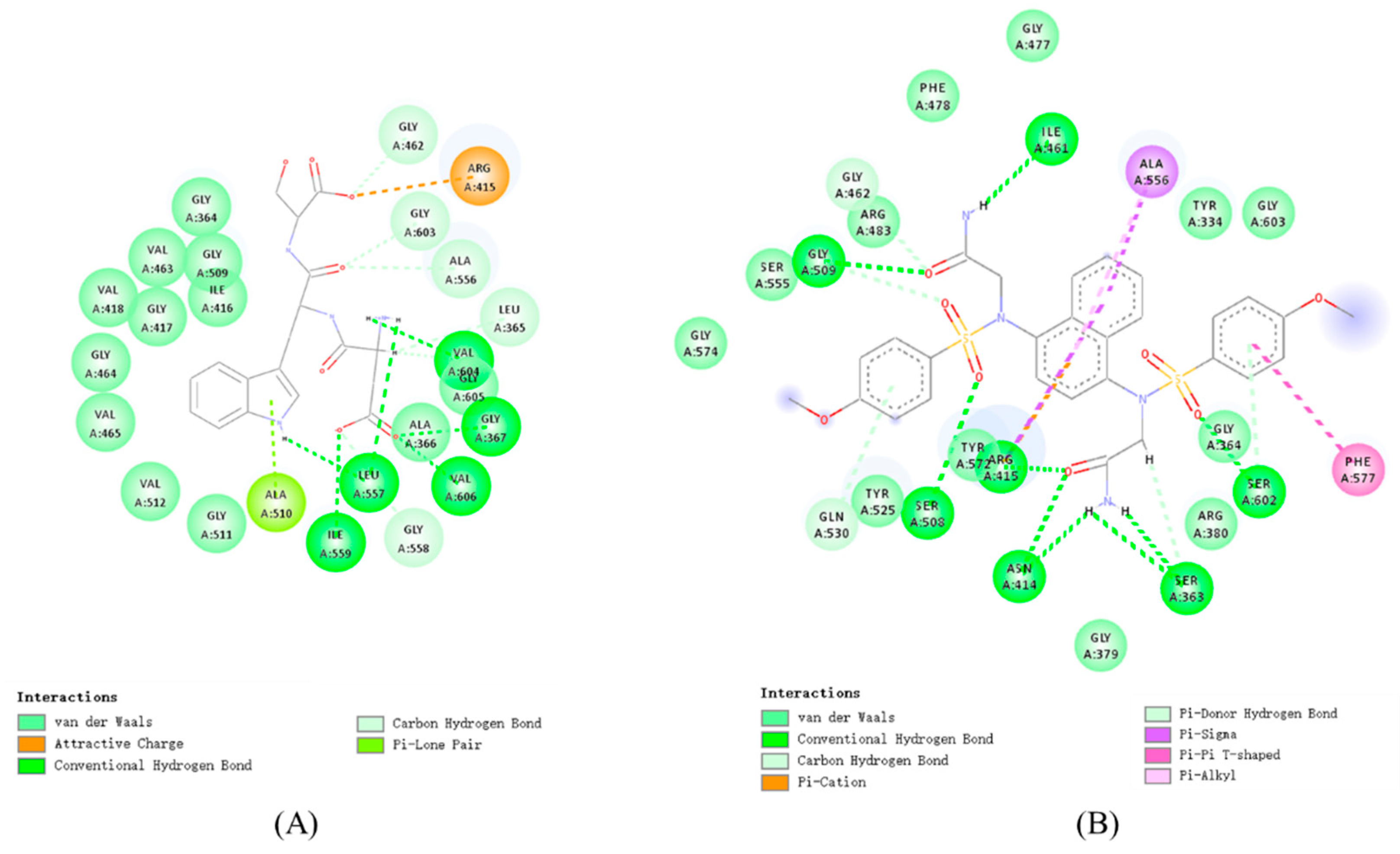
3.2. Molecular Docking Analysis
3.3. Molecular Dynamics Simulation Analysis
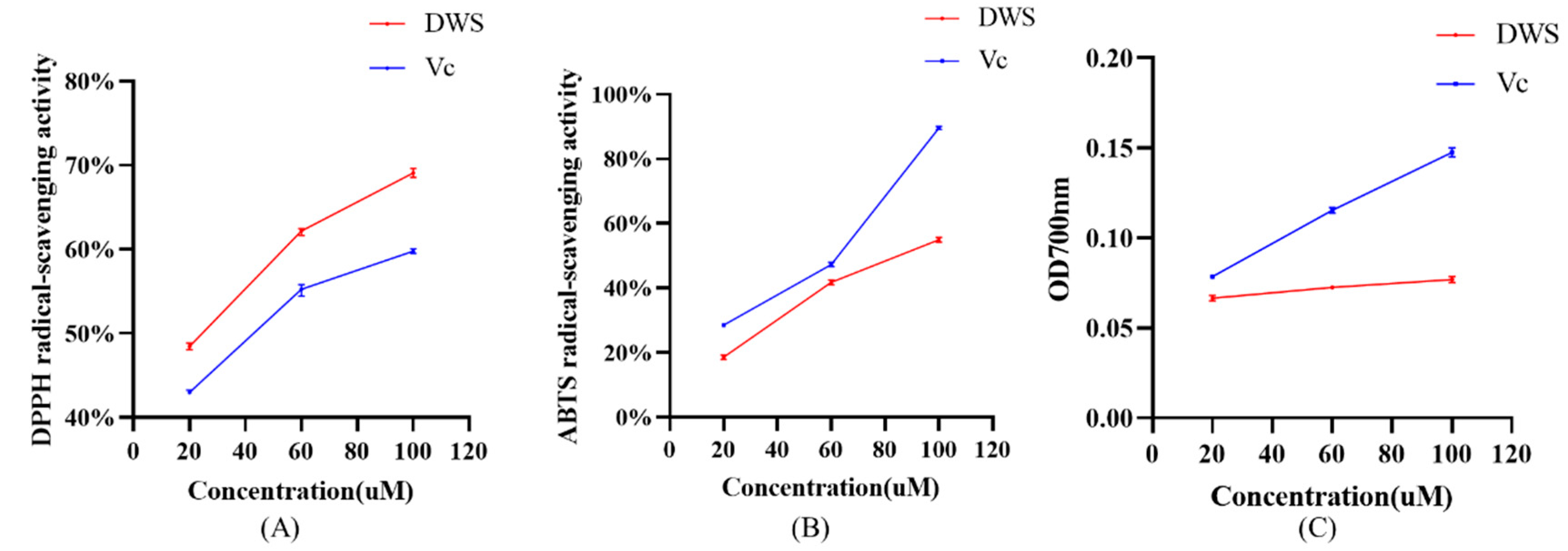
3.4. DPPH and ABTS Radical Scavenging and Reducing Capacity Analysis
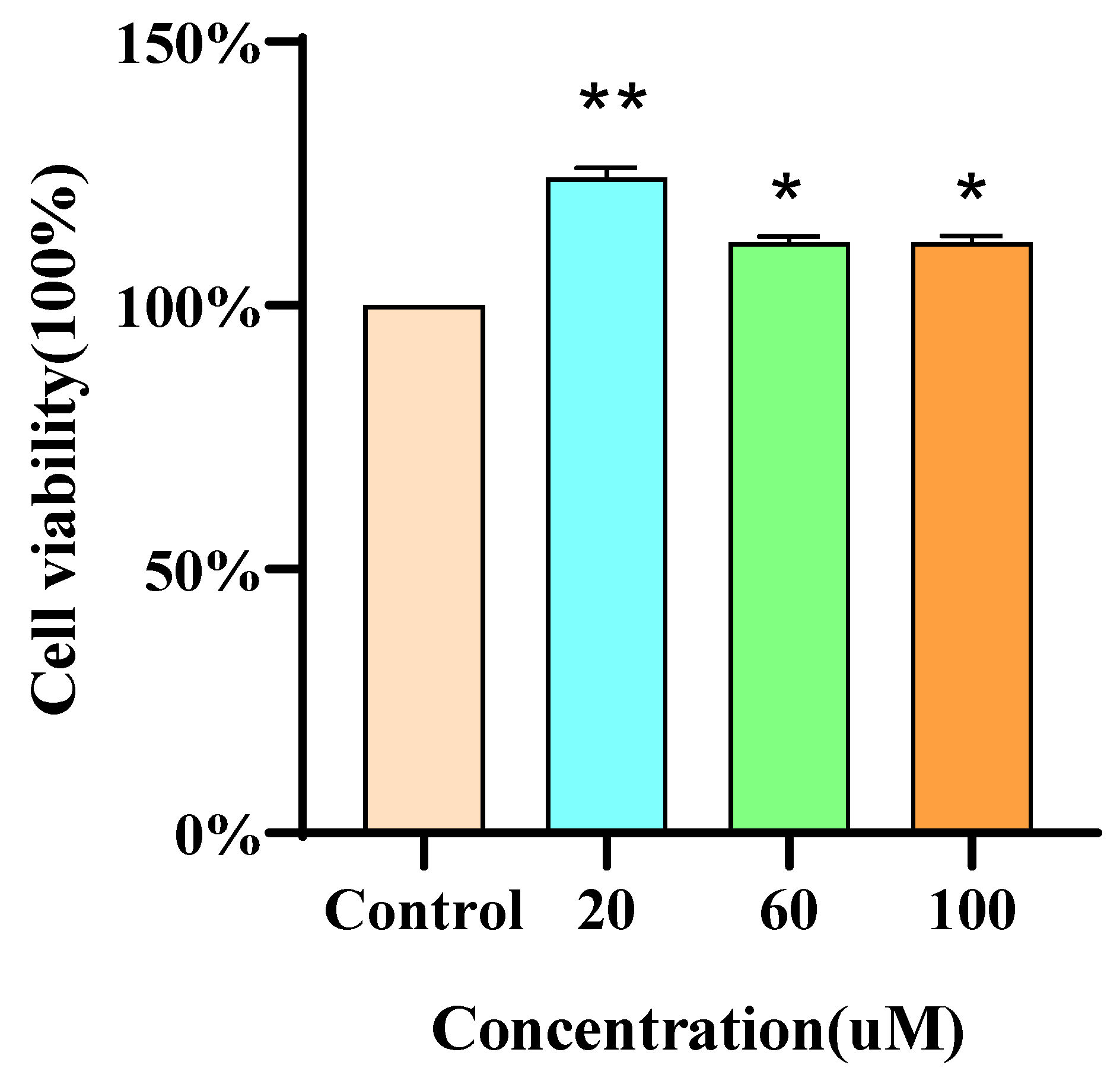
3.5. RAW264.7 Cell Viability
3.6. Measurement of NO Levels
3.7. Effect of DWS on Antioxidant Enzyme Activities and MDA Content in LPS-Induced RAW264.7 Cells
3.8. Effects of DWS on mRNA Expression of Antioxidant and Anti-Inflammation-Related Genes in RAW264.7 Cells
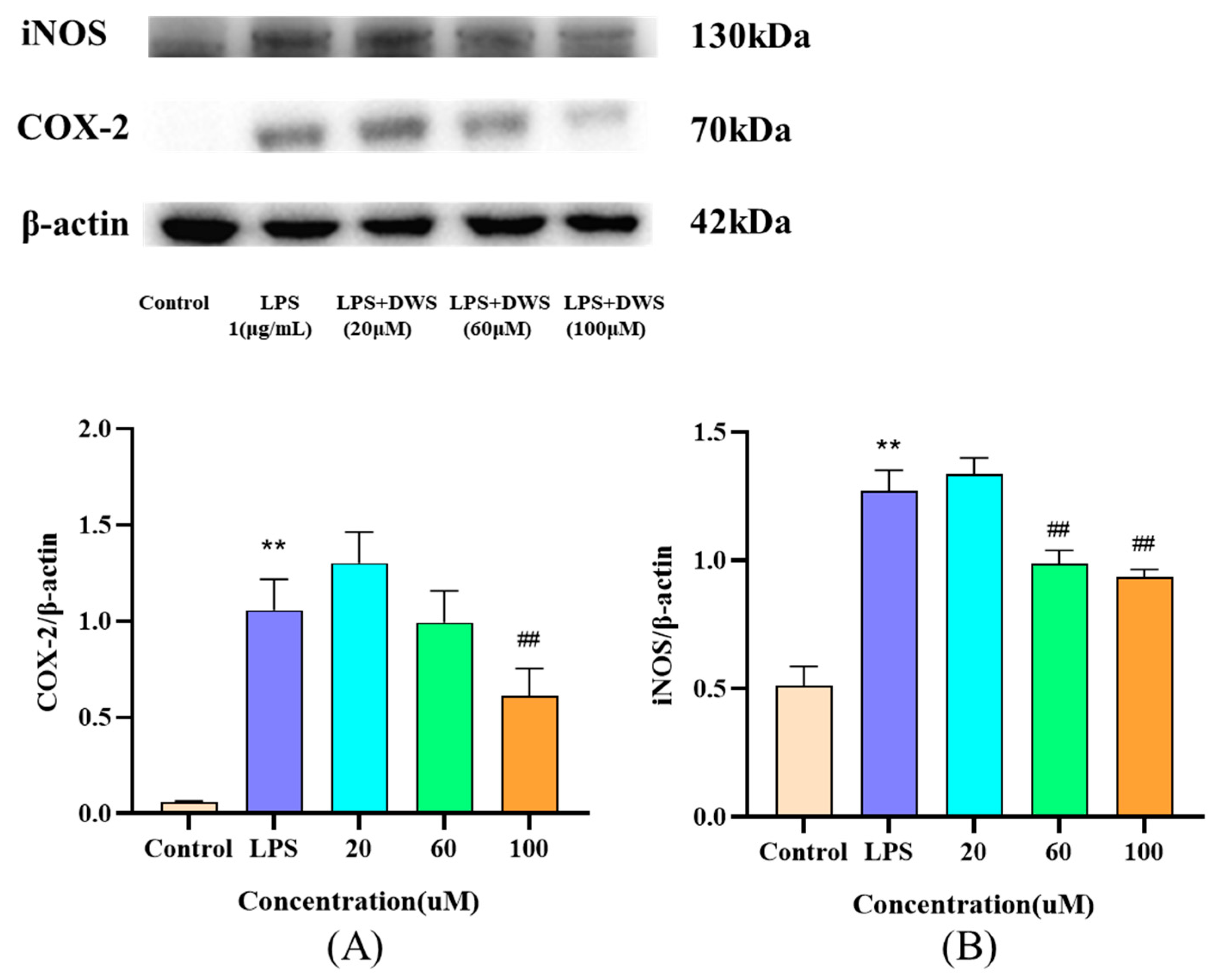
3.9. Western Blot Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Z.; Xu, X.; Leng, X.; He, M.; Wang, J.; Cheng, S.; Wu, H. Roles of reactive oxygen species in cell signaling pathways and immune responses to viral infections. Arch. Virol. 2017, 162, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Torres, I.; Castrejón-Téllez, V.; Soto, M.E.; Rubio-Ruiz, M.E.; Manzano-Pech, L.; Guarner-Lans, V. Oxidative Stress, Plant Natural Antioxidants, and Obesity. Int. J. Mol. Sci. 2021, 22, 1786. [Google Scholar] [CrossRef]
- Estévez, M. Protein carbonyls in meat systems: A review. Meat Sci. 2011, 89, 259–279. [Google Scholar] [CrossRef]
- Lieber, M.R.; Karanjawala, Z.E. Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell Biol. 2004, 5, 69–75. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.; de la Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating NRF2 in Disease: Timing Is Everything. Annu. Rev. Pharmacol. 2019, 59, 555–575. [Google Scholar] [CrossRef]
- Taguchi, K.; Yamamoto, M. The KEAP1-NRF2 System as a Molecular Target of Cancer Treatment. Cancers 2020, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Cloer, E.W.; Siesser, P.F.; Cousins, E.M.; Goldfarb, D.; Mowrey, D.D.; Harrison, J.S.; Weir, S.J.; Dokholyan, N.V.; Major, M.B. p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol. Cell. Biol. 2018, 38, e00644-17. [Google Scholar] [CrossRef]
- Saadi, S.; Saari, N.; Anwar, F.; Abdul Hamid, A.; Ghazali, H.M. Recent advances in food biopeptides: Production, biological functionalities and therapeutic applications. Biotechnol. Adv. 2015, 33, 80–116. [Google Scholar] [CrossRef]
- Taniguchi, M.; Aida, R.; Saito, K.; Ochiai, A.; Takesono, S.; Saitoh, E.; Tanaka, T. Identification and characterization of multifunctional cationic peptides from traditional Japanese fermented soybean Natto extracts. J. Biosci. Bioeng. 2018, 127, 472–478. [Google Scholar] [CrossRef]
- Du, Y.; Esfandi, R.; Willmore, W.G.; Tsopmo, A. Antioxidant Activity of Oat Proteins Derived Peptides in Stressed Hepatic HepG2 Cells. Antioxidants 2016, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.X.; Liu, X.L.; Zheng, X.Q.; Wang, X.J.; He, J.F. Preparation of antioxidative corn protein hydrolysates, purification and evaluation of three novel corn antioxidant peptides. Food Chem. 2016, 204, 427–436. [Google Scholar] [CrossRef]
- Samaei, S.P.; Ghorbani, M.; Tagliazucchi, D.; Martini, S.; Gotti, R.; Themelis, T.; Tesini, F.; Gianotti, A.; Gallina Toschi, T.; Babini, E. Functional, nutritional, antioxidant, sensory properties and comparative peptidomic profile of faba bean (Vicia faba, L.) seed protein hydrolysates and fortified apple juice. Food Chem. 2020, 330, 127120. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Fei, Y.; Xu, Q.; Lei, T.; Mo, X.; Wang, Z.; Zhang, L.; Mou, X.; Li, H. Isolation and identification of antioxidant peptides from tartary buckwheat albumin (Fagopyrum tataricum Gaertn.) and their antioxidant activities. J. Food Sci. 2020, 85, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, T.J.; Han, L.J.; Gui, L.S.; Hou, S.Z.; Sun, S.G.; Zhao, C.Z. Simulated enzymatic screening and activity validation of antioxidant peptides from Tibetan sheep hemoglobin. Food Ferment. Ind. 2024, 1–14. [Google Scholar]
- Chen, Y.N.; Qiu, Z.J.; Zhang, B. Virtual Screening and Molecular Docking of Antioxidant Peptides Derived from Walnut Proteins. Agric. Prod. Process. 2024, 43, 112–118. [Google Scholar]
- Li, J.C.; Gao, X.Y.; Jiang, C.Y.; Yang, Y.S.; Saimaiti, A.; Liu, Y.N.; Batuer, A. Simulation of Enzymolysis to Optimize the Preparation of Pigeon Hemoglobin Antioxidant Peptides. Food Res. Dev. 2023, 44, 108–115. [Google Scholar]
- Wani, T.A.; Zargar, S.; Hussain, A. Spectroscopic, Thermodynamic and Molecular Docking Studies on Molecular Mechanisms of Drug Binding to Proteins. Molecules 2022, 27, 8405. [Google Scholar] [CrossRef]
- Tuckerman, M.E.; Martyna, G.J. Understanding modern molecular dynamics: Techniques and applications. J. Phys. Chem. B 2001, 105, 7598. [Google Scholar] [CrossRef]
- Khan, A.; Ali, S.S.; Khan, M.T.; Saleem, S.; Ali, A.; Suleman, M.; Babar, Z.; Shafiq, A.; Khan, M.; Wei, D.Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro). J. Biomol. Struct. Dyn. 2021, 39, 4659–4670. [Google Scholar] [CrossRef]
- Xu, Y.; Song, D.; Su, Y.; Chen, J.; Wu, L.; Lian, H.; Hai, N.; Li, J.; Jiang, J.; Zhao, J.; et al. Pharmacology-based molecular docking of 4-methylcatechol and its role in RANKL-mediated ROS/Keap1/Nrf2 signalling axis and osteoclastogenesis. Biomed. Pharmacother. 2023, 159, 114101. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Huang, W.; Jie, F.; Wang, M.; Zhong, Y.; Chen, Q.; Lu, B. Discovery of Keap1-Nrf2 small-molecule inhibitors from phytochemicals based on molecular docking. Food Chem. Toxicol. 2019, 133, 110758. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; He, Q.; Huang, C.; Wei, S.; Gong, Y.; Li, X.; Liu, W.; Xu, Z.; Wu, H.; Zheng, C.; et al. Panaxatriol saponin ameliorates myocardial infarction-induced cardiac fibrosis by targeting Keap1/Nrf2 to regulate oxidative stress and inhibit cardiac-fibroblast activation and proliferation. Free Radic. Biol. Med. 2022, 190, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Tonolo, F.; Moretto, L.; Grinzato, A.; Fiorese, F.; Folda, A.; Scalcon, V.; Ferro, S.; Arrigoni, G.; Bellamio, M.; Feller, E.; et al. Fermented Soy-Derived Bioactive Peptides Selected by a Molecular Docking Approach Show Antioxidant Properties Involving the Keap1/Nrf2 Pathway. Antioxidants 2020, 9, 1306. [Google Scholar] [CrossRef]
- Ding, L.; Luan, R.; Huang, B. The taxonomical study on Capsosiphonaceae (Ulvales, Chlorophyta) from Huanghai-Bohai Seas of China. Acta Oceanol. Sin. 2008, 30, 168–174. [Google Scholar]
- Jiang, X.L.; Zhou, X.J.; Lin, J.N.; Kang, Z.J.; Liu, Q. Research progress in the ecological consequences of Ulva prolifera green tides in the Yellow Sea. Mar. Environ. Sci. 2021, 40, 647–652. [Google Scholar]
- Gao, S.; Chen, X.; Yi, Q.; Wang, G.; Pan, G.; Lin, A.; Peng, G. Astrategy for the proliferation of Ulva prolifera, main causative species of tides, with formation of sporangia by fragmentation. PLoS ONE 2010, 5, e8571. [Google Scholar] [CrossRef]
- Li, J.Y.; Yang, F.; Jin, L.; Wang, Q.; Yin, J.; He, P.; Chen, Y. Safety and quality of the green tide algal species Ulva prolifera for option of human consumption: A nutrition and contamination study. Chemosphere 2018, 210, 1021–1028. [Google Scholar] [CrossRef]
- Dhargalkar, V.K.; Pereira, N. Seaweed: Promising plant of the millennium. Sci. Cul. 2005, 71, 60–66. [Google Scholar]
- Echave, J.; Fraga-Corral, M.; Garcia-Perez, P.; Popović-Djordjević, J.; Avdović, E.H.; Radulović, M.; Xiao, J.; Prieto, M.A.; Simal-Gandara, J. Seaweed Protein Hydrolysates and Bioactive Peptides: Extraction, Purification, and Applications. Mar. Drugs 2021, 19, 500. [Google Scholar] [CrossRef]
- Pimentel, F.B.; Cermeño, M.; Kleekayai, T.; Harnedy-Rothwell, P.A.; Fernandes, E.; Alves, R.C.; Oliveira, M.B.P.P.; FitzGerald, R.J. Enzymatic Modification of Porphyra dioica-Derived Proteins to Improve their Antioxidant Potential. Molecules 2020, 25, 2838. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, Y.; He, H.; Zhou, W.; Li, M.; Lu, A.; Che, T.; Shen, S. Purification identification and function analysis of ACE inhibitory peptide from Ulva prolifera protein. Food Chem. 2023, 401, 134127. [Google Scholar] [CrossRef]
- Wiederschain, G.Y. The proteomics protocols handbook. Biochemistry (Moscow) 2006, 71, 969. [Google Scholar]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar]
- Liu, J.; Wang, C.; Wang, Z.; Zhang, C.; Lu, S.; Liu, J. The antioxidant and free-radical scavenging activities of extract and fractions from corn silk (Zea mays L.) and related flavone glycosides. Food Chem. 2011, 126, 261–269. [Google Scholar] [CrossRef]
- Liu, R.; Zheng, W.; Li, J.; Wang, L.; Wu, H.; Wang, X.; Shi, L. Rapid identification of bioactive peptides with antioxidant activity from the enzymatic hydrolysate of Mactra veneriformis by UHPLC-Q-TOF mass spectrometry. Food Chem. 2015, 167, 484–489. [Google Scholar] [CrossRef]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros-Zevallos, L.; Byrne, D.H. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compos. Anal. 2012, 19, 669–675. [Google Scholar] [CrossRef]
- Ferreira, L.L.G.; Andricopulo, A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today 2019, 24, 1157–1165. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y. ADME evaluation in drug discovery.8. The prediction of human intestinal absorption by a support vector machine. J. Chem. Inf. Model. 2007, 47, 2408–2415. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Kan, R.; Wu, S.; Guo, H.; Zhao, W.; Ding, L.; Zheng, F.; Liu, J. Xanthine oxidase inhibitory peptides derived from tuna protein: Virtual screening, inhibitory activity, and molecular mechanisms. J. Sci. Food Agric. 2021, 101, 1349–1354. [Google Scholar] [CrossRef]
- Duni, A.; Liakopoulos, V.; Roumeliotis, S.; Peschos, D.; Dounousi, E. Oxidative Stress in the Pathogenesis and Evolution of Chronic Kidney Disease: Untangling Ariadne’s Thread. Int. J. Mol. Sci. 2019, 20, 3711. [Google Scholar] [CrossRef] [PubMed]
- Stenvinkel, P.; Chertow, G.M.; Devarajan, P.; Levin, A.; Andreoli, S.P.; Bangalore, S.; Warady, B.A. Chronic Inflammation in Chronic Kidney Disease Progression: Role of Nrf2. Kidney Int. Rep. 2021, 6, 1775–1787. [Google Scholar] [CrossRef]
- Chen, J.; Yu, X.; Chen, Q.; Wu, Q.; He, Q. Screening and mechanisms of novel angiotensin-I-converting enzyme inhibitory peptides from rabbit meat proteins: A combined in silico and in vitro study. Food Chem. 2022, 370, 131070. [Google Scholar] [CrossRef]
- Dinesh, S.; Sharma, S.; Chourasiya, R. Therapeutic Applications of Plant and Nutraceutical-Based Compounds for the Management of Type 2 Diabetes Mellitus: A Narrative Review. Curr. Diabetes Rev. 2024, 20, e050523216593. [Google Scholar] [CrossRef]
- Suzuki, T.; Yamamoto, M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015, 88, 93–100. [Google Scholar] [CrossRef]
- Zhuang, C.; Narayanapillai, S.; Zhang, W.; Sham, Y.Y.; Xing, C. Rapid identification of Keap1-Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med. Chem. 2014, 57, 1121–1126. [Google Scholar] [CrossRef]
- Jain, A.D.; Potteti, H.; Richardson, B.G.; Kingsley, L.; Luciano, J.P.; Ryuzoji, A.F.; Lee, H.; Krunic, A.; Mesecar, A.D.; Reddy, S.P.; et al. Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators. Eur. J. Med. Chem. 2015, 103, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef]
- Liu, W.; Liu, R.; Qin, Q.; Wang, H.; Zhang, X.; Meng, G. Molecular docking and molecular dynamics simulation of wheat gluten-derived antioxidant peptides acting through the Keap1-Nrf2 pathway. J. Sci. Food. Agric. 2024, 104, 8150–8161. [Google Scholar] [CrossRef]
- Wilson, C.J.; Chang, M.; Karttunen, M.; Choy, W.Y. KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability. Int. J. Mol. Sci. 2021, 22, 5408. [Google Scholar] [CrossRef]
- Huang, D.; Ou, B.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef]
- Yang, X.R.; Zhao, Y.Q.; Qiu, Y.T.; Chi, C.F.; Wang, B. Preparation and characterization of gelatin and antioxidant peptides from gelatin hydrolysate of Skipjack Tuna (Katsuwonus pelamis) bone stimulated by in vitro gastrointestinal digestion. Mar. Drugs 2019, 17, 78. [Google Scholar] [CrossRef]
- Farzaneh, R.S.A. Bioactive properties of Agaricus bisporus and Terfezia claveryi proteins hydrolyzed by gastrointestinal proteases. Lebensm. Wiss. Technol. 2018, 91, 322–329. [Google Scholar] [CrossRef]
- Chen, J.; Yan, Y.; Zhang, L.; Zheng, J.; Guo, J.; Li, R.; Zeng, J. Purification of novel antioxidant peptides from myofibrillar protein hydrolysate of chicken breast and their antioxidant potential in chemical and H2O2-stressed cell systems. Food Funct. 2021, 12, 4897–4908. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Liceaga, A.M. Cellular antioxidant effect of bioactive peptides and molecular mechanisms underlying: Beyond chemical properties. Int. J. Food Sci. Technol. 2021, 56, 2193–2204. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Yu, Q.; Tao, Y.; Huang, Y.; Zogona, D.; Wu, T.; Liu, R.; Pan, S.; Xu, X. Aged Pericarpium Citri Reticulatae ‘Chachi’ Attenuates Oxidative Damage Induced by tert-Butyl Hydroperoxide (t-BHP) in HepG2 Cells. Foods 2022, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, Y.; Yang, H.Z.; Wang, Y.J.; Chen, Y. HMGB1 regulates ferroptosis through Nrf2 pathway in mesangial cells in response to high glucose. Biosci. Rep. 2021, 41, BSR20202924. [Google Scholar] [CrossRef]
- Zhou, M.; Tang, Y.; Liao, L.; Liu, M.; Deng, Y.; Zhao, X.; Li, Y. Phillygenin inhibited LPS-induced RAW 264.7 cell inflammation by NF-κB pathway. Eur. J. Pharmacol. 2021, 899, 174043. [Google Scholar] [CrossRef]
- Huang, D.; Chen, Y.; Chen, W.; Liu, Y.; Yao, F.; Xue, D.; Sun, L. Anti-inflammatory effects of the extract of Gnaphalium affine D Don in vivo and in vitro. J. Ethnopharmacol. 2015, 176, 356–364. [Google Scholar] [CrossRef]
- Boscá, L.; Zeini, M.; Través, P.G.; Hortelano, S. Nitric oxide and cell viability in inflammatory cells: A role for NO in macrophage function and fate. Toxicology 2005, 208, 249–258. [Google Scholar] [CrossRef]
- Beutler, B.; Cerami, A. Cachectin: More than a tumor necrosis factor. N. Engl. J. Med. 1987, 316, 379–385. [Google Scholar] [CrossRef]
- Moreira-Tabaka, H.; Peluso, J.; Vonesch, J.L.; Hentsch, D.; Kessler, P.; Reimund, J.M.; Dumont, S.; Muller, C.D. Unlike for human monocytes after LPS activation, release of TNF-α by THP-1 cells is produced by a TACE catalytically different from constitutive TACE. PLoS ONE 2012, 7, e34184. [Google Scholar] [CrossRef][Green Version]
- Hirano, T. Interleukin 6 in autoimmune and inflammatory diseases: A personal memoir. Proceedings of the Japan Academy. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 717–730. [Google Scholar] [CrossRef]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Wang, J.; Guo, L.; Xu, Y.; Hu, L.; Mao, H.; Miao, L.; Zhang, H.; Chai, L. Neobavaisoflavone ameliorates LPS-induced RAW264.7 cell inflammations by suppressing the activation of NF-κB and MAPKs signaling pathways. Iran J. Basic Med. Sci. 2022, 25, 1021–1027. [Google Scholar] [PubMed]
- Zhang, X.; Xiong, H.; Li, H.; Yu, L.; Deng, X. Effects of florfenicol on LPS-induced nitric oxide and prostaglandin E2 production in RAW 264.7 macrophages. Fundam. Clin. Pharmacol. 2011, 25, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Jiao, Q.; Liu, T.; Tan, S.J.; Zhou, H.S.; You, Q.D.; Jiang, Z.Y. Discovery of a head-to-tail cyclic peptide as the Keap1-Nrf2 protein-protein interaction inhibitor with high cell potency. Eur. J. Med. Chem. 2018, 143, 1578–1589. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, X.; Zhang, X.L.; Xie, Q.F. Purification and identification of anti-inflammatory peptides derived from simulated gastrointestinal digests of velvet antler protein (Cervus elaphus Linnaeus). J. Food Drug Anal. 2016, 24, 376–384. [Google Scholar] [CrossRef]
- Abeer, M.M.; Trajkovic, S.; Brayden, D.J. Measuring the oral bioavailability of protein hydrolysates derived from food sources: A critical review of current bioassays. Biomed. Pharmacother. 2021, 144, 112275. [Google Scholar] [CrossRef]




| Gene | Forward/Reverse | Primer Sequence |
|---|---|---|
| β-actin | Forward | CGGTTCCGATGCCCTGAGGCTCTT |
| Reverse | CGTCACACTTCATGATGGAATTGA | |
| NQO-1 | Forward | TTCTCTGGCCGATTCAGAGTG |
| Reverse | TCAGCTGGAATGGACTTGCC | |
| HO-1 | Forward | CCCACCAAGTTCAAACAGCTC |
| Reverse | AGGAAGGCGGTCTTAGCCTC | |
| GCLM | Forward | ATGACCCGAAAGAACTGCTC |
| Reverse | CTGCTCTTCACGATGACCGA | |
| TNF-α | Forward | ACCCTCACACTCACAAACCA |
| Reverse | ATAGCAAATCGGCTGACGGT | |
| IL-6 | Forward | GGTGACAACCACGGCCTTCCC |
| Reverse | AAGCCTCCGACTTGTGAAGTGGT |
| Peptide | Activity Score | Water Solubility | Peptide | Activity Score | Water Solubility | Peptide | Activity Score | Water Solubility |
|---|---|---|---|---|---|---|---|---|
| FR | 0.985719 | good | FK | 0.860392 | good | EF | 0.598976 | good |
| WR | 0.977891 | good | MR | 0.849148 | good | LR | 0.569984 | good |
| WK | 0.866241 | good | PR | 0.787626 | good | CQ | 0.540359 | good |
| CR | 0.865233 | good | GR | 0.766288 | good | YR | 0.525242 | good |
| FMR | 0.980845 | good | DDF | 0.748981 | good | LGR | 0.620526 | good |
| FPR | 0.976464 | good | DQF | 0.736087 | good | YDC | 0.609016 | good |
| MDF | 0.959246 | good | NDF | 0.723384 | good | DPR | 0.599321 | good |
| FDP | 0.930726 | good | FTR | 0.706612 | good | FNK | 0.592861 | good |
| GCR | 0.902065 | good | AWK | 0.698525 | good | MGK | 0.584511 | good |
| FIR | 0.893444 | good | CDG | 0.68045 | good | NFK | 0.577538 | good |
| DFL | 0.889906 | good | YGR | 0.676333 | good | GPK | 0.567438 | good |
| DWR | 0.876857 | good | SWK | 0.666979 | good | FEL | 0.566207 | good |
| FKP | 0.871158 | good | MIR | 0.66231 | good | EFR | 0.557213 | good |
| YDF | 0.870185 | good | GSP | 0.653894 | good | FSK | 0.556465 | good |
| LDF | 0.839471 | good | QCR | 0.643288 | good | MSR | 0.554004 | good |
| FSR | 0.831241 | good | CKP | 0.642963 | good | DVF | 0.551408 | good |
| GGR | 0.81127 | good | DWS | 0.637383 | good | GDL | 0.546664 | good |
| FLK | 0.805333 | good | MAR | 0.636319 | good | FER | 0.529433 | good |
| YMR | 0.780662 | good | GCK | 0.632386 | good | DGL | 0.525785 | good |
| FYK | 0.764839 | good | IPR | 0.629111 | good | PGK | 0.517877 | good |
| YPR | 0.754834 | good | FAK | 0.621611 | good | SGR | 0.508729 | good |
| PWFR | 0.990371 | good | QPPR | 0.762773 | good | MYHR | 0.587398 | good |
| PWYR | 0.942612 | good | FQPK | 0.72783 | good | FLIK | 0.582359 | good |
| FIPR | 0.912571 | good | YQWD | 0.687401 | good | FNYK | 0.577253 | good |
| IPFR | 0.906606 | good | GSMR | 0.675175 | good | FHYK | 0.572896 | good |
| PGMR | 0.900324 | good | DMHP | 0.639519 | good | TFPK | 0.561414 | good |
| CDGC | 0.839955 | good | DCYR | 0.631674 | good | KPEF | 0.557678 | good |
| FMTR | 0.821677 | good | CCDA | 0.625188 | good | GGNR | 0.546288 | good |
| KPDF | 0.810801 | good | FGNK | 0.601468 | good | LGPK | 0.516678 | good |
| HCPR | 0.800373 | good | FKPK | 0.592118 | good | FIIK | 0.506281 | good |
| FKPDL | 0.802916 | good | LDPCR | 0.727549 | good | DDSMC | 0.586729 | good |
| DWIGR | 0.789988 | good | FISGR | 0.723567 | good | NKPVF | 0.583004 | good |
| QFPNR | 0.772362 | good | SCPQR | 0.680029 | good | YECGL | 0.543409 | good |
| EGCAW | 0.761261 | good | FAPAK | 0.672649 | good | NDPGR | 0.531025 | good |
| WSPEL | 0.736445 | good | AMDGL | 0.640645 | good | GPAAR | 0.515691 | good |
| FSPIK | 0.730555 | good | LDPAL | 0.595302 | good | YGPTR | 0.514815 | good |
| DDYHC | 0.504222 | good | FDGNMP | 0.863932 | good | FLPPNK | 0.744351 | good |
| SWNTPR | 0.739203 | good | WAKPGH | 0.688682 | good | SGMCHD | 0.559215 | good |
| FLKPDL | 0.711564 | good | GRPADM | 0.636649 | good | SGKPAL | 0.540218 | good |
| AWDSYR | 0.692275 | good | PEPWAK | 0.578204 | good | DGLNPR | 0.50385 | good |
| Peptides | Caco2- | BBB+ | HIA+ | Metabolism or Inhibitor | Toxicity |
|---|---|---|---|---|---|
| Probability | |||||
| MR | 0.6739 | 0.7091 | 0.8599 | non | non |
| PR | 0.7649 | 0.7597 | 0.6889 | non | non |
| LR | 0.752 | 0.698 | 0.8271 | non | non |
| YR | 0.8827 | 0.6121 | 0.6163 | non | non |
| FR | 0.7607 | 0.7849 | 0.7912 | non | non |
| WR | 0.762 | 0.8368 | 0.9644 | non | non |
| WK | 0.7574 | 0.8534 | 0.9708 | non | non |
| FK | 0.7064 | 0.8011 | 0.8238 | non | non |
| AWK | 0.8394 | 0.6041 | 0.9441 | non | non |
| DWS | 0.8544 | 0.7165 | 0.7926 | non | non |
| DWR | 0.8101 | 0.6244 | 0.7047 | non | non |
| MAR | 0.7039 | 0.5787 | 0.5799 | non | non |
| MGK | 0.685 | 0.683 | 0.6725 | non | non |
| FAK | 0.8181 | 0.6068 | 0.7042 | non | non |
| FYK | 0.8468 | 0.5196 | 0.7024 | non | non |
| FEL | 0.8278 | 0.5648 | 0.5695 | non | non |
| PWFR | 0.822 | 0.5349 | 0.8572 | non | non |
| PWYR | 0.861 | 0.6014 | 0.8955 | non | non |
| YQWD | 0.8771 | 0.5461 | 0.6572 | non | non |
| Peptides | CDOCKER-ENERGY (kcal/mol) | Peptides | CDOCKER-ENERGY (kcal/mol) |
|---|---|---|---|
| DWS | 88.4352 | LR | 71.0558 |
| DWR | 87.7048 | YR | 71.0558 |
| MGK | 87.4723 | FR | 68.3871 |
| FAK | 87.175 | WR | 67.3156 |
| FEL | 80.9024 | MR | 67.1224 |
| FYK | 79.266 | PR | 42.9183 |
| MAR | 73.7247 | PWFR | No |
| FK | 73.5543 | PWYR | No |
| AWK | 72.8721 | YQWD | No |
| WK | 71.1864 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Li, Z.; Gu, H.; Shen, S. Antioxidant Activity In Vitro and Protective Effects Against Lipopolysaccharide-Induced Oxidative Stress and Inflammation in RAW264.7 Cells of Ulva prolifera-Derived Bioactive Peptides Identified by Virtual Screening, Molecular Docking, and Dynamics Simulations. Foods 2025, 14, 2202. https://doi.org/10.3390/foods14132202
Liu J, Li Z, Gu H, Shen S. Antioxidant Activity In Vitro and Protective Effects Against Lipopolysaccharide-Induced Oxidative Stress and Inflammation in RAW264.7 Cells of Ulva prolifera-Derived Bioactive Peptides Identified by Virtual Screening, Molecular Docking, and Dynamics Simulations. Foods. 2025; 14(13):2202. https://doi.org/10.3390/foods14132202
Chicago/Turabian StyleLiu, Jiasi, Zhiyong Li, Huiyue Gu, and Songdong Shen. 2025. "Antioxidant Activity In Vitro and Protective Effects Against Lipopolysaccharide-Induced Oxidative Stress and Inflammation in RAW264.7 Cells of Ulva prolifera-Derived Bioactive Peptides Identified by Virtual Screening, Molecular Docking, and Dynamics Simulations" Foods 14, no. 13: 2202. https://doi.org/10.3390/foods14132202
APA StyleLiu, J., Li, Z., Gu, H., & Shen, S. (2025). Antioxidant Activity In Vitro and Protective Effects Against Lipopolysaccharide-Induced Oxidative Stress and Inflammation in RAW264.7 Cells of Ulva prolifera-Derived Bioactive Peptides Identified by Virtual Screening, Molecular Docking, and Dynamics Simulations. Foods, 14(13), 2202. https://doi.org/10.3390/foods14132202






