Bioinformatics-Assisted Discovery of Antioxidant Cyclic Peptides from Corn Gluten Meal
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. In Silico Digestion of Protein of CGM and Enzyme Selection
2.3. Pretreatment of CGM
2.4. Hydrolysis of Protein of CGM and Cyclization of Linear Peptide Precursors
2.5. Determination of Antioxidant Activity
2.5.1. DPPH Radical Scavenging Activity
2.5.2. ABTS Radical Scavenging Activity
2.5.3. Hydroxyl Radical Scavenging Activity
2.6. Qualitative and Quantitative TLC Analyses of Cyclic Peptides
2.7. Separation and Purification of the Antioxidant Cyclic Peptide from Protein Hydrolysate of CGM
2.7.1. Silica Gel Column Chromatography
2.7.2. Semi-Preparative Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.8. Structural Characterization of Cyclic Peptides
2.8.1. UPLC/MS and MS/MS Analysis
2.8.2. Detection of the Cyclization Sites of Cyclic Peptides
2.9. Molecular Docking Analysis
2.10. Data Statistical Analysis
3. Results
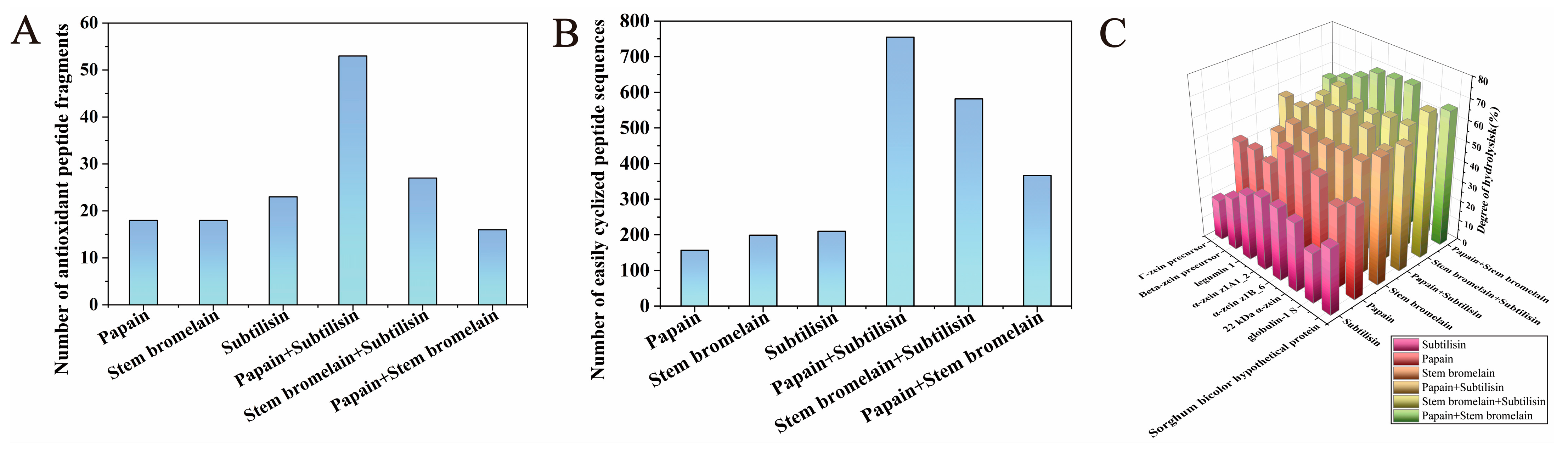
3.1. Selection of Enzyme or Enzyme Combination Using a Bioinformatics Approach
3.2. Purification and Identification of Antioxidant Cyclic Peptide from Hydrolysate
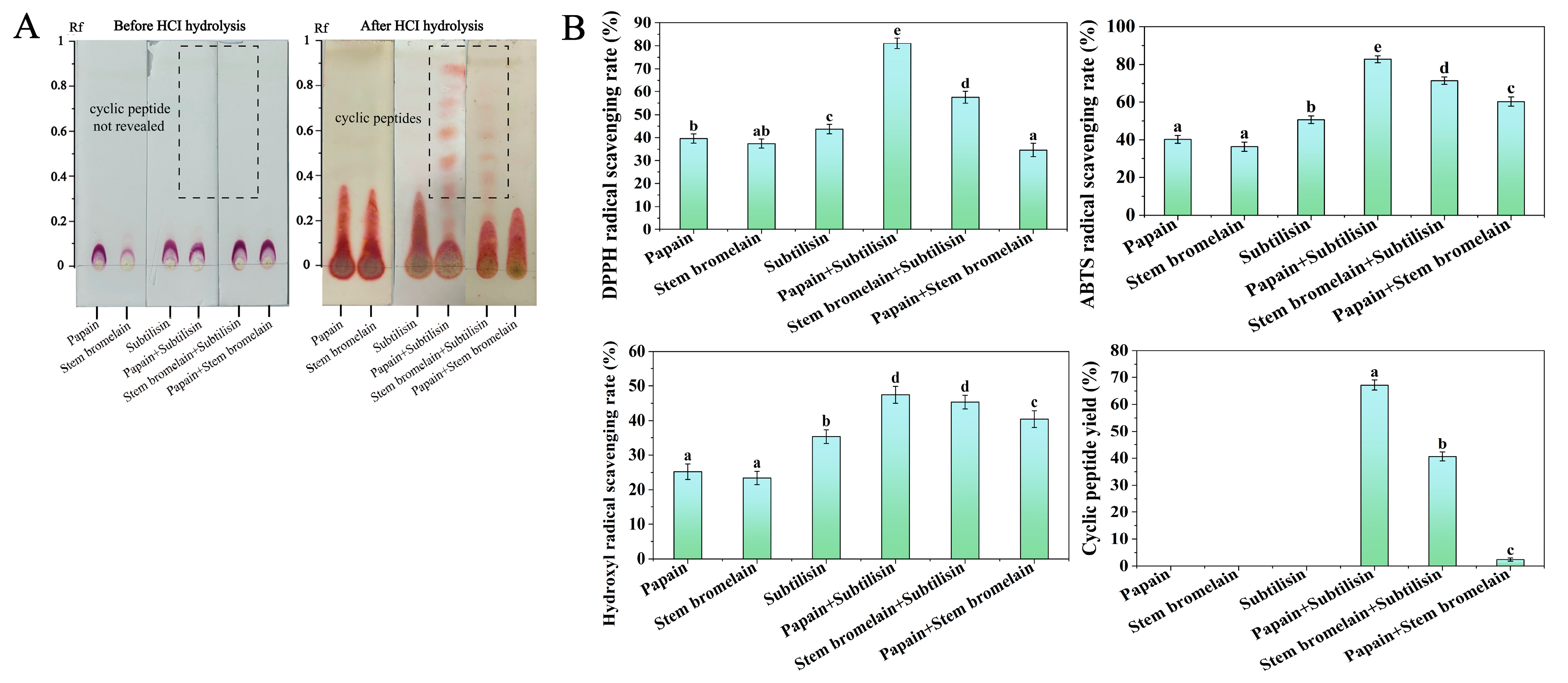
3.2.1. Silica Gel Column Separation and Purification
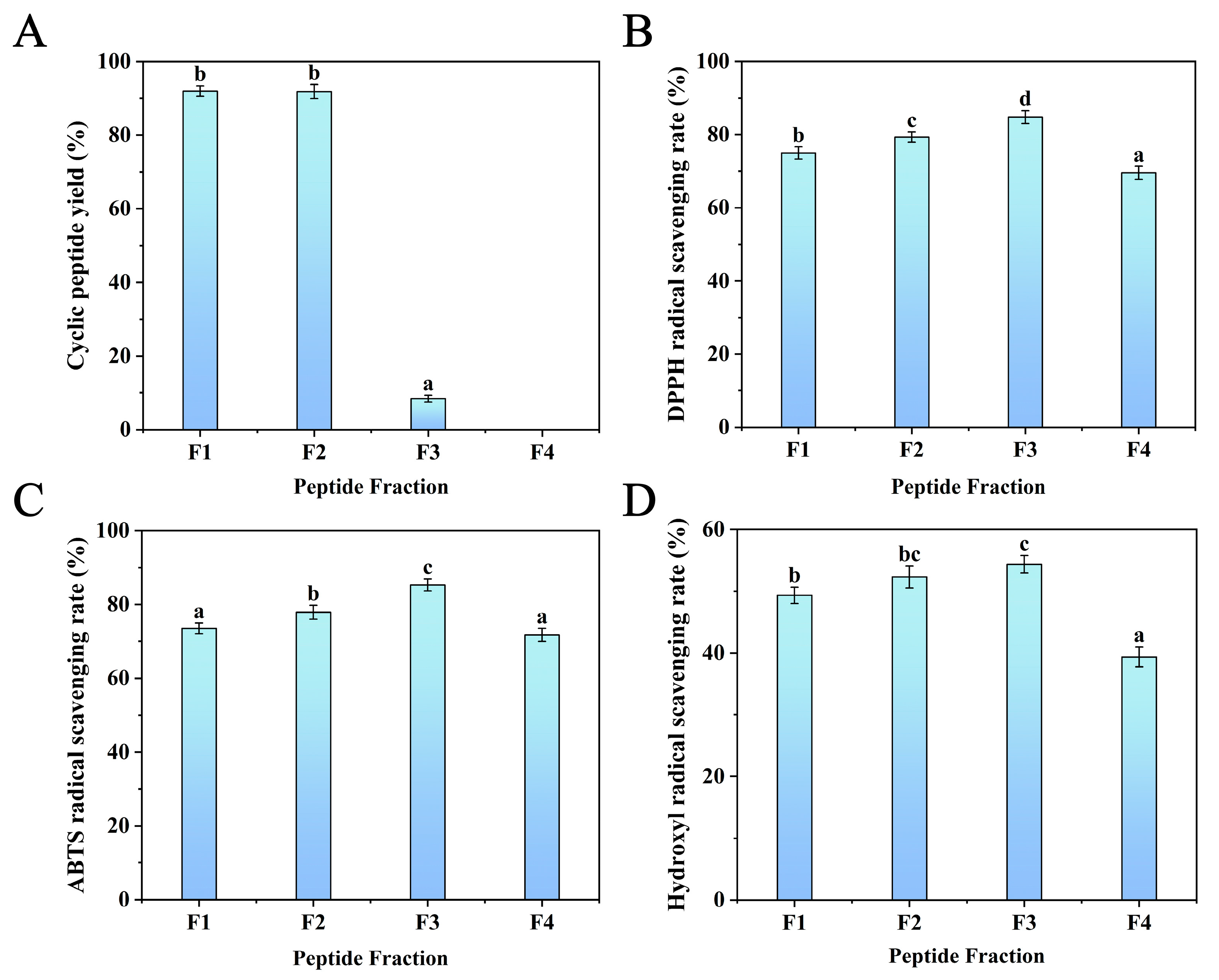
3.2.2. Separation and Purification by Semi-Preparative RP-HPLC
3.2.3. Sequence Identification of Antioxidant Cyclic Peptides Using UPLC/MS and MS/MS
3.2.4. Identification of the Cyclization Sites of the Antioxidant Cyclic Peptide
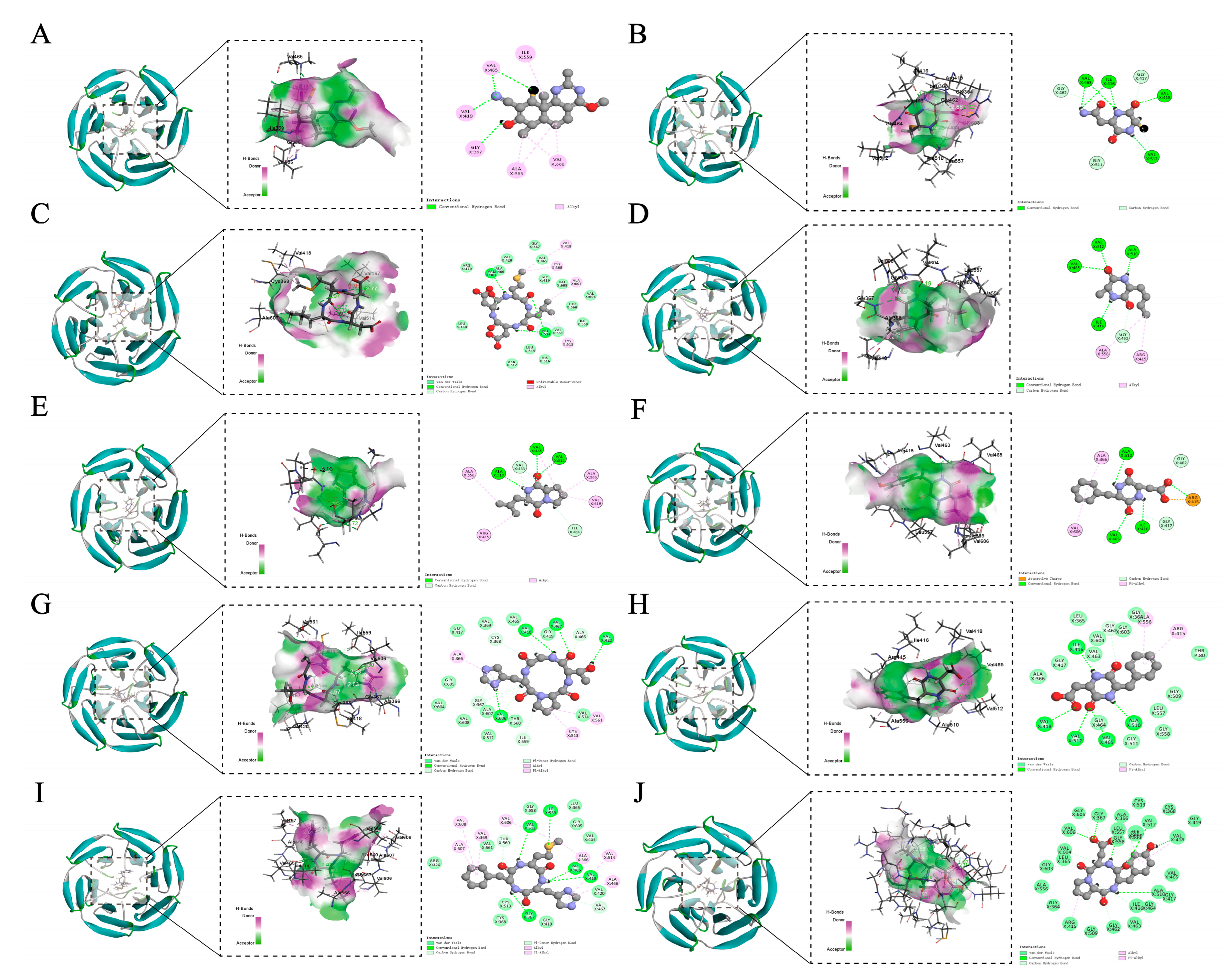
3.3. Molecular Docking with Keap1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CGM | Corn gluten meal |
| Keap1 | Kelch-like ECH-associated protein 1 |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
References
- Lv, R.; Dong, Y.; Bao, Z.; Zhang, S.; Lin, S.; Sun, N. Advances in the activity evaluation and cellular regulation pathways of food-derived antioxidant peptides. Trends Food Sci. Technol. 2022, 122, 171–186. [Google Scholar] [CrossRef]
- Fan, H.; Liu, H.; Zhang, Y.; Zhang, S.; Liu, T.; Wang, D. Review on plant-derived bioactive peptides: Biological activities, mechanism of action and utilizations in food development. J. Future Foods 2022, 2, 143–159. [Google Scholar] [CrossRef]
- Wang, C.K.; Craik, D.J. Cyclic peptide oral bioavailability: Lessons from the past. Biopolymers 2016, 106, 901–909. [Google Scholar] [CrossRef]
- Hayes, H.C.; Luk, L.Y.P.; Tsai, Y.H. Approaches for peptide and protein cyclisation. Org. Biomol. Chem. 2021, 19, 3983–4001. [Google Scholar] [CrossRef] [PubMed]
- Prasad, C. Bioactive cyclic dipeptides. Peptides 1995, 16, 151–164. [Google Scholar] [CrossRef]
- Routhu, S.R.; Ragi, N.C.; Yedla, P.; Shaik, A.B.; Venkataraman, G.; Cheemalamarri, C.; Chityala, G.K.; Amanchy, R.; Sripadi, P.; Kamal, A. Identification, characterization and evaluation of novel antifungal cyclic peptides from Neobacillus drentensis. Bioorg Chem. 2021, 115, 105180. [Google Scholar] [CrossRef]
- Zhao, J.; Gong, L.; Wu, L.; She, S.; Liao, Y.; Zheng, H.; Zhao, Z.; Liu, G.; Yan, S. Immunomodulatory effects of fermented fig (Ficus carica L.) fruit extracts on cyclophosphamide-treated mice. J. Funct. Foods 2020, 75, 104219. [Google Scholar] [CrossRef]
- Fan, H.; Liu, H.; Dong, X.; Zhang, Y. Separation, Purification, Structural Identification and Bioactivities of Corn Cyclo (His-Pro). Food Sci. 2016, 37, 53–60. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Wang, Q.; Miao, Z.; Zheng, X. Anti-adhesive activity peptides against Helicobacter pylori from corn protein: Properties, identification, structural characterization and molecular docking in vitro and in silico. Food Biosci. 2024, 59, 104267. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Fan, X.; Zhang, T.; Wang, H.; Ye, K. Novel antioxidant peptides from bovine blood: Purification, identification and mechanism of action. LWT 2024, 205, 116499. [Google Scholar] [CrossRef]
- Chi, Z.; Feng, Y.; Wei, X.; Yang, H.; Fang, X.; Cheng, B.; Li, Z.; Shi, B.; Gu, Z. A co-fermentation technology based on small peptide production and protein structure modification increases the edible value of corn gluten meal. LWT 2024, 198, 116054. [Google Scholar] [CrossRef]
- Piazza, S.; Bani, C.; Colombo, F.; Mercogliano, F.; Pozzoli, C.; Martinelli, G.; Petroni, K.; Roberto Pilu, S.; Sonzogni, E.; Fumagalli, M.; et al. Pigmented corn as a gluten-free source of polyphenols with anti-inflammatory and antioxidant properties in CaCo-2 cells. Food Res. Int. 2024, 191, 114640. [Google Scholar] [CrossRef] [PubMed]
- Jakas, A.; Vlahovicek-Kahlina, K.; Ljolic-Bilic, V.; Horvat, L.; Kosalec, I. Design and synthesis of novel antimicrobial peptide scaffolds. Bioorg Chem. 2020, 103, 104178. [Google Scholar] [CrossRef]
- Kurnianto, M.A.; Defri, I.; Syahbanu, F.; Aulia, S.S. Fish-derived bioactive peptide: Bioactivity potency, structural characteristics, and conventional and bioinformatics approaches for identification. Future Foods 2024, 9, 100386. [Google Scholar] [CrossRef]
- Peredo-Lovillo, A.; Hernandez-Mendoza, A.; Vallejo-Cordoba, B.; Romero-Luna, H.E. Conventional and in silico approaches to select promising food-derived bioactive peptides: A review. Food Chem. X 2022, 13, 100183. [Google Scholar] [CrossRef] [PubMed]
- Kandemir-Cavas, C.; Perez-Sanchez, H.; Mert-Ozupek, N.; Cavas, L. In Silico Analysis of Bioactive Peptides in Invasive Sea Grass Halophila stipulacea. Cells 2019, 8, 557. [Google Scholar] [CrossRef]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef]
- Filiz, E.; Kurt, F. Antimicrobial peptides Snakin/GASA gene family in sorghum (Sorghum bicolor): Genome-wide identification and bioinformatics analyses. Gene Rep. 2020, 20, 100766. [Google Scholar] [CrossRef]
- Lin, H.; Zhao, J.; Xie, Y.; Tang, J.; Wang, Q.; Zhao, J.; Xu, M.; Liu, P. Identification and molecular mechanisms of novel antioxidant peptides from fermented broad bean paste: A combined in silico and in vitro study. Food Chem. 2024, 450, 139297. [Google Scholar] [CrossRef]
- Ngoh, Y.Y.; Gan, C.Y. Identification of Pinto bean peptides with inhibitory effects on alpha-amylase and angiotensin converting enzyme (ACE) activities using an integrated bioinformatics-assisted approach. Food Chem. 2018, 267, 124–131. [Google Scholar] [CrossRef]
- Pan, C.; Ma, J.; Tao, F.; Ji, C.; Zhao, Y.; Chen, S.; Yang, X. Novel insight into the antioxidant proteins derived from laver (Porphyra haitanensis) by proteomics analysis and protein based bioinformatics. Food Biosci. 2021, 42, 101134. [Google Scholar] [CrossRef]
- Dharmisthaben, P.; Basaiawmoit, B.; Sakure, A.; Das, S.; Maurya, R.; Bishnoi, M.; Kondepudi, K.K.; Hati, S. Exploring potentials of antioxidative, anti-inflammatory activities and production of bioactive peptides in lactic fermented camel milk. Food Biosci. 2021, 44, 101404. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides exhibit antioxidant activity through the Keap1-Nrf2 signaling pathway. J. Funct. Foods 2020, 64, 103696. [Google Scholar] [CrossRef]
- Liu, W.; Liu, R.; Qin, Q.; Wang, H.; Zhang, X.; Meng, G. Molecular docking and molecular dynamics simulation of wheat gluten-derived antioxidant peptides acting through the Keap1–Nrf2 pathway. J. Sci. Food Agric. 2024, 104, 8150–8161. [Google Scholar] [CrossRef] [PubMed]
- Tonolo, F.; Moretto, L.; Grinzato, A.; Fiorese, F.; Folda, A.; Scalcon, V.; Ferro, S.; Arrigoni, G.; Bellamio, M.; Feller, E.; et al. Fermented soy-derived bioactive peptides selected by a molecular docking approach show antioxidant properties involving the Keap1/Nrf2 pathway. Antioxidants 2020, 9, 1306. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, E.; Martínez-Villaluenga, C.; Paterson, S.; Hernández-Ledesma, B.; Gutiérrez, L.-F. Antidiabetic and Immunomodulatory Properties of Peptide Fractions from Sacha Inchi Oil Press-Cake. Foods 2025, 14, 1231. [Google Scholar] [CrossRef]
- Tang, Y.; Tian, G.; Ye, Y. Research progress on the synthesis methods of cyclic peptides. Chem. J. Chin. Univ. 2000, 21, 1056–1063, (in Chinese with English abstract). [Google Scholar] [CrossRef]
- Liu, H.; Fan, H.; Teng, X.; Sun, T.; Zhang, S.; Wang, N.; Zhang, X.; Liu, T.; Zhang, Y.; Wang, D. Exploring novel antioxidant cyclic peptides in corn protein hydrolysate: Preparation, identification and molecular docking analysis. Food Chem. 2025, 464 Pt 2, 141747. [Google Scholar] [CrossRef]
- Wijethunga, A.M.; Wang, H.; Sun, X. Starch removal treatment, not the specific thermal processing techniques, improved protein digestibility of corn (Zea mays) gluten meal. Food Hydrocoll. 2024, 149, 109619. [Google Scholar] [CrossRef]
- Koo, K.B.; Suh, H.J.; Ra, K.S.; Choi, J.W. Protective effect of cyclo(his-pro) on streptozotocin-induced cytotoxicity and apoptosis in vitro. J. Microbiol. Biotechnol. 2011, 21, 218–227. [Google Scholar] [CrossRef]
- Wang, J.; Li, X. Complex cyclic peptide synthesis via serine/threonine ligation chemistry. Bioorg Med. Chem. Lett. 2021, 54, 128430. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Ohmura, R.; Toshima, S.; Park, H.; Narasako, Y.; Hirano, T.; Otani, M.; Kunitake, H. Changes in polyphenols, anthocyanins, and DPPH radical-scavenging activities in sweetpotato (Ipomoea batatas L.) during tuber growth. Sci. Hortic. 2021, 284, 110100. [Google Scholar] [CrossRef]
- Wang, J.; Yang, G.; Li, H.; Zhang, T.; Sun, D.; Peng Lu, W.; Zhang, W.; Wang, Y.; Ma, M.; Cao, X.; et al. Preparation and identification of novel antioxidant peptides from camel bone protein. Food Chem. 2023, 424, 136253. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, S.; Rong, Y.; Zeng, W.; Hu, Z.; Ma, X.; Feng, S. Identification and evaluation of antioxidant peptides from highland barley distiller’s grains protein hydrolysate assisted by molecular docking. Food Chem. 2024, 434, 137441. [Google Scholar] [CrossRef]
- Galarce-Bustos, O.; Pavon-Perez, J.; Henriquez-Aedo, K.; Aranda, M. An improved method for a fast screening of alpha-glucosidase inhibitors in cherimoya fruit (Annona cherimola Mill.) applying effect-directed analysis via high-performance thin-layer chromatography-bioassay-mass spectrometry. J. Chromatogr. A 2019, 1608, 460415. [Google Scholar] [CrossRef]
- Sefler, A.M.; Lauri, G.; Bartlett, P.A. A convenient method for determining cyclic peptide conformation from 1D 1H-NMR information. Int. J. Pept. Protein Res. 1996, 48, 129–138. [Google Scholar] [CrossRef]
- Cai, L.; Wu, S.; Jia, C.; Cui, C.; Sun-Waterhouse, D. Active peptides with hypoglycemic effect obtained from hemp (Cannabis sativa L) protein through identification, molecular docking, and virtual screening. Food Chem. 2023, 429, 136912. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Z.; Xu, M.; Zhang, X.; Cao, S.; Hu, Y.; Luan, G. Physicochemical properties and protein structure of extruded corn gluten meal: Implication of temperature. Food Chem. 2023, 399, 133985. [Google Scholar] [CrossRef]
- Wu, D.; Cao, Y.; Su, D.; Karrar, E.; Zhang, L.; Chen, C.; Deng, N.; Zhang, Z.; Liu, J.; Li, G. Preparation and identification of antioxidant peptides from Quasipaa spinosa skin through two-step enzymatic hydrolysis and molecular simulation. Food Chem. 2024, 445, 138801. [Google Scholar] [CrossRef]
- Cui, Z.; Yu, Y.; Zhou, T.; Qi, C.; Gu, J.; Zhang, N.; Feng, X.; Zhang, Z.; Zhu, Y.; Zhang, Y.; et al. Cyclization: A potential effective modification strategy for umami peptides. Food Chem. 2024, 469, 142457. [Google Scholar] [CrossRef]
- Liu, H.; Bai, L.; Jiang, X. Recent progress on total synthesis of cyclic peptides. Tetrahedron Lett. 2024, 151, 155314. [Google Scholar] [CrossRef]
- Joo, S.H. Cyclic peptides as therapeutic agents and biochemical tools. Biomol. Ther. 2012, 20, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, H.; Wang, L.; Guo, X.; Wang, X.; Yao, H. Isolation and identification of antioxidative peptides from rice endosperm protein enzymatic hydrolysate by consecutive chromatography and MALDI-TOF/TOF MS/MS. Food Chem. 2010, 119, 226–234. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, J.; Chen, Y.; Sun, H.; Liu, Y. Preparation and characterization of lipophilic antioxidative peptides derived from mung bean protein. Food Chem. 2022, 395, 133535. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Chelliah, R.; Banan-Mwine Daliri, E.; Sultan, G.; Madar, I.H.; Kim, N.; Shabbir, U.; Oh, D.H. Antioxidant activities of novel peptides from Limosilactobacillus reuteri fermented brown rice: A combined in vitro and in silico study. Food Chem. 2023, 404 Pt B, 134747. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Mohandas, C.; Nambisan, B. Purification of an antifungal compound, cyclo(l-Pro-d-Leu) for cereals produced by Bacillus cereus subsp. thuringiensis associated with entomopathogenic nematode. Microbiol. Res. 2013, 168, 278–288. [Google Scholar] [CrossRef]
- Conrad, U.; Taichrib, A.; Neususs, C.; Scriba, G.K. Capillary electrophoresis analysis of the degradation of the aspartyl tripeptide Phe-Asp-GlyOH at pH 2.0 and 7.4 under forced conditions. J. Pharm. Biomed. Anal. 2010, 51, 640–648. [Google Scholar] [CrossRef]
- Doucet, D.; Otter, D.E.; Gauthier, S.F.; Foegeding, E.A. Enzyme-induced gelation of extensively hydrolyzed whey proteins by Alcalase: Peptide identification and determination of enzyme specificity. J. Agric. Food Chem. 2003, 51, 6300–6308. [Google Scholar] [CrossRef]
- Iegre, J.; Krajcovicova, S.; Gunnarsson, A.; Wissler, L.; Kack, H.; Luchniak, A.; Tangefjord, S.; Narjes, F.; Spring, D.R. A cell-active cyclic peptide targeting the Nrf2/Keap1 protein-protein interaction. Chem. Sci. 2023, 14, 10800–10805. [Google Scholar] [CrossRef]
- Huerta, C.; Jiang, X.; Trevino, I.; Bender, C.F.; Ferguson, D.A.; Probst, B.; Swinger, K.K.; Stoll, V.S.; Thomas, P.J.; Dulubova, I.; et al. Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding. Biochim. Biophys. Acta 2016, 1860, 2537–2552. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.L.; Shen, L.H.; Feng, L.J.; Zhou, Q. Inhibition mechanism of diacylated anthocyanins from purple sweet potato (Ipomoea batatas L.) against alpha-amylase and alpha-glucosidase. Food Chem. 2021, 359, 129934. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | Enzyme Addition (U/g) | pH | Time (h) | Temperature (℃) |
|---|---|---|---|---|
| Single-enzyme hydrolysis | ||||
| Papain | 5000 | 6.5 | 8 | 65 |
| Stem bromelain | 5000 | 6.8 | 8 | 50 |
| Subtilisin | 5000 | 7.5 | 8 | 60 |
| Compound-enzyme hydrolysis (added in a 1:1 ratio) | ||||
| Papain+subtilisin | 2500 + 2500 | 7.0 | 8 | 63 |
| Stem bromelain+subtilisin | 2500 + 2500 | 7.2 | 8 | 55 |
| Papain+stem bromelain | 2500 + 2500 | 6.7 | 8 | 58 |
| Fraction | Peak No. | RT (min) | Cyclic Peptide | Parent Ions ([M+H]+) m/z | Theoretical Mw (Da) | MS/MS |
|---|---|---|---|---|---|---|
| F1a | 1 | 2.295 | Cyclo(Asp-Leu-Asn-Met) | 474.25 | 473.56 | 360.17, 343.16, 247.14, 132.18 |
| 2 | 3.012 | Cyclo(Ala-Met) | 203.14 | 202.29 | 131.00 | |
| 3 | 3.634 | Cyclo(Gln-Leu-Asp-Leu-Ser-Thr-Lys) | 786.45 | 785.91 | 673.33, 657.82, 600.73, 571.47, 557.46, 471.75, 444.90, 343.35, 315.51 | |
| 4 | 4.170 | Cyclo(Asp-Phe) | 263.24 | 262.28 | 147.70 | |
| 4 | 4.170 | Cyclo(Pro-Asp-Tyr) | 376.22 | 375.40 | 263.14, 166.09 | |
| 5 | 5.527 | Cyclo(Asn-Pro-Ala-Pro-Asn-Cys-Thr) | 698.84 | 697.78 | 681.24, 663.30, 641.37, 584.52, 560.12, 470.39, 431.03, 331.12, 240.08 | |
| 6 | 18.868 | Cyclo(Cys-Asn) | 218.21 | 217.26 | 103.96 | |
| 7 | 19.963 | Cyclo(Val-Tyr) | 263.24 | 262.32 | 99.09 | |
| 8 | 24.458 | Cyclo(Met-His-Phe) | 416.21 | 415.53 | 285.11, 147.72 | |
| F2a | 1 | 2.994 | Cyclo(Leu-Pro) | 211.14 | 210.29 | 113.87 |
| 1 | 2.994 | Cyclo(Pro-His-Thr) | 336.32 | 335.38 | 239.01, 137.99 | |
| 2 | 9.906 | Cyclo(Cys-Asn) | 218.21 | 217.26 | 103.96 | |
| 3 | 12.335 | Cyclo(Val-Tyr) | 263.24 | 262.32 | 99.09 |
| Sequence of Linear Peptide Precursors | Molecular Weight (Da) | Sequence of Cyclic Peptide | Molecular Weight (Da) | Cyclization Site |
|---|---|---|---|---|
| Asn-Leu-Asp-Met | 493.84 | Cyclo(Asp-Leu-Asn-Met) | 474.25 | Asn, Met |
| Ala-Met | 221.74 | Cyclo(Ala-Met) | 203.14 | Ala, Met |
| Ser-Thr-Lys-Gln-Leu-Asp-Leu | 804.12 | Cyclo(Gln-Leu-Asp-Leu-Ser-Thr-Lys) | 786.45 | Ser, Leu |
| Asp-Phe | 281.87 | Cyclo(Asp-Phe) | 263.24 | Asp, Phe |
| Pro-Asp-Tyr | 394.82 | Cyclo(Pro-Asp-Tyr) | 376.22 | Pro, Tyr |
| Ala-Pro-Asn-Cys-Thr-Asn-Pro | 716.99 | Cyclo(Asn-Pro-Ala-Pro-Asn-Cys-Thr) | 698.84 | Ala, Pro |
| Cys-Asn | 236.69 | Cyclo(Cys-Asn) | 218.21 | Cys, Asn |
| Val-Tyr | 281.79 | Cyclo(Val-Tyr) | 263.24 | Val, Tyr |
| His-Phe-Met | 434.63 | Cyclo(Met-His-Phe) | 416.21 | His, Met |
| Leu-Pro | 229.72 | Cyclo(Leu-Pro) | 211.14 | Leu, Pro |
| Pro-His-Thr | 354.79 | Cyclo(Pro-His-Thr) | 336.32 | Pro, Thr |
| No. | Ligand | IECD (kcal/mol) | Hydrogen Bond | Hydrophobic Bond | ||
|---|---|---|---|---|---|---|
| Number | Amino Acid Residues | Number | Amino Acid Residues | |||
| 1 | TX6 | −29.7613 | 4 | Val465, Val418, Gly367 | 5 | Ile559, Ala366, Val606 |
| 2 | Cyclo(Cys-Asn) | −75.6207 | 10 | Gly462, Val463, Ile416, Gly417, Val418, Val512, Gly511 | – | – |
| 3 | Cyclo(Asn-Leu-Asp-Met) | −79.0846 | 4 | Val467, Val514 | 4 | Val418, Cys368, Ala607, Cys513 |
| 4 | Cyclo(Ala-Met) | −79.3505 | 5 | Val465, Val512, Ala510, Ile416, Gly462 | 2 | Ala556, Arg415 |
| 5 | Cyclo(Leu-Pro) | −84.7523 | 6 | Ala510, Val463, Val465, Val512, Ile416 | 5 | Ala556, Arg415, Val418, Ala366 |
| 6 | Cyclo(Val-Tyr) | −107.268 | 6 | Ala510, Gly462, Gly417, Ile416, Val465 | 2 | Ala366, Val606 |
| 7 | Cyclo(Pro-His-Thr) | −111.776 | 11 | Cys368, Val418, Val467, Ala466, Val420, Ile559, Val606, Gly367 | 4 | Ala366, Val606, Cys513, Val561 |
| 8 | Cyclo(Asp-Phe) | −113.424 | 6 | Ile416, Gly462, Ala510, Val465, Val512, Val418 | 2 | Ala556, Arg415 |
| 9 | Cyclo(Met-His-Phe) | −121.817 | 6 | Thr560, Val512, Ile559, Val465, Val418, Val467, Gly367 | 7 | Ala607, Val608, Val369, Val606, Ala366, Val517, Ala466 |
| 10 | Cyclo(Pro-Asp-Tyr) | −138.385 | 13 | Gly603, Val604, Val606, Gly605, Gly367, Ala366, Leu557, Val512, Gly559, Val418, Ala510, Ile416, Gly462 | 3 | Ala366, Val418, Arg415 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Sun, T.; Gao, H.; Liu, X.; Zhang, S.; Liu, T.; Wang, D.; Fan, H.; Zhang, Y. Bioinformatics-Assisted Discovery of Antioxidant Cyclic Peptides from Corn Gluten Meal. Foods 2025, 14, 1709. https://doi.org/10.3390/foods14101709
Liu H, Sun T, Gao H, Liu X, Zhang S, Liu T, Wang D, Fan H, Zhang Y. Bioinformatics-Assisted Discovery of Antioxidant Cyclic Peptides from Corn Gluten Meal. Foods. 2025; 14(10):1709. https://doi.org/10.3390/foods14101709
Chicago/Turabian StyleLiu, Hongcheng, Tong Sun, He Gao, Xiaolong Liu, Shanshan Zhang, Tingting Liu, Dawei Wang, Hongxiu Fan, and Yanrong Zhang. 2025. "Bioinformatics-Assisted Discovery of Antioxidant Cyclic Peptides from Corn Gluten Meal" Foods 14, no. 10: 1709. https://doi.org/10.3390/foods14101709
APA StyleLiu, H., Sun, T., Gao, H., Liu, X., Zhang, S., Liu, T., Wang, D., Fan, H., & Zhang, Y. (2025). Bioinformatics-Assisted Discovery of Antioxidant Cyclic Peptides from Corn Gluten Meal. Foods, 14(10), 1709. https://doi.org/10.3390/foods14101709





