CRISPR-Cas12a/Aurora Deoxyribozyme Cascade: A Label-Free Ultrasensitive Platform for Rapid Salmonella Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Bacterial Culture and Extraction
2.3. RPA Amplification and Validation
2.4. Label-Free Detection of Salmonella by Aurora-Based CRISPR/Cas12a Sensor
2.5. Preparation of Spiked Samples
3. Results and Discussion
3.1. Novel Strategy for Highly Sensitive Detection of Salmonella typhimurium Based on Synergistic Action of CRISPR-Cas12a and Aurora Fluorescence System
3.2. Comprehensive Validation of the CRISPR-Cas12a/Aurora-Based Detection System for Salmonella typhimurium
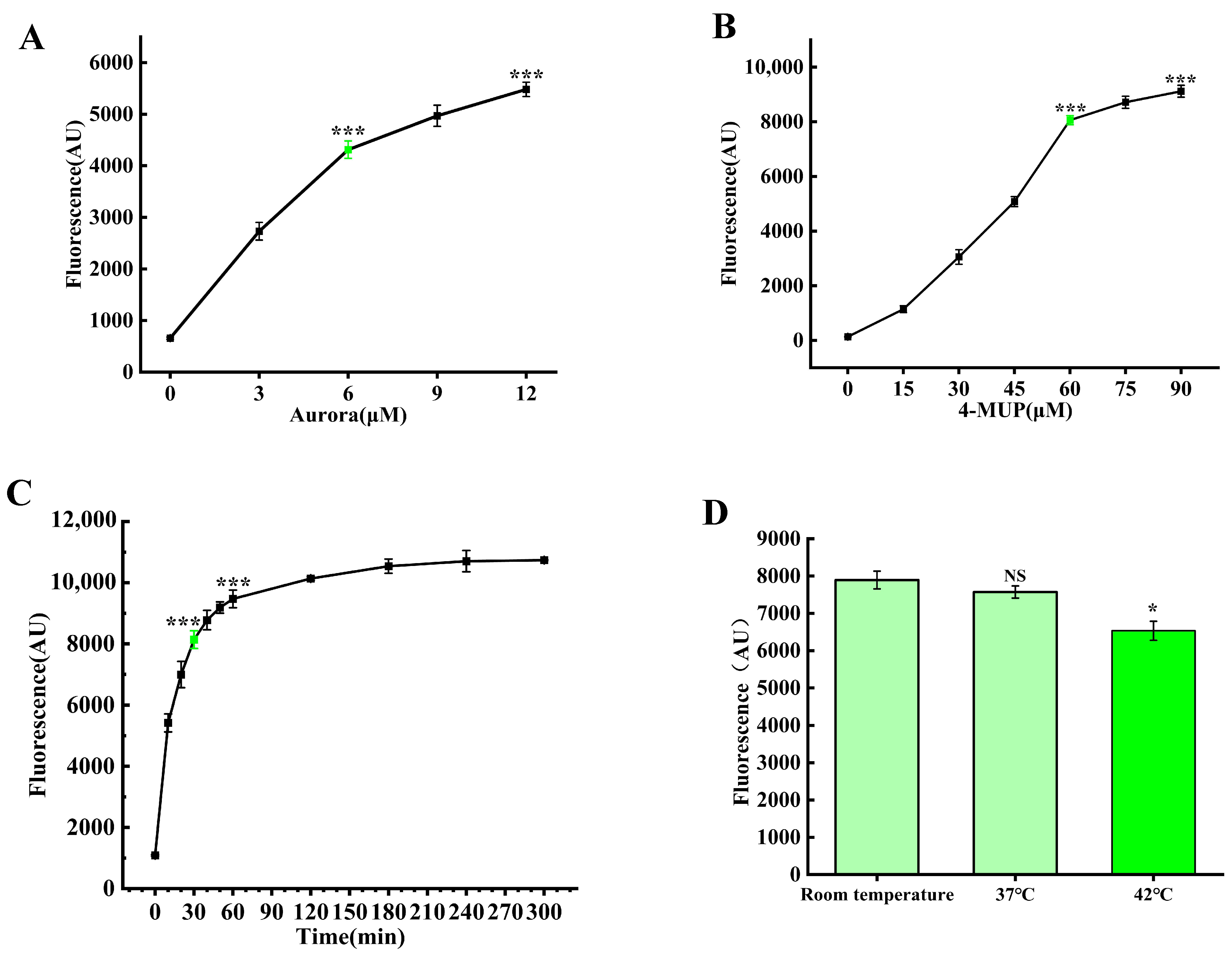
3.3. Optimization of Reaction Conditions
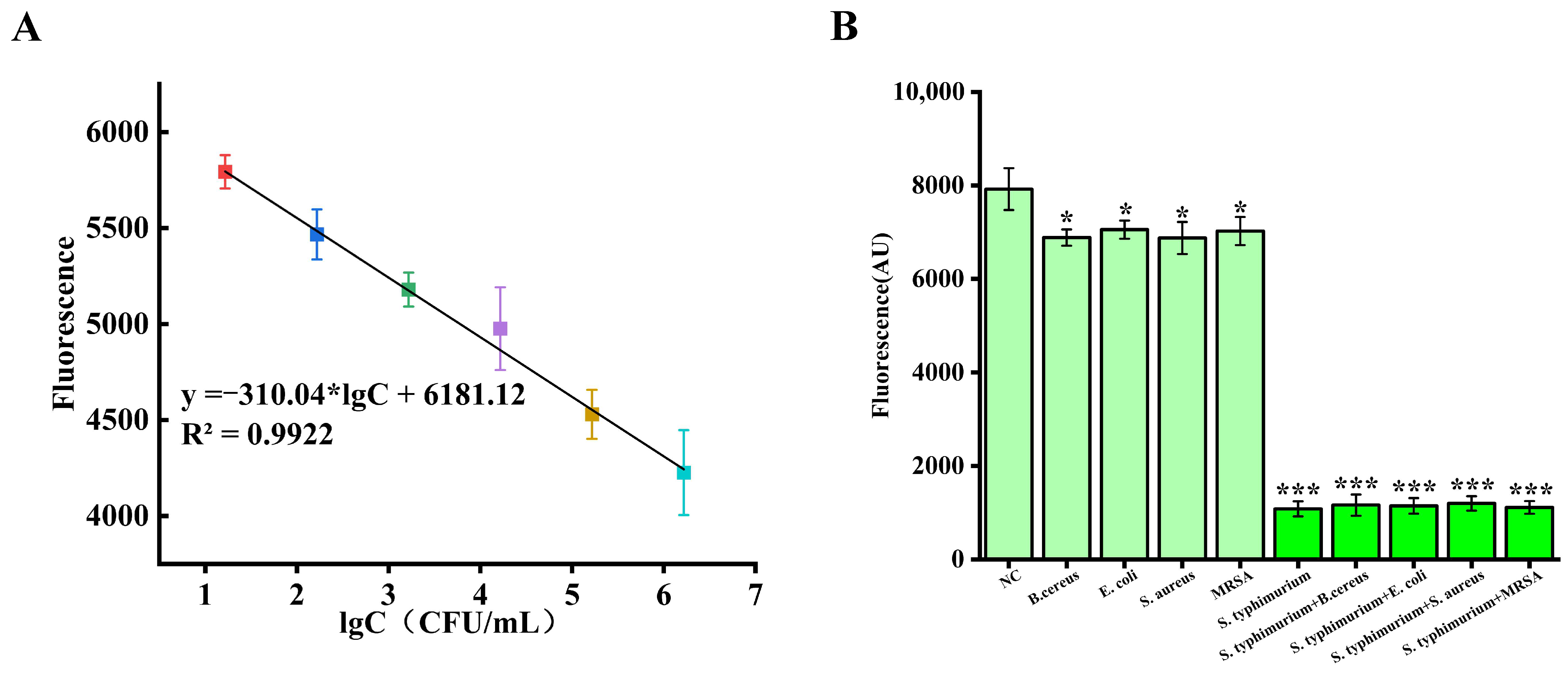
3.4. Label-Free Detection of Salmonella Using Aurora-Based CRISPR-Cas12a Sensors
3.5. Application in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maskery, B.; Ochiai, R.L.; Lee, J.S.; Mogasale, V.V.; Ramani, E.; Kim, Y.E.; Park, J.K.; Wierzba, T.F. Burden of typhoid fever in low-income and middle-income countries: A systematic, literature-based update with risk-factor adjustment. Lancet Glob. Health 2014, 2, e570–e580. [Google Scholar]
- Li, G.; Huang, Y.; Duan, M.; Xing, K.; You, X.; Zhou, H.; Liu, Y.; Liu, C.; Liu, D.; Lai, W. Biosensing multiplexer based on immunochromatographic assay for rapid and high-throughput classification of Salmonella serogroups. Sens. Actuators B Chem. 2019, 282, 317–321. [Google Scholar] [CrossRef]
- Xiang, X.; Shang, Y.; Ye, Q.; Li, F.; Zhang, J.; Zhou, B.; Suo, H.; Chen, M.; Gu, Q.; Ding, Y.; et al. A Salmonella serogroup rapid identification system for food safety based on high throughput microfluidic chip combined with recombinase aided amplification. Sens. Actuators B Chem. 2022, 357, 131402. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, G.; Li, X.; Xu, Z.; Lei, H.; Shen, X. Development of rapid and easy detection of Salmonella in food matrics using RPA-CRISPR/Cas12a method. LWT 2022, 162, 113443. [Google Scholar] [CrossRef]
- Duan, M.; Li, B.; Zhao, Y.; Liu, Y.; Liu, Y.; Dai, R.; Li, X.; Jia, F. A CRISPR/Cas12a-mediated, DNA extraction and amplification-free, highly direct and rapid biosensor for Salmonella Typhimurium. Biosens. Bioelectron. 2023, 219, 114823. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O'Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jia, F.; Sun, Y.; Fang, W.; Li, Y.; Chen, J.; Fu, Y. An electrochemical sensing method based on CRISPR/Cas12a system and hairpin DNA probe for rapid and sensitive detection of Salmonella Typhimurium. Sens. Actuators B Chem. 2022, 369, 132301. [Google Scholar] [CrossRef]
- Chen, L.; Hu, M.; Zhou, X. Trends in developing one-pot CRISPR diagnostics strategies. Trends Biotechnol. 2024, 43, 98–110. [Google Scholar] [CrossRef]
- Chen, B.; Li, Y.; Xu, F.; Yang, X. Powerful CRISPR-based biosensing techniques and their integration with microfluidic platforms. Front. Bioeng. Biotechnol. 2022, 10, 851712. [Google Scholar] [CrossRef]
- Liu, P.; Lin, Y.; Zhuo, X.; Zeng, J.; Chen, B.; Zou, Z.; Liu, G.; Xiong, E.; Yang, R. Universal crRNA Acylation Strategy for Robust Photo-Initiated One-Pot CRISPR–Cas12a Nucleic Acid Diagnostics. Angew. Chem. Int. Ed. 2024, 63, e202401486. [Google Scholar] [CrossRef]
- Qin, C.; Liu, J.; Zhu, W.; Zeng, M.; Xu, K.; Ding, J.; Zhou, H.; Zhu, J.; Ke, Y.; Li, L.Y.; et al. One-pot visual detection of african swine fever virus using CRISPR-Cas12a. Front. Vet. Sci. 2022, 9, 962438. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Macaluso, N.C.; Pizzano, B.L.; Cash, M.N.; Spacek, J.; Karasek, J.; Miller, M.R.; Lednicky, J.A.; Dinglasan, R.R.; Salemi, M.; et al. A thermostable Cas12b from Brevibacillus leverages one-pot discrimination of SARS-CoV-2 variants of concern. EBioMedicine 2022, 77, 103926. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Duan, J.-J.; Wei, X.-Y.; Hu, H.; Wang, Y.-B.; Jia, P.-P.; Pei, D.-S. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci. Total Environ. 2022, 838, 156048. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zong, N.; Ye, F.; Mei, Y.; Qu, J.; Jiang, X. Dual-CRISPR/Cas12a-assisted RT-RAA for ultrasensitive SARS-CoV-2 detection on automated centrifugal microfluidics. Anal. Chem. 2022, 94, 9603–9609. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, S.; Xu, Y.; Li, A.; Wu, W.; Liang, M.; Niu, G.; Wang, Z.; Wang, T. CRISPR/Cas12a technology combined with RPA for rapid and portable SFTSV detection. Front. Microbiol. 2022, 13, 754995. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The biology of CRISPR-Cas: Backward and forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef]
- Smith, C.W. Development of Nucleic Acid Diagnostics for Targeted and Non-Targeted Biosensing. Ph.D. Thesis, State University of New York, Albany, NY, USA, 2022. [Google Scholar]
- Yang, H.; Gao, P.; Rajashankar, K.R.; Patel, D.J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 2016, 167, 1814–1828. [Google Scholar] [CrossRef]
- Sohail, M.; Xie, S.; Zhang, X.; Li, B. Methodologies in visualizing the activation of CRISPR/Cas: The last mile in developing CRISPR-Based diagnostics and biosensing–a review. Anal. Chim. Acta 2022, 1205, 339541. [Google Scholar] [CrossRef]
- Yan, F.; Wang, W.; Zhang, J. CRISPR-Cas12 and Cas13: The lesser known siblings of CRISPR-Cas9. Cell Biol. Toxicol. 2019, 35, 489–492. [Google Scholar] [CrossRef]
- Volek, M.; Kurfürst, J.; Drexler, M.; Svoboda, M.; Srb, P.; Veverka, V.; A Curtis, E. Aurora: A fluorescent deoxyribozyme for high-throughput screening. Nucleic Acids Res. 2024, 52, 9049–9061. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, C. Designing in vivo active DNAzymes. Nat. Chem. 2021, 13, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Sintim, H.O. Biomolecule detection with peroxidase-mimicking DNAzymes; expanding detection modality with fluorogenic compounds. Mol. Biosyst. 2009, 6, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, Q.; Chen, J.; Huang, L.; Zhang, H.; Rong, K.; Dong, S. Biomimetic design for enhancing the peroxidase mimicking activity of hemin. Nanoscale 2019, 11, 12603–12609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Y.; Wu, Y.; Chang, Y.; Liu, M. CRISPR-Cas systems: From gene scissors to programmable biosensors. TrAC Trends Anal. Chem. 2021, 137, 116210. [Google Scholar] [CrossRef]
- Jing, S.; Liu, Q.; Jin, Y.; Li, B. Dimeric G-quadruplex: An effective nucleic acid scaffold for lighting up thioflavin T. Anal. Chem. 2020, 93, 1333–1341. [Google Scholar] [CrossRef]
- Zheng, Q.; Mikš-Krajnik, M.; Yang, Y.; Xu, W.; Yuk, H.-G. Real-time PCR method combined with immunomagnetic separation for detecting healthy and heat-injured Salmonella Typhimurium on raw duck wings. Int. J. Food Microbiol. 2014, 186, 6–13. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Pan, D.; Li, J.; Song, S.; Rong, M.; Li, Z.; Gao, J.; Lu, J. Gold nanoparticle-based enzyme-linked antibody-aptamer sandwich assay for detection of Salmonella Typhimurium. ACS Appl. Mater. Interfaces 2014, 6, 16974–16981. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Q.; Zhou, M.; Li, C.; Yan, C.; Huang, L.; Qin, P. Development of a CRISPR/Cas9-integrated lateral flow strip for rapid and accurate detection of Salmonella. Food Control 2022, 142, 109203. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, K.; Yin, H.; Li, Q.; Wang, L.; Liu, Z. Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification method. Food Sci. Hum. Wellness 2015, 4, 75–79. [Google Scholar] [CrossRef]
- Jia, F.; Li, B.; He, Y.; Shen, Y.; Chen, J.; Li, X.; Li, Y. An amplification-free CRISPR-SERS biosensor for specific, sensitive and rapid detection of Salmonella Typhimurium in poultry. LWT 2023, 189, 115476. [Google Scholar] [CrossRef]
- Yin, L.; Duan, N.; Chen, S.; Yao, Y.; Liu, J.; Ma, L. Ultrasensitive pathogenic bacteria detection by a smartphone-read G-quadruplex-based CRISPR-Cas12a bioassay. Sens. Actuators B Chem. 2021, 347, 130586. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, R.; Qiao, Z.; Yang, W. Single-digit Salmonella detection with the naked eye using bio-barcode immunoassay coupled with recombinase polymerase amplification and a CRISPR-Cas12a system. Analyst 2021, 146, 5271–5279. [Google Scholar] [CrossRef] [PubMed]


| Method | LOD (CFU/mL) | DNA Amplification | Time | Samples | References |
|---|---|---|---|---|---|
| PCR | 1 × 103 | PCR | 10 h | Raw duck wing | [28] |
| ELISA | 1 × 103 | _ | <3 h | Milk | [29] |
| Traditional CRISPR | 1×102 | PCR | <100 min | Milk | [30] |
| LAMP | 2 × 101 | LAMP | 45–70 min | Pork | [31] |
| Electrochemical CRISPR | 5.5 × 101 | PCR | <2.5 h | Chicken | [7] |
| CRISPR-SERS biosensor | 1.1 × 102 | - | 2 h | Chicken | [32] |
| smartphone-read G-quadruplex-based CRISPR-Cas12a bioassay | 1 | RPA | 3 h | Bear and Juice | [33] |
| BCA-RPA-Cas 12 a | 1 | RPA | 60 min | Milk | [34] |
| This work | 1.29 | RPA | 100 min | Chicken, Milk, Vegetable | This work |
| Samples | C(S. typhimurium) (CFU/mL) | Results (CFU/mL) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Milk | 0 | ND a | — | — |
| Milk | 1.65 × 106 | 1.57 × 106 | 95.15 | 3.55 |
| Milk | 1.65 × 105 | 1.63 × 105 | 98.79 | 3.59 |
| Chicken | 0 | ND | — | — |
| Chicken | 1.65 × 106 | 1.49 × 106 | 93.74 | 3.60 |
| Chicken | 1.65 × 105 | 1.59 × 105 | 99.60 | 4.14 |
| Lettuce | 0 | ND | — | — |
| Lettuce | 1.65 × 106 | 1.50 × 106 | 90.91 | 4.72 |
| Lettuce | 1.65 × 105 | 1.64 × 105 | 99.40 | 3.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Tan, H.; Yu, Z.; Li, W.; Man, Y.; Zhang, Q. CRISPR-Cas12a/Aurora Deoxyribozyme Cascade: A Label-Free Ultrasensitive Platform for Rapid Salmonella Detection. Foods 2025, 14, 1892. https://doi.org/10.3390/foods14111892
Shi C, Tan H, Yu Z, Li W, Man Y, Zhang Q. CRISPR-Cas12a/Aurora Deoxyribozyme Cascade: A Label-Free Ultrasensitive Platform for Rapid Salmonella Detection. Foods. 2025; 14(11):1892. https://doi.org/10.3390/foods14111892
Chicago/Turabian StyleShi, Cong, Huimin Tan, Zhou Yu, Weilin Li, Yan Man, and Qinghai Zhang. 2025. "CRISPR-Cas12a/Aurora Deoxyribozyme Cascade: A Label-Free Ultrasensitive Platform for Rapid Salmonella Detection" Foods 14, no. 11: 1892. https://doi.org/10.3390/foods14111892
APA StyleShi, C., Tan, H., Yu, Z., Li, W., Man, Y., & Zhang, Q. (2025). CRISPR-Cas12a/Aurora Deoxyribozyme Cascade: A Label-Free Ultrasensitive Platform for Rapid Salmonella Detection. Foods, 14(11), 1892. https://doi.org/10.3390/foods14111892






