From Isolation to Pilot-Scale Production: Enterococcus faecium YC07 with Urate-Lowering Potential from Fermented Food Jiangshui
Abstract
1. Introduction
2. Materials and Methods
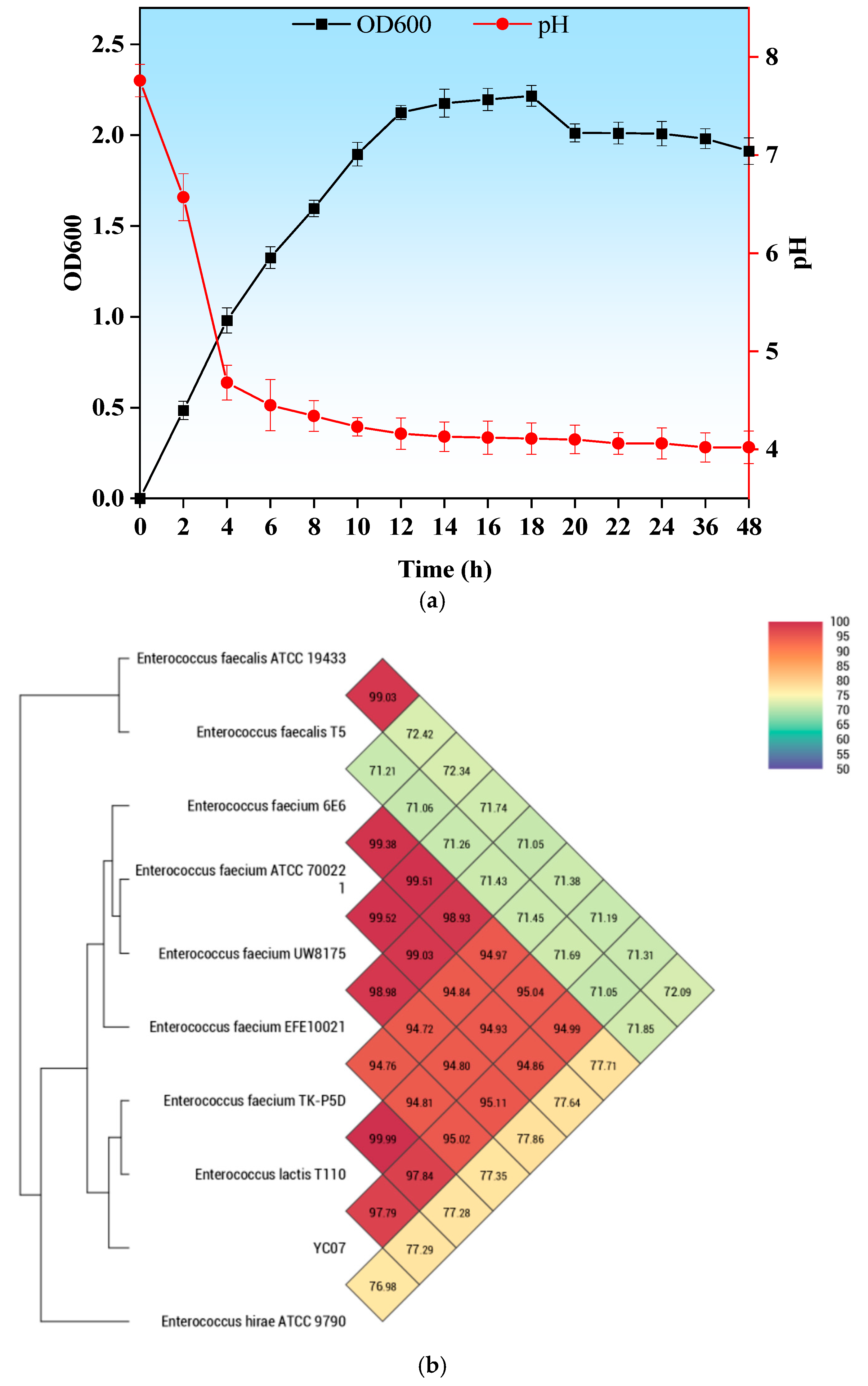
2.1. Isolation of the Bacterial Strain and Growth Conditions
2.2. Whole Genome Sequencing and Genome Annotation
2.3. Identification of YC07
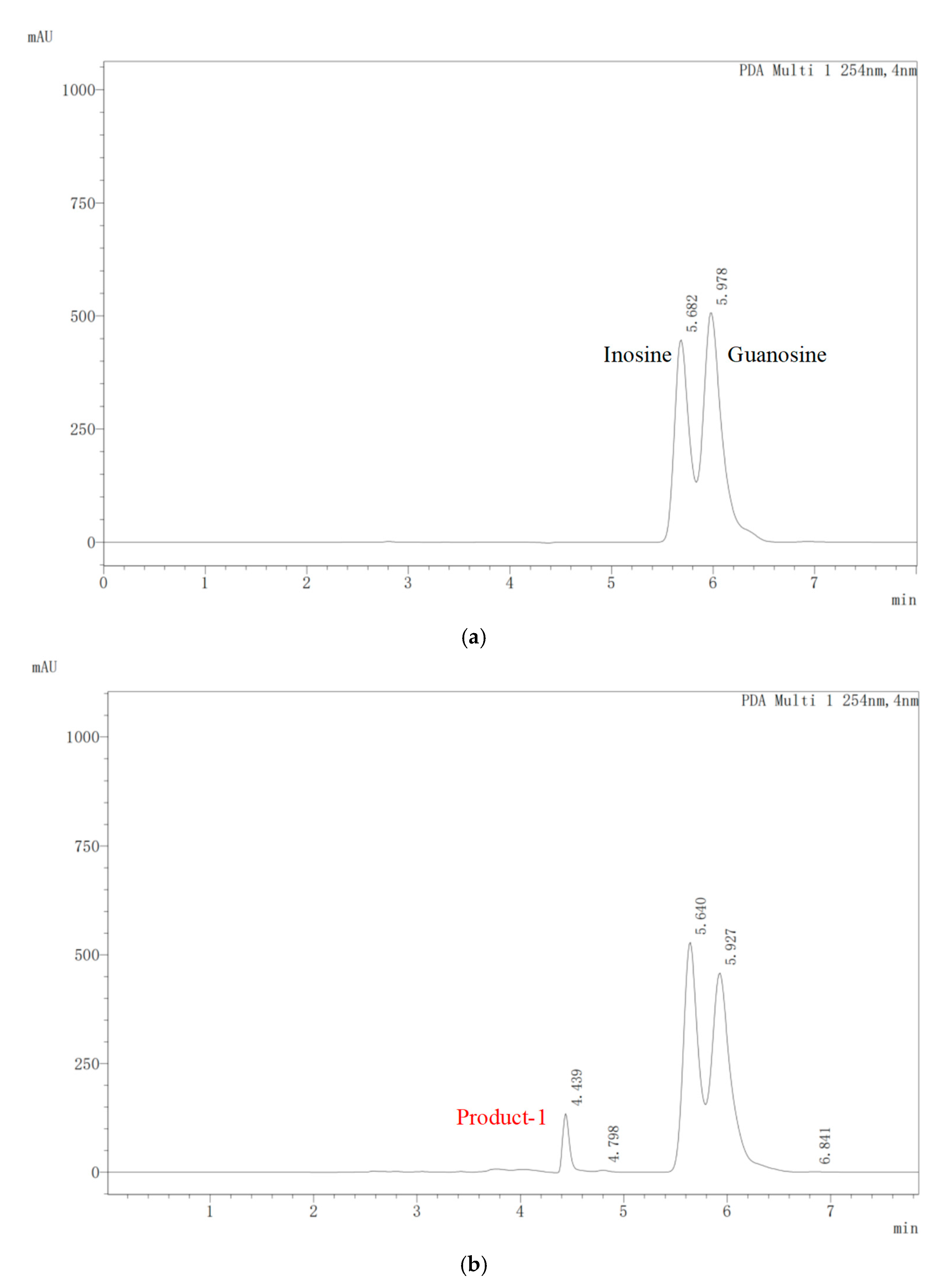
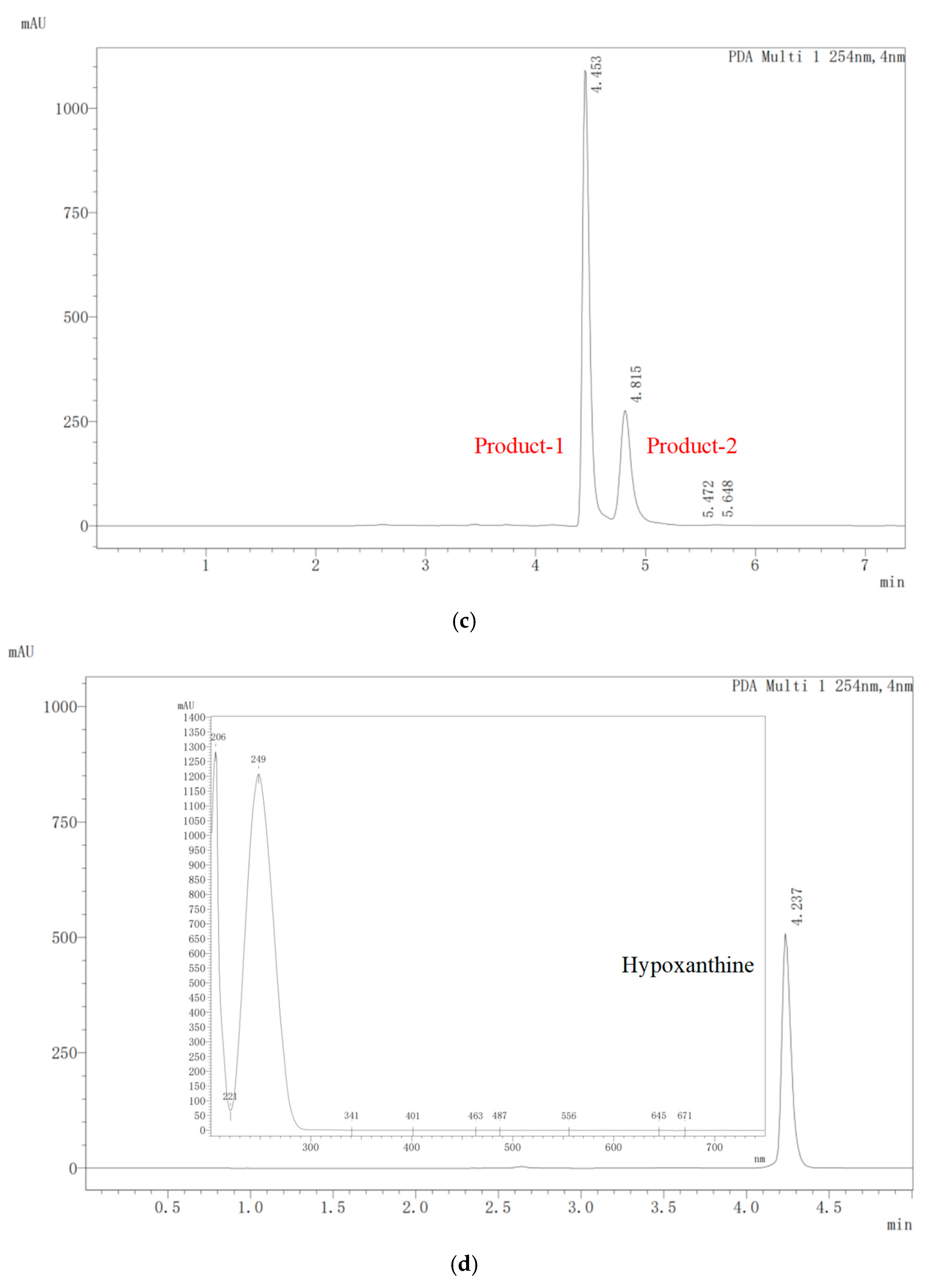
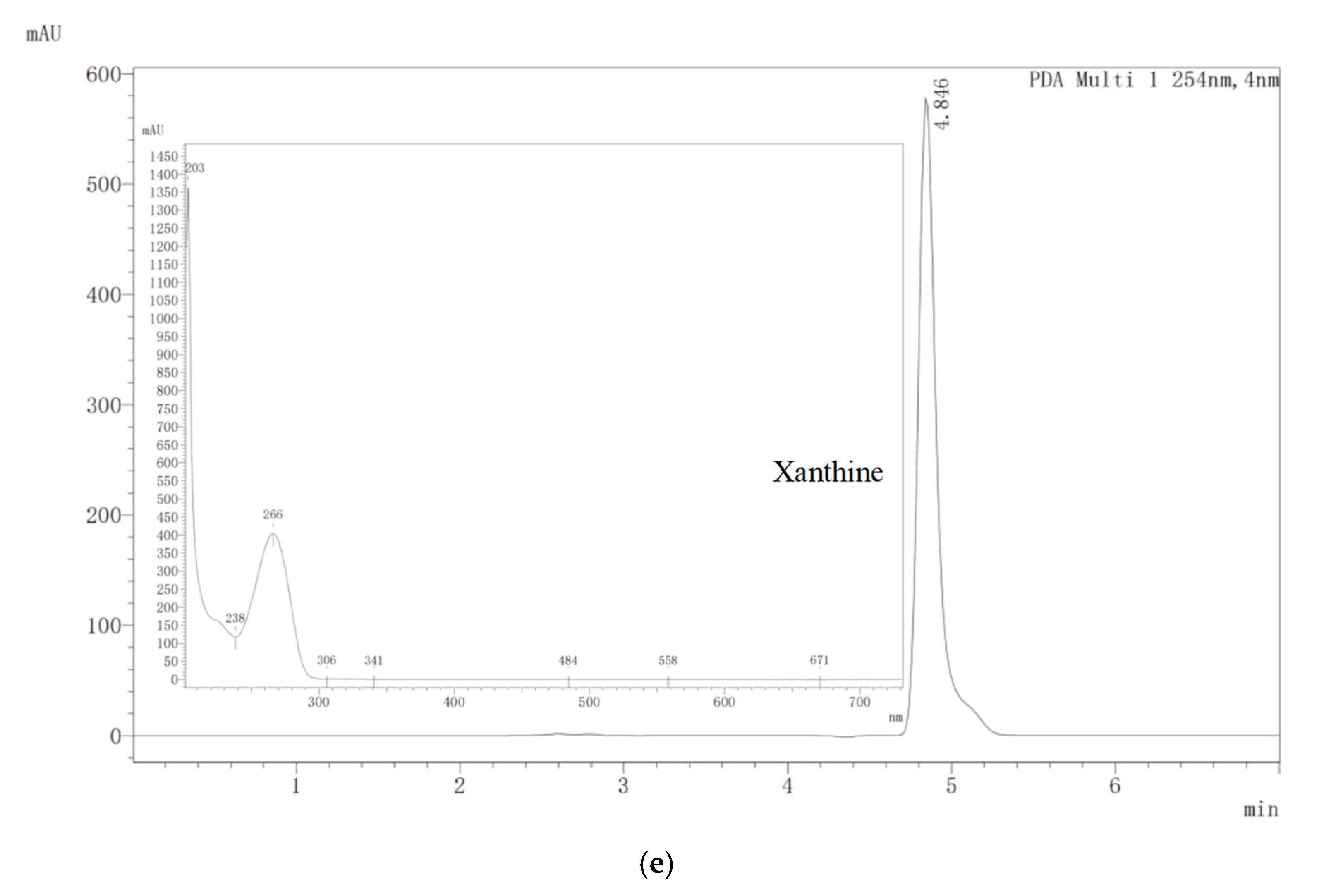
2.4. Biodegradation of Nucleosides by E. faecium YC07
2.5. Probiotic Characteristics
2.5.1. Tolerance of Simulated Gastrointestinal Fluids
2.5.2. Antimicrobial Activity
2.5.3. Auto-Aggregation and Cell Surface Hydrophobicity
2.5.4. Evaluation of the Antioxidant Activity
2.6. Safety Assessment
2.6.1. Hemolytic Activity
2.6.2. Antibiotic Resistance
2.6.3. Biogenic Amine Production Assay
2.6.4. Gelatinase Activity
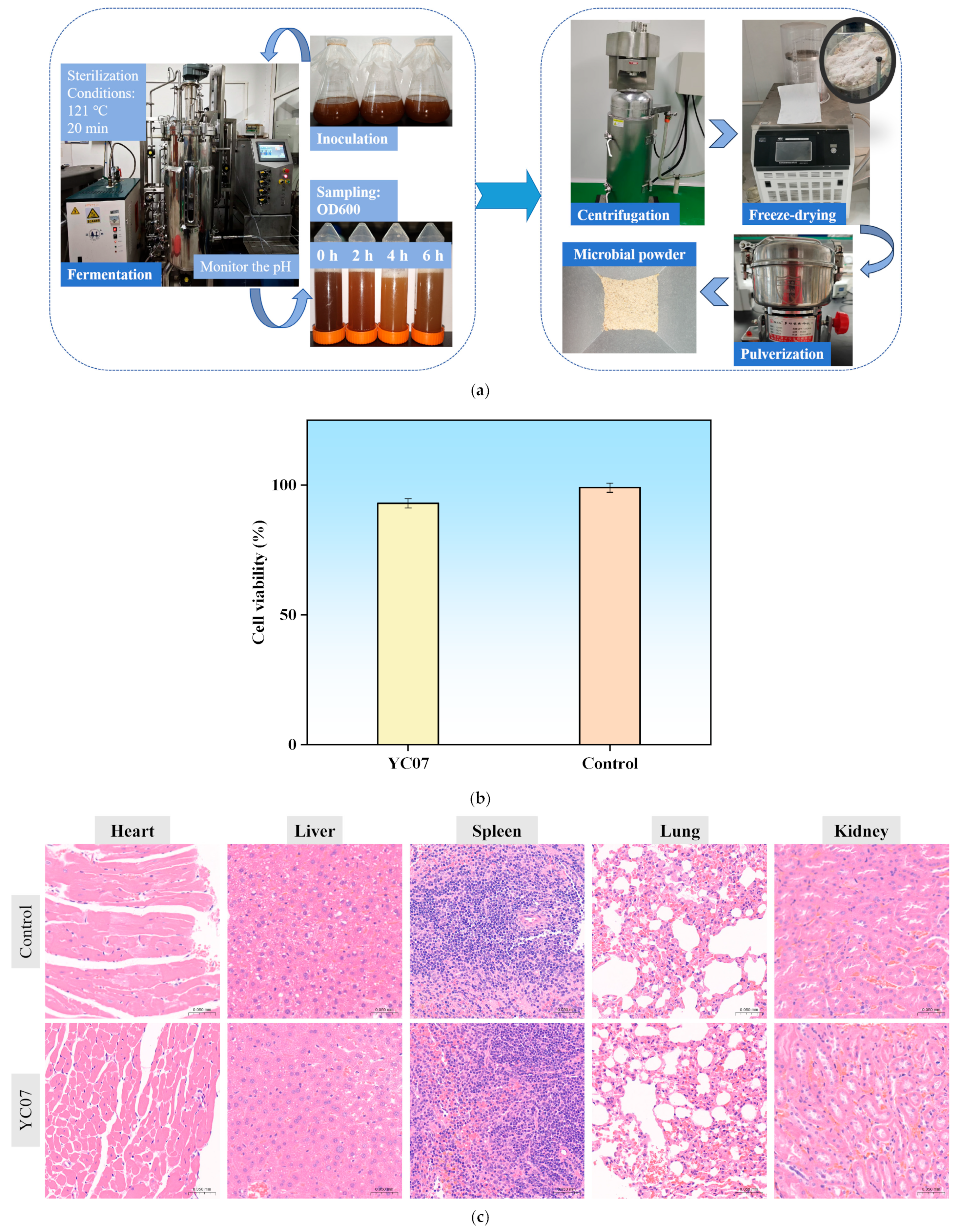
2.6.5. Acute Oral Toxicology Test and Cytotoxicity Assay
2.7. Statistical Analysis
3. Results and Discussion
3.1. Isolation of Nucleoside Biodegrading Strain YC07 and Product Identification
3.2. Genome Feature of E. faecium YC07
3.3. Simulated Gastrointestinal Fluid Tolerance Assays
3.4. Inhibition Ability of YC07 to Pathogens
3.5. Auto-Aggregation and Cell Surface Hydrophobicity
3.6. Antioxidant Activity
3.7. Safety Properties
3.7.1. Safety-Related Gene Profile
3.7.2. Antimicrobial Susceptibility Test
3.8. In Vitro Safety Assessment of E. faecium YC07
3.9. Acute Oral Toxicology and Cytotoxicity Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Wang, S.; Wang, L.; Lu, H.; Zhang, T.; Zeng, W. Characterization of Genomic, Physiological, and Probiotic Features of Lactiplantibacillus plantarum JS21 Strain Isolated from Traditional Fermented Jiangshui. Foods 2024, 13, 1082. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ye, Z.; Feng, P.; Li, R.; Chen, X.; Tian, X.; Han, R.; Kakade, A.; Liu, P.; Li, X. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Feng, P.; Hu, X.; Cao, W.; Liu, P.; Han, H.; Jin, W.; Li, X. Probiotic Limosilactobacillus fermentum GR-3 ameliorates human hyperuricemia via degrading and promoting excretion of uric acid. iScience 2022, 25, 105198. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Aga, L.; Tang, L.; Li, H.; Wang, N.; Yang, L.; Zhang, N.; Wang, X.; Wang, X. Lacticaseibacillus paracasei JS-3 Isolated from “Jiangshui” Ameliorates Hyperuricemia by Regulating Gut Microbiota and iTS Metabolism. Foods 2024, 13, 1317. [Google Scholar] [CrossRef]
- El Ridi, R.; Tallima, H. Physiological functions and pathogenic potential of uric acid: A review. J. Adv. Res. 2017, 8, 487–493. [Google Scholar] [CrossRef]
- Maiuolo, J.; Oppedisano, F.; Gratteri, S.; Muscoli, C.; Mollace, V. Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 2016, 213, 8–14. [Google Scholar] [CrossRef]
- Bardin, T.; Richette, P. Definition of hyperuricemia and gouty conditions. Curr. Opin. Rheumatol. 2014, 26, 186–191. [Google Scholar] [CrossRef]
- Ragab, G.; Elshahaly, M.; Bardin, T. Gout: An old disease in new perspective—A review. J. Adv. Res. 2017, 8, 495–511. [Google Scholar] [CrossRef]
- Sun, L.; Ni, C.; Zhao, J.; Wang, G.; Chen, W. Probiotics, bioactive compounds and dietary patterns for the effective management of hyperuricemia: A review. Crit. Rev. Food Sci. Nutr. 2022, 64, 2016–2031. [Google Scholar] [CrossRef]
- Strilchuk, L.; Fogacci, F.; Cicero, A.F. Safety and tolerability of available urate-lowering drugs: A critical review. Expert. Opin. Drug Saf. 2019, 18, 261–271. [Google Scholar] [CrossRef]
- Shahid, H.; Singh, J.A. Investigational drugs for hyperuricemia. Expert. Opin. Investig. Drugs 2015, 24, 1013–1030. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, J.L.; Zhou, Q. Targets and mechanisms of dietary anthocyanins to combat hyperglycemia and hyperuricemia: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2022, 62, 1119–1143. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, N.K.; Bhattacharya, S.K.; Sesikeran, B.; Nair, G.B.; Ramakrishna, B.S.; Sachdev, H.P.S.; Batish, V.K.; Kanagasabapathy, A.S.; Muthuswamy, V.; Kathuria, S.C. ICMR-DBT Guidelines for Evaluation of Probiotics in Food. Indian J. Med. Res. 2011, 134, 22–25. [Google Scholar]
- Zhao, H.; Lu, Z.; Lu, Y. The potential of probiotics in the amelioration of hyperuricemia. Food Funct. 2022, 13, 2394–2414. [Google Scholar] [CrossRef]
- Javed, A.; Masud, T.; ul Ain, Q.; Imran, M.; Maqsood, S. Enterocins of Enterococcus faecium, emerging natural food preservatives. Ann. Microbiol. 2011, 61, 699–708. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Nascimento, L.C.S.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Probiotic potential and safety of enterococci strains. Ann. Microbiol. 2019, 69, 241–252. [Google Scholar] [CrossRef]
- Fu, X.; Lyu, L.; Wang, Y.; Zhang, Y.; Guo, X.; Chen, Q.; Liu, C. Safety assessment and probiotic characteristics of Enterococcus lactis JDM1. Microb. Pathog. 2022, 163, 105380. [Google Scholar] [CrossRef]
- Du, X.; Jiang, Y.; Sun, Y.; Cao, X.; Zhang, Y.; Xu, Q.; Yan, H. Biodegradation of Inosine and Guanosine by Bacillus paranthracis YD01. Int. J. Mol. Sci. 2023, 24, 4462. [Google Scholar] [CrossRef]
- Çetin, B.; Aktaş, H. Monitoring probiotic properties and safety evaluation of antilisterial Enterococcus faecium strains with cholesterol-lowering potential from raw Cow’s milk. Food Biosci. 2024, 61, 104532. [Google Scholar] [CrossRef]
- Mu, Y.; Zhang, C.; Jin, C.-Z.; Li, T.; Jin, F.-J.; Lee, H.-G.; Jin, L. Antibacterial activity and action mode of crude bacteriocin C2-1 from Ligilactobacillus salivarius C2-1 against Listeria monocytogenes CICC 21633. LWT 2024, 193, 115765. [Google Scholar] [CrossRef]
- Ait Seddik, H.; Bendali, F.; Cudennec, B.; Drider, D. Anti-pathogenic and probiotic attributes of Lactobacillus salivarius and Lactobacillus plantarum strains isolated from feces of Algerian infants and adults. Res. Microbiol. 2017, 168, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Park, S.; Sathiyaseelan, A.; Wang, M.-H. Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects. Fermentation 2023, 9, 760. [Google Scholar] [CrossRef]
- Khan, A.; Miller, W.R.; Axell-House, D.; Munita, J.M.; Arias, C.A.; Humphries, R.M. Antimicrobial Susceptibility Testing for Enterococci. J. Clin. Microbiol. 2022, 60, e0084321. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-p.; Liu, D.-m.; Zhao, S.; Huang, Y.-y.; Yu, J.-j.; Zhou, Q.-y. Assessing the safety and probiotic characteristics of Bacillus coagulans 13002 based on complete genome and phenotype analysis. LWT 2022, 155, 112847. [Google Scholar] [CrossRef]
- M’hir, S.; Minervini, F.; Di Cagno, R.; Chammem, N.; Hamdi, M. Technological, functional and safety aspects of enterococci in fermented vegetable products: A mini-review. Ann. Microbiol. 2011, 62, 469–481. [Google Scholar] [CrossRef]
- Gołaś-Prądzyńska, M.; Łuszczyńska, M.; Rola, J.G. Dairy Products: A Potential Source of Multidrug-Resistant Enterococcus faecalis and Enterococcus faecium Strains. Foods 2022, 11, 4116. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef]
- Yamada, N.; Iwamoto, C.; Kano, H.; Yamaoka, N.; Fukuuchi, T.; Kaneko, K.; Asami, Y. Evaluation of purine utilization by Lactobacillus gasseri strains with potential to decrease the absorption of food-derived purines in the human intestine. Nucleosides Nucleotides Nucleic Acids 2016, 35, 670–676. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, X.; Meng, F.; Zhou, L.; Pang, X.; Lu, Z.; Lu, Y. Ameliorative effect of Lacticaseibacillus rhamnosus Fmb14 from Chinese yogurt on hyperuricemia. Food Sci. Hum. Wellness 2023, 12, 1379–1390. [Google Scholar] [CrossRef]
- Meng, Y.P.; Hu, Y.S.; Wei, M.; Wang, K.M.; Wang, Y.Y.; Wang, S.L.; Hu, Q.; Wei, H.; Zhang, Z.H. Amelioration of hyperuricemia by Lactobacillus acidophilus F02 with uric acid-lowering ability via modulation of NLRP3 inflammasome and gut microbiota homeostasis. J. Funct. Foods 2023, 111, 105903. [Google Scholar] [CrossRef]
- Lu, L.; Liu, T.; Liu, X.; Wang, C. Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice-flour noodles. Food Sci. Hum. Wellness 2022, 11, 1402–1408. [Google Scholar] [CrossRef]
- Xu, J.; Tu, M.; Fan, X.; Guo, Y.; Zhang, T.; Zeng, X.; Cai, Z.; Wu, Z.; Pan, D. A novel strain of Levilactobacillus brevis PDD-5 isolated from salty vegetables has beneficial effects on hyperuricemia through anti-inflammation and improvement of kidney damage. Food Sci. Hum. Wellness 2024, 13, 898–908. [Google Scholar] [CrossRef]
- Li, T.; Lyu, L.; Zhang, Y.; Dong, K.; Li, Q.; Guo, X.; Zhu, Y. A newly isolated E. thailandicus strain d5B with exclusively antimicrobial activity against C. difficile might be a novel therapy for controlling CDI. Genomics 2021, 113, 475–483. [Google Scholar] [CrossRef]
- Li, M.; Wu, X.; Guo, Z.; Gao, R.; Ni, Z.; Cui, H.; Zong, M.; Van Bockstaele, F.; Lou, W. Lactiplantibacillus plantarum enables blood urate control in mice through degradation of nucleosides in gastrointestinal tract. Microbiome 2023, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; De Souza, C.; Ramachandran, M.; Wang, S.L.; Yi, H.X.; Ma, Z.; Zhang, L.W.; Lin, K. Precise oral delivery systems for probiotics: A review. J. Control. Release 2022, 352, 371–384. [Google Scholar] [CrossRef]
- Singhal, N.; Maurya, A.K.; Mohanty, S.; Kumar, M.; Virdi, J.S. Evaluation of Bile Salt Hydrolases, Cholesterol-Lowering Capabilities, and Probiotic Potential of Enterococcus faecium Isolated From Rhizosphere. Front. Microbiol. 2019, 10, 1567. [Google Scholar] [CrossRef] [PubMed]
- Nami, Y.; Vaseghi Bakhshayesh, R.; Mohammadzadeh Jalaly, H.; Lotfi, H.; Eslami, S.; Hejazi, M.A. Probiotic Properties of Enterococcus Isolated From Artisanal Dairy Products. Front. Microbiol. 2019, 10, 300. [Google Scholar] [CrossRef]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum Strains as a Bio-Control Strategy against Food-Borne Pathogenic Microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef]
- Wilkins, T.; Sequoia, J. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician 2017, 96, 170–178. [Google Scholar]
- Wang, Y.; Wu, Y.P.; Wang, Y.Y.; Xu, H.; Mei, X.Q.; Yu, D.Y.; Wang, Y.B.; Li, W.F. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Mishra, V.; Shah, C.; Mokashe, N.; Chavan, R.; Yadav, H.; Prajapati, J. Probiotics as Potential Antioxidants: A Systematic Review. J. Agric. Food Chem. 2015, 63, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Lin, J.H.; Kuo, Y.W.; Chiang, P.F.R.; Ho, H.H. Probiotics and their Metabolites Reduce Oxidative Stress in Middle-Aged Mice. Curr. Microbiol. 2022, 79, 104. [Google Scholar] [CrossRef] [PubMed]
- Ahadaf, S.; Azzouz, S.; Galiou, O.E.; Errahmouni, M.A.; Mentag, R.; Arakrak, A.; Laglaoui, A. Genomic Insights Into Enterococcus mundtii 203: A Promising Probiotic Candidate Isolated From Camel Feces. Probiotics Antimicrob. Proteins 2024, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria. Molecules 2019, 24, 3430. [Google Scholar] [CrossRef]
- Wójcik, W.; Lukasiewicz, M.; Puppel, K. Biogenic amines: Formation, action and toxicity—A review. J. Sci. Food Agric. 2021, 101, 2634–2640. [Google Scholar] [CrossRef]
- Erkekoglu, P.; Baydar, T. Evaluation of the protective effect of ascorbic acid on nitrite- and nitrosamine-induced cytotoxicity and genotoxicity in human hepatoma line. Toxicol. Mech. Methods 2010, 20, 45–52. [Google Scholar] [CrossRef]
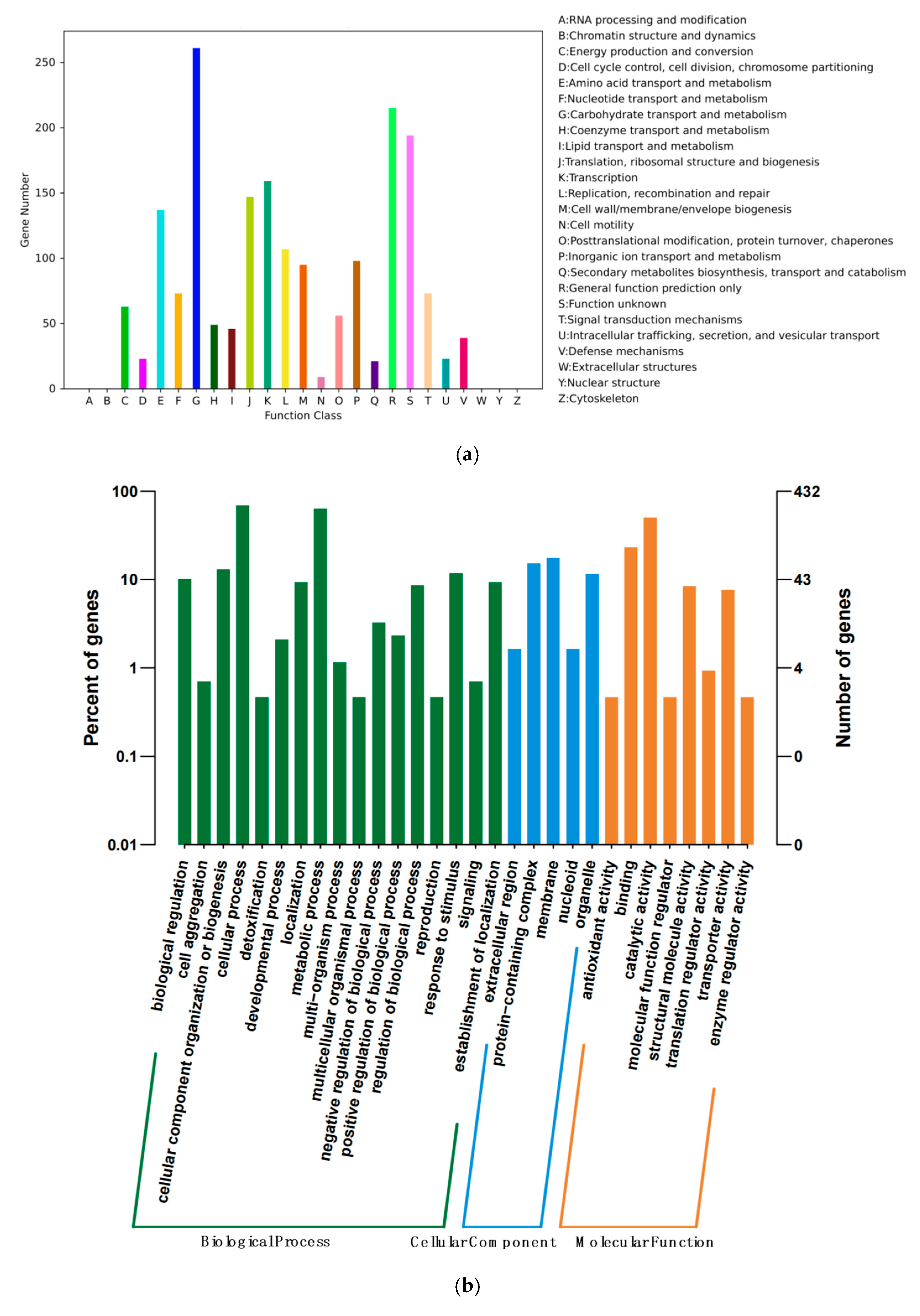
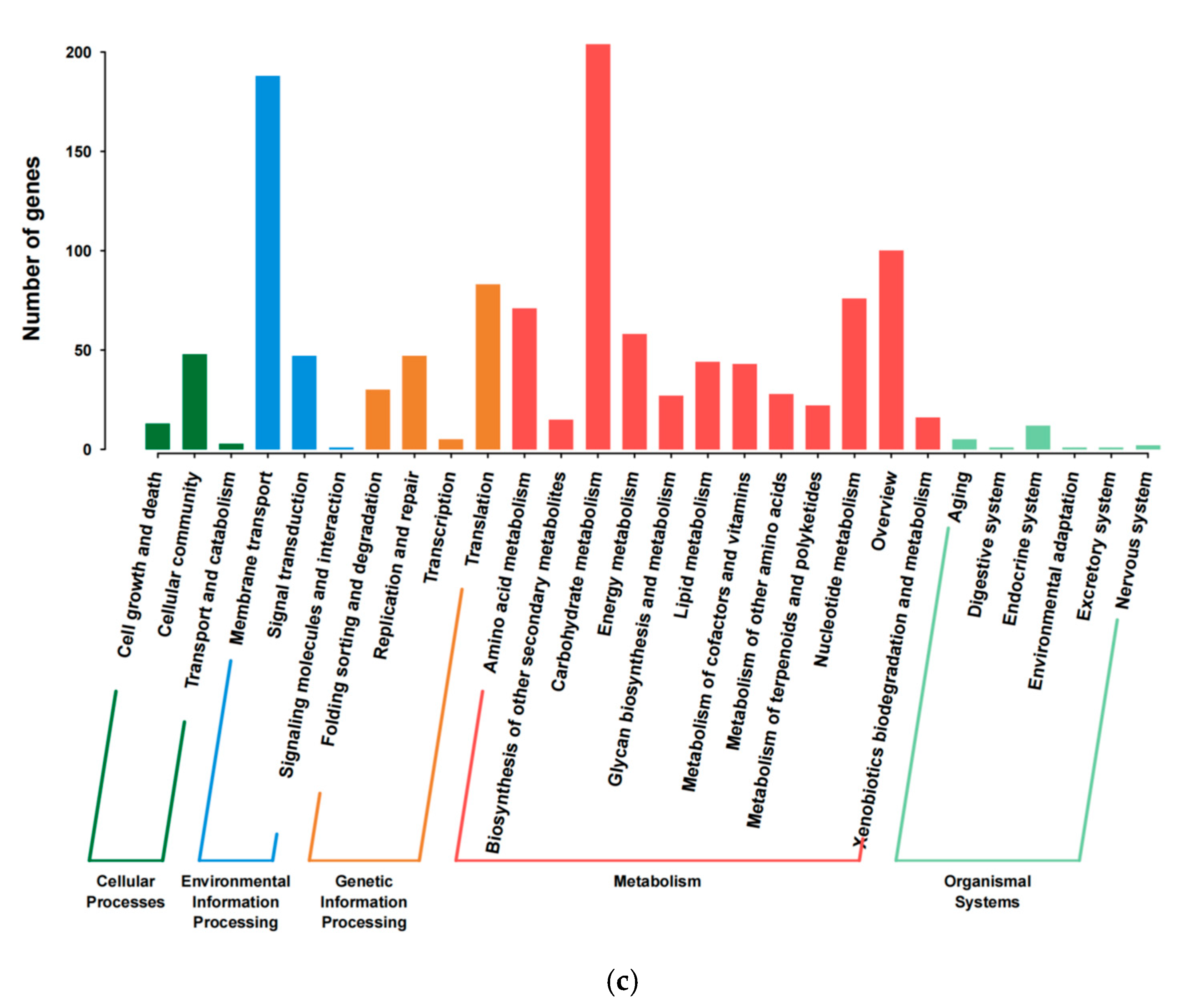
| Gene ID | Location | Gene Name | Product |
|---|---|---|---|
| Antimicrobial activity | |||
| gene0082 | Contig1 | msrA | Peptide methionine sulfoxide reductase MsrA |
| gene0413 | Contig3 | - | hypothetical protein |
| gene1641 | Contig13 | btuD | Vitamin B12 import ATP-binding protein BtuD |
| gene2177 | Contig23 | - | hypothetical protein |
| gene2464 | Contig43 | - | hypothetical protein |
| gene2465 | Contig43 | lagD | Lactococcin-G-processing and transport ATP-binding protein LagD |
| Acid resistance | |||
| gene1045 | Contig6 | atpC | ATP synthase epsilon chain |
| gene1046 | Contig6 | atpD | ATP synthase subunit beta |
| gene1047 | Contig6 | atpG | ATP synthase gamma chain |
| gene1048 | Contig6 | atpA | ATP synthase subunit alpha |
| gene1049 | Contig6 | atpH | ATP synthase subunit delta |
| gene1050 | Contig6 | atpF | ATP synthase subunit b |
| gene1051 | Contig6 | atpE-1 | ATP synthase subunit c |
| gene1052 | Contig6 | atpB | ATP synthase subunit a |
| gene1648 | Contig13 | atpE-2 | V-type proton ATPase subunit E |
| gene0004 | Contig1 | tyrS1 | Tyrosine--tRNA ligase 1 |
| gene0001 | Contig1 | nhaC-1 | Na(+)/H(+) antiporter NhaC |
| gene1755 | Contig15 | nhaC-2 | Na(+)/H(+) antiporter NhaC |
| gene1075 | Contig7 | asd | Aspartate-semialdehyde dehydrogenase |
| gene0065 | Contig1 | panP | Aspartate 1-decarboxylase |
| gene0261 | Contig2 | panT-1 | Pantothenate transporter PanT |
| gene1759 | Contig15 | panE | 2-dehydropantoate 2-reductase |
| gene1799 | Contig15 | panT-2 | Pantothenic acid transporter PanT |
| gene1146 | Contig7 | arcD1-1 | Arginine/ornithine antiporter ArcD1 |
| gene1477 | Contig11 | arcC1-1 | Carbamate kinase 1 |
| gene1478 | Contig11 | arcB | Ornithine carbamoyltransferase, catabolic |
| gene1479 | Contig11 | arcA | Arginine deiminase |
| gene1767 | Contig15 | arcD1-2 | Arginine/ornithine antiporter ArcD1 |
| gene2459 | Contig41 | arcC1-2 | Carbamate kinase 1 |
| gene0857 | Contig5 | argR-1 | Arginine repressor |
| gene1101 | Contig7 | argR-2 | Arginine repressor |
| gene1472 | Contig11 | argR-3 | Arginine repressor |
| gene1475 | Contig11 | argS | Arginine-tRNA ligase |
| Bile salt tolerance | |||
| gene1519 | Contig11 | cbh | Choloylglycine hydrolase |
| gene0588 | Contig3 | ecsA | ABC-type transporter ATP-binding protein EcsA |
| gene1078 | Contig7 | patA | Putative N-acetyl-LL-diaminopimelate aminotransferase |
| gene1438 | Contig10 | patB | Cystathionine beta-lyase PatB |
| Oxidative stress | |||
| gene1079 | Contig7 | npr | NADH peroxidase |
| gene0033 | Contig1 | nox-1 | NADH oxidase |
| gene0234 | Contig2 | nox-2 | NADH oxidase |
| gene1628 | Contig13 | nox-3 | NADH oxidase |
| gene0777 | Contig5 | sodA | Superoxide dismutase [Mn] |
| gene0988 | Contig6 | trxB | Thioredoxin reductase |
| gene1639 | Contig13 | trxA | Thioredoxin |
| gene2439 | Contig38 | tpx | Thiol peroxidase |
| gene0367 | Contig2 | ahpC | Alkyl hydroperoxide reductase C |
| gene0368 | Contig2 | ahpF | NADH dehydrogenase |
| Adhesion and aggregation | |||
| gene0290 | Contig2 | yloA | putative protein YloA |
| gene1275 | Contig8 | tuf | Elongation factor Tu |
| gene1324 | Contig9 | pdhA | Pyruvate dehydrogenase E1 component subunit alpha |
| gene1325 | Contig9 | pdhB | Pyruvate dehydrogenase E1 component subunit beta |
| gene1327 | Contig9 | pdhD | Dihydrolipoyl dehydrogenase |
| gene1674 | Contig13 | eno | Enolase |
| gene0219 | Contig2 | gap-1 | Glyceraldehyde-3-phosphate dehydrogenase |
| gene1671 | Contig13 | gap-2 | Glyceraldehyde-3-phosphate dehydrogenase |
| gene1673 | Contig13 | tpiA | Triosephosphate isomerase |
| Ionic and heavy metal stress resistance | |||
| gene0779 | Contig5 | czcD | Cadmium, cobalt and zinc/H(+)-K(+) antiporter |
| gene0984 | Contig6 | zur | Zinc-specific metallo-regulatory protein |
| gene1571 | Contig12 | znuA | High-affinity zinc uptake system binding-protein ZnuA |
| gene1572 | Contig12 | znuC | High-affinity zinc uptake system ATP-binding protein ZnuC |
| gene1573 | Contig12 | znuB | High-affinity zinc uptake system membrane protein ZnuB |
| gene2300 | Contig28 | fetB | putative iron export permease protein FetB |
| gene1167 | Contig7 | iscU | Iron-sulfur cluster assembly scaffold protein IscU |
| Temperature stress | |||
| gene2279 | Contig27 | hslO | 33 kDa chaperonin |
| gene1820 | Contig16 | ctsR | Transcriptional regulator CtsR |
| gene0238 | Contig2 | dnaD | DNA replication protein DnaD |
| gene0606 | Contig4 | dnaK | Chaperone protein DnaK |
| gene0607 | Contig4 | dnaJ-1 | Chaperone protein DnaJ |
| gene0698 | Contig4 | dnaB | Replication initiation and membrane attachment protein |
| gene0699 | Contig4 | dnaI | Primosomal protein DnaI |
| gene1067 | Contig7 | dnaJ-2 | Chaperone protein DnaJ |
| gene2032 | Contig19 | dnaE | DNA polymerase III subunit alpha |
| gene2059 | Contig20 | dnaA | Chromosomal replication initiator protein DnaA |
| gene2060 | Contig20 | dnaN | Beta sliding clamp |
| gene2070 | Contig20 | dnaC | Replicative DNA helicase |
| gene0605 | Contig4 | grpE | Protein GrpE |
| gene0604 | Contig4 | hrcA | Heat-inducible transcription repressor HrcA |
| gene0265 | Contig2 | csp-1 | Cold shock-like protein |
| gene0776 | Contig5 | cspD | Cold shock protein CspD |
| gene0942 | Contig6 | cspLA-1 | Cold shock-like protein CspLA |
| gene2106 | Contig21 | cspLA-2 | Cold shock-like protein CspLA |
| gene2214 | Contig25 | cspLA-3 | Cold shock-like protein CspLA |
| gene2237 | Contig25 | csp-2 | Cold shock protein 1 |
| gene1054 | Contig6 | rnr | Ribonuclease R |
| Lactate synthesis | |||
| gene0302 | Contig2 | ldhB | L-lactate dehydrogenase 2 |
| gene2269 | Contig27 | ldh | L-lactate dehydrogenase |
| Gene ID | Location | Gene Name | Antibiotics | Product | Identify (%) |
|---|---|---|---|---|---|
| gene1371 | Contig10 | AAC(6′)-Ii | aminoglycoside | chromosomal-encoded aminoglycoside acetyltransferase | 98.9 |
| gene1139 | Contig7 | efmA | fluoroquinolone; macrolide | MFS transporter permease | 74.5 |
| Gene ID | Location | Gene Name | Predicted Functions | Identify (%) |
|---|---|---|---|---|
| gene0412 | Contig3 | acm | collagen adhesin precursor | 95.5 |
| gene2228 | Contig25 | sgrA | surface protein from Gram-positive cocci, anchor region | 93.5 |
| gene1872 | Contig17 | bopD | sugar-binding transcriptional regulator, LacI family | 86.3 |
| gene2420 | Contig35 | bopD | sugar-binding transcriptional regulator, LacI family | 84.5 |
| gene2112 | Contig21 | clpP | ATP-dependent Clp protease proteolytic subunit | 81.1 |
| gene1467 | Contig11 | cpsA | undecaprenyl diphosphate synthase | 79.4 |
| gene1519 | Contig11 | bsh | bile salt hydrolase | 78.1 |
| gene0411 | Contig3 | acm | collagen adhesin precursor | 75.1 |
| gene0412 | Contig3 | acm | collagen adhesin precursor | 95.5 |
| Sex | Dose (mL/kg BW) | Test Animals (n) | Weight (X ± SD) (g) | Death of Animals (n) | Death Rate (%) | ||
|---|---|---|---|---|---|---|---|
| 0 Day | 7 Day | 14 Day | |||||
| Male | 20.0 | 6 | 22.01 ± 0.09 | 22.02 ± 0.06 | 21.93 ± 0.09 | 0 | 0 |
| Female | 20.0 | 6 | 21.98 ± 0.14 | 21.99 ± 0.07 | 22.10 ± 0.11 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Xu, Q.; Zhang, Y.; Yan, H. From Isolation to Pilot-Scale Production: Enterococcus faecium YC07 with Urate-Lowering Potential from Fermented Food Jiangshui. Foods 2025, 14, 2076. https://doi.org/10.3390/foods14122076
Cao X, Xu Q, Zhang Y, Yan H. From Isolation to Pilot-Scale Production: Enterococcus faecium YC07 with Urate-Lowering Potential from Fermented Food Jiangshui. Foods. 2025; 14(12):2076. https://doi.org/10.3390/foods14122076
Chicago/Turabian StyleCao, Xiaoyu, Qianqian Xu, Yu Zhang, and Hai Yan. 2025. "From Isolation to Pilot-Scale Production: Enterococcus faecium YC07 with Urate-Lowering Potential from Fermented Food Jiangshui" Foods 14, no. 12: 2076. https://doi.org/10.3390/foods14122076
APA StyleCao, X., Xu, Q., Zhang, Y., & Yan, H. (2025). From Isolation to Pilot-Scale Production: Enterococcus faecium YC07 with Urate-Lowering Potential from Fermented Food Jiangshui. Foods, 14(12), 2076. https://doi.org/10.3390/foods14122076







