Antioxidant Peptides from the Fruit Source of the Oil Crop Litsea cubeba Ameliorate FFA-Induced Oxidative Stress Injury: Based on Nrf2/Keap1 Pathway and Molecular Dynamics Simulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Analyzing and Screening of Antioxidant Peptides
2.3. FFA-Induced Lipid Accumulation in HepG2 Cells
2.4. CCK8 Experiments
2.5. Peptide Grouping and Administration
2.6. Oil Red O Staining
2.7. TG and TC Content
2.8. Antioxidant Kit Content Assay
2.9. ROS Staining
2.10. JC-1 Staining
2.11. RT-qPCR Analysis
2.12. Molecular Docking and Molecular Dynamics Simulation
2.13. Statistical Analysis
3. Results
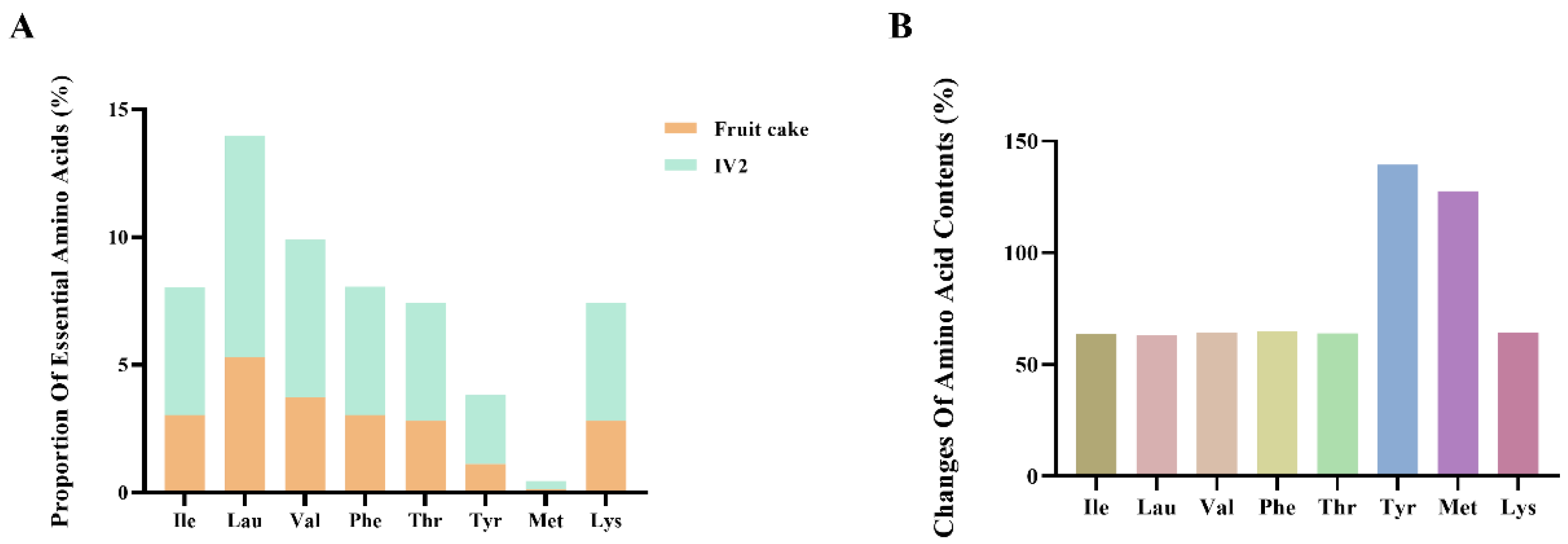
3.1. Identification of Antioxidant Peptides
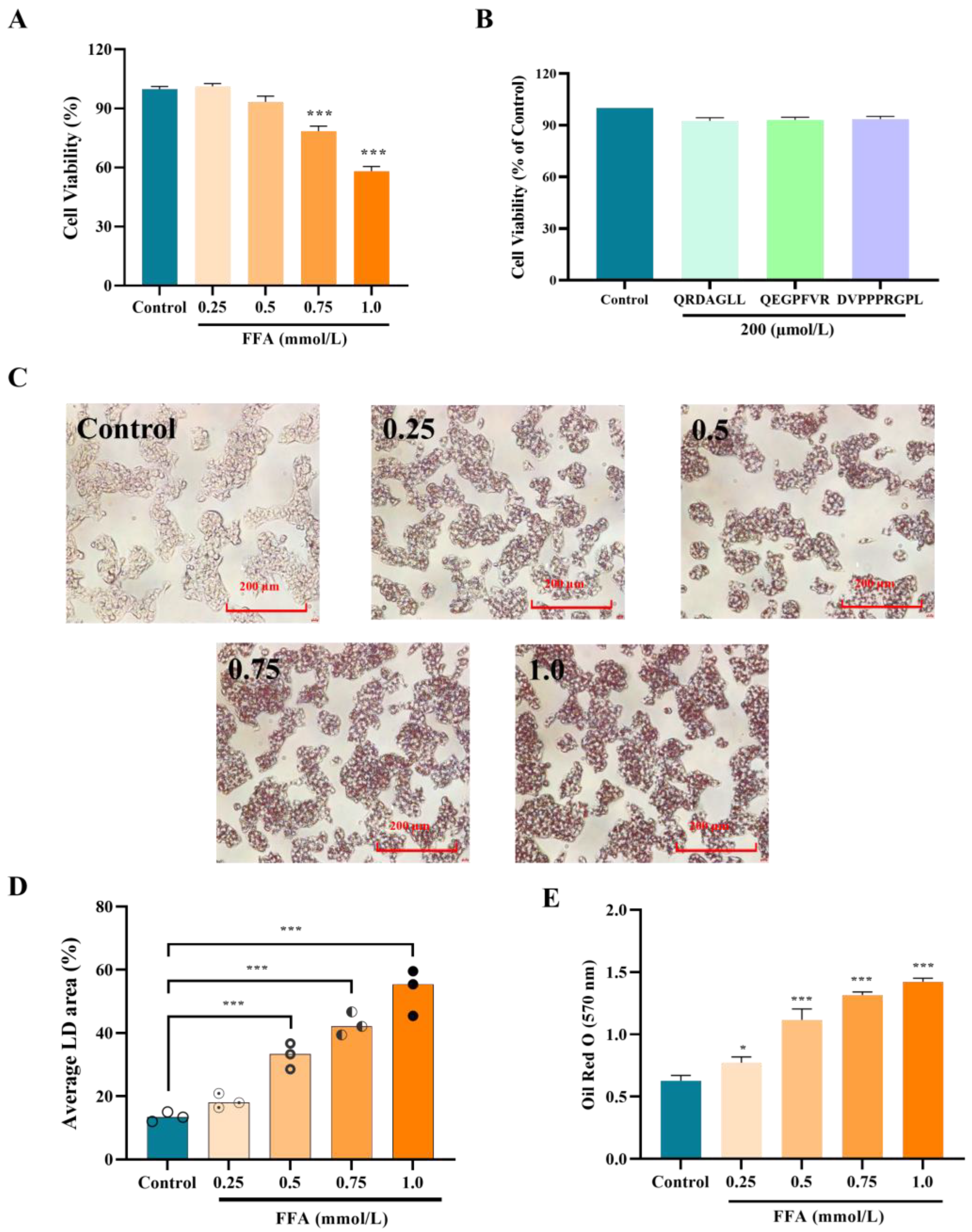
3.2. Effect of Concentration of FFAs and Peptides on the Activity of HepG2 Cells
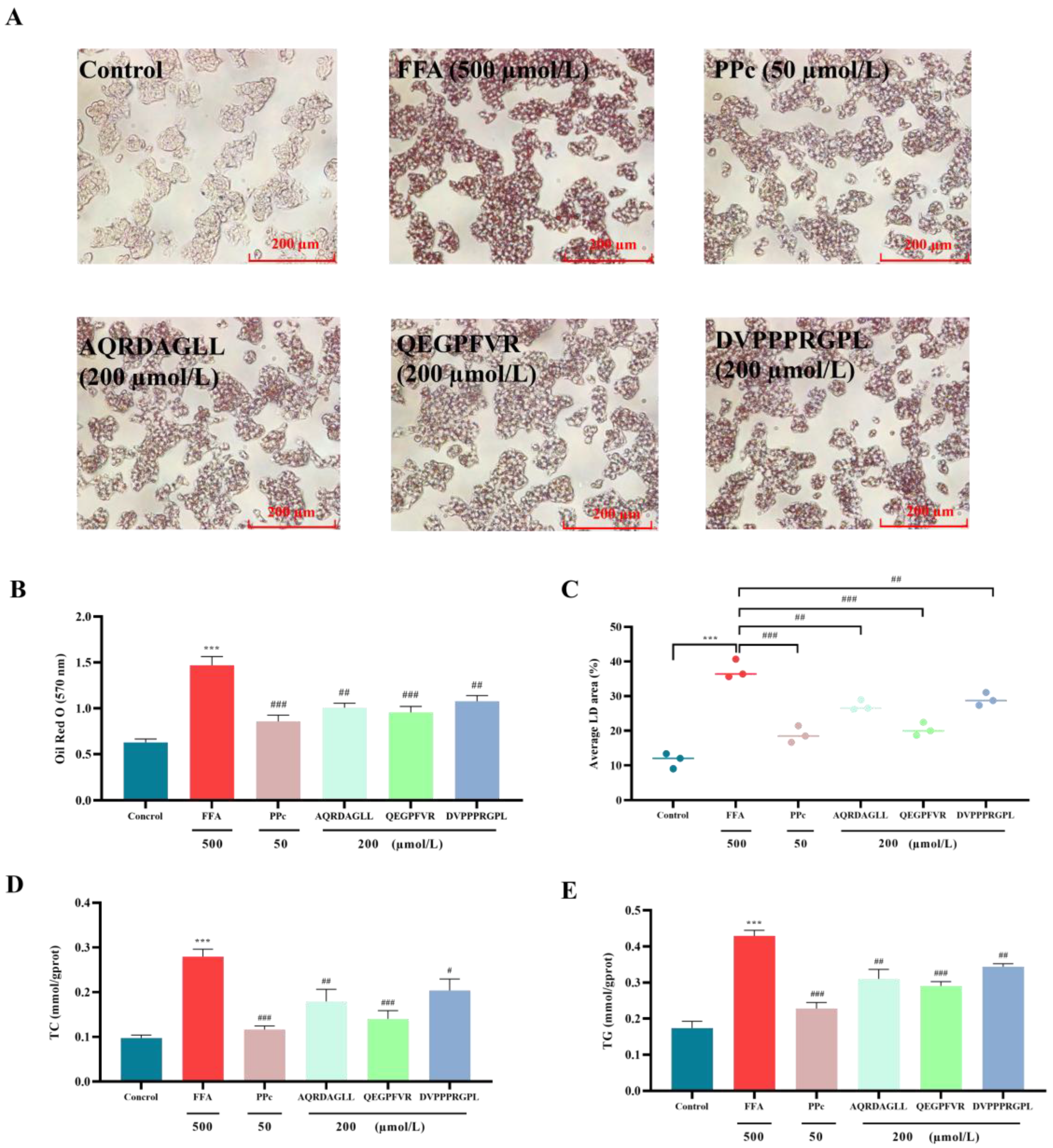
3.3. Antioxidant Peptides Improve Lipid Accumulation in HepG2 Cells
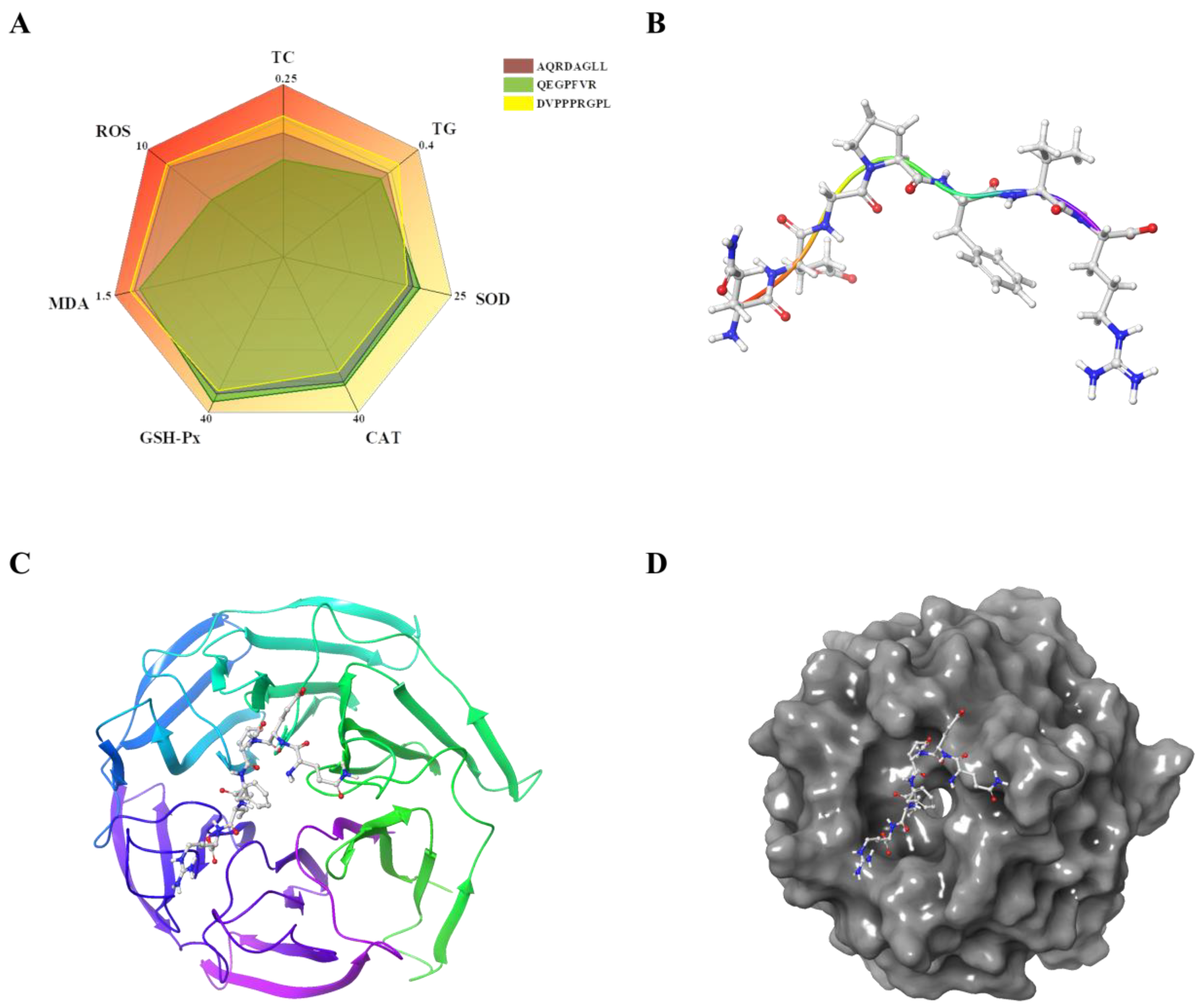
3.4. Antioxidant Peptides Ameliorate Oxidative Damage Induced by Lipid Accumulation
3.5. Effects of Antioxidant Peptides on ROS Levels in HepG2 Cells
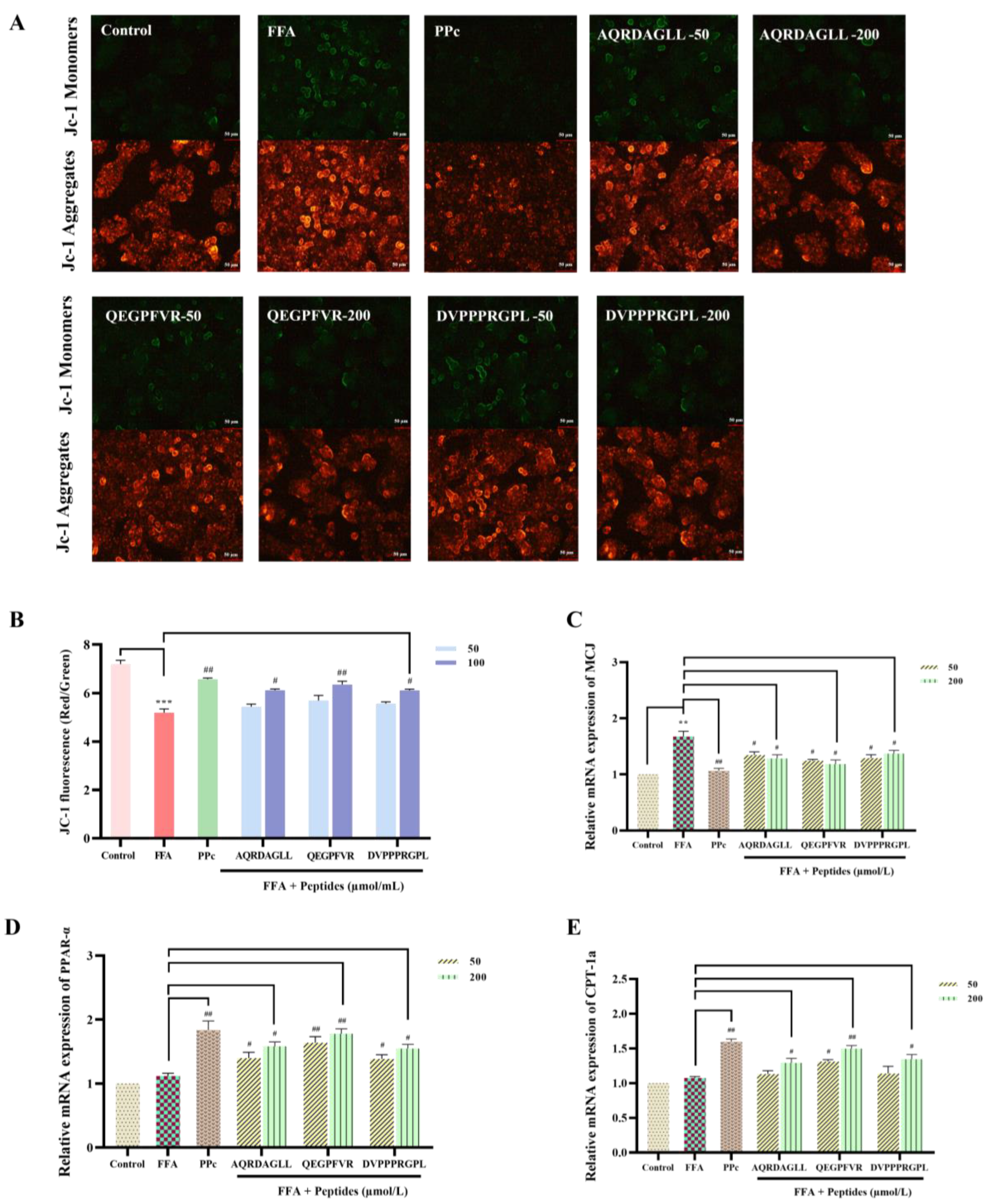
3.6. Protective Effects of Antioxidant Peptides on Mitochondria
3.7. Molecular Docking of QEGPFVR with Keap1
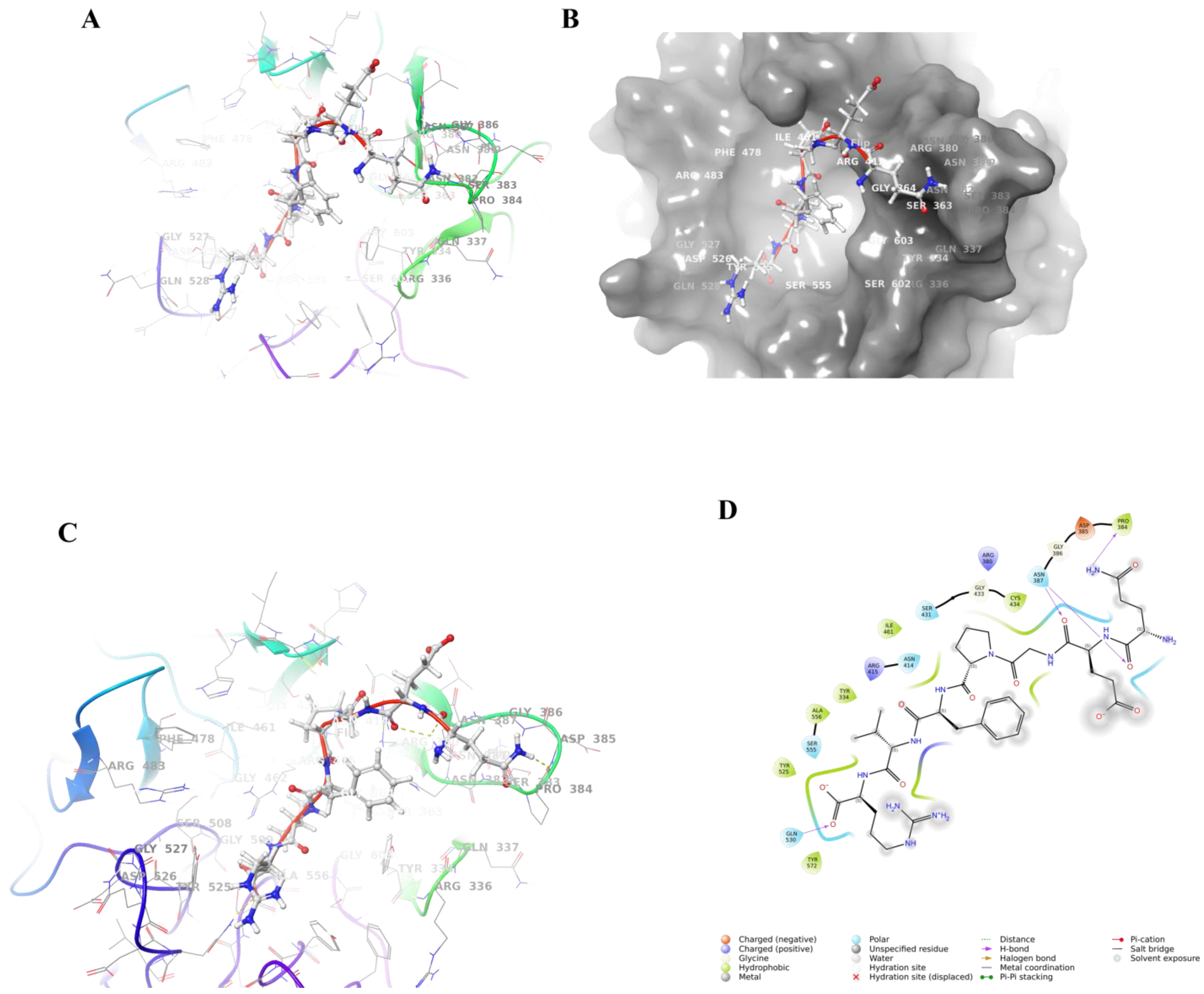
3.7.1. Molecular Docking Results
3.7.2. Combined Model Analysis
3.8. Molecular Dynamics Simulation of Keap1 Protein with QEGPFVR
Stability Analysis of QEGPFVR–Keap1 Protein Complexes
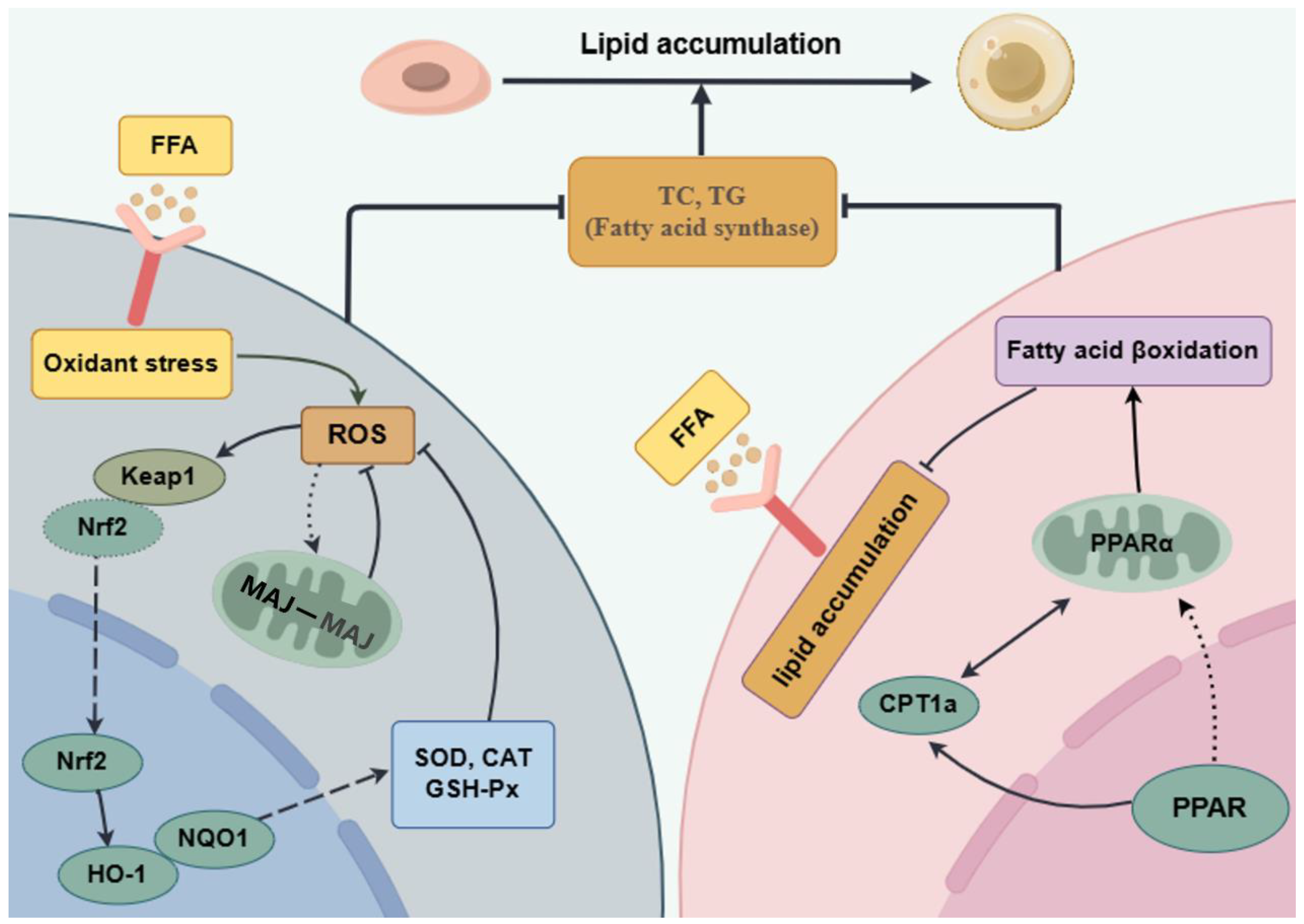
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ahmad, T.; Belwal, T.; Li, L.; Ramola, S.; Aadil, R.M.; Abdullah; Xu, Y.; Zisheng, L. Utilization of wastewater from edible oil industry, turning waste into valuable products: A review. Trends Food Sci. Technol. 2020, 99, 21–33. [Google Scholar] [CrossRef]
- Xu, L.; Wei, Z.; Guo, B.; Bai, R.; Liu, J.; Li, Y.; Sun, W.; Jiang, X.; Li, X.; Pi, Y. Flaxseed Meal and Its Application in Animal Husbandry: A Review. Agriculture 2022, 12, 2027. [Google Scholar] [CrossRef]
- Yong, K.J.; Wu, T.Y. Second-generation bioenergy from oilseed crop residues: Recent technologies, techno-economic assessments and policies. Energy Convers. Manag. 2022, 267, 115869. [Google Scholar] [CrossRef]
- Kotecka-Majchrzak, K.; Sumara, A.; Fornal, E.; Montowska, M. Oilseed proteins—Properties and application as a food ingredient. Trends Food Sci. Technol. 2020, 106, 160–170. [Google Scholar] [CrossRef]
- Shao, J.; Zhang, G.; Fu, J.; Zhang, B. Advancement of the preparation methods and biological activity of peptides from sesame oil byproducts: A review. Int. J. Food Prop. 2020, 23, 2189–2200. [Google Scholar] [CrossRef]
- Maestri, E.; Marmiroli, M.; Marmiroli, N. Bioactive peptides in plant-derived foodstuffs. J. Proteom. 2016, 147, 140–155. [Google Scholar] [CrossRef]
- Görgüç, A.; Gençdağ, E.; Yılmaz, F.M. Bioactive peptides derived from plant origin by-products: Biological activities and techno-functional utilizations in food developments—A review. Food Res. Int. 2020, 136, 109504. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, K.; Li, H.; Sun, H.; Liu, X. Improvement of active peptide yield, antioxidant activity and anti-aging capacity of rapeseed meal fermented with YY-112 pure fermentation and co-fermentation. Food Biosci. 2022, 49, 101938. [Google Scholar] [CrossRef]
- Han, R.; Hernández Álvarez, A.J.; Maycock, J.; Murray, B.S.; Boesch, C. Comparison of alcalase- and pepsin-treated oilseed protein hydrolysates—Experimental validation of predicted antioxidant, antihypertensive and antidiabetic properties. Curr. Res. Food Sci. 2021, 4, 141–149. [Google Scholar] [CrossRef]
- Ashaolu, T.J. Health Applications of Soy Protein Hydrolysates. Int. J. Pept. Res. Ther. 2020, 26, 2333–2343. [Google Scholar] [CrossRef]
- Kim, I.-S.; Yang, W.-S.; Kim, C.-H. Beneficial Effects of Soybean-Derived Bioactive Peptides. Int. J. Mol. Sci. 2021, 22, 8570. [Google Scholar] [CrossRef] [PubMed]
- González-Montoya, M.; Hernández-Ledesma, B.; Silván, J.M.; Mora-Escobedo, R.; Martínez-Villaluenga, C. Peptides derived from in vitro gastrointestinal digestion of germinated soybean proteins inhibit human colon cancer cells proliferation and inflammation. Food Chem. 2018, 242, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Tonolo, F.; Coletta, S.; Fiorese, F.; Grinzato, A.; Albanesi, M.; Folda, A.; Ferro, S.; De Mario, A.; Piazza, I.; Mammucari, C.; et al. Sunflower seed-derived bioactive peptides show antioxidant and anti-inflammatory activity: From in silico simulation to the animal model. Food Chem. 2024, 439, 138124. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.-T.; Wu, C.-H.; Li, L.-H.; Hung, D.-Y.; Chiu, H.-W.; Hsu, H.-T.; Ho, C.-L.; Chernikov, O.V.; Cheng, S.-M.; Yang, S.-P.; et al. The leaves of the seasoning plant Litsea cubeba inhibit the NLRP3 inflammasome and ameliorate dextran sulfate sodium-induced colitis in mice. Front. Nutr. 2022, 9, 871325. [Google Scholar] [CrossRef]
- Kamle, M.; Mahato, D.K.; Lee, K.E.; Bajpai, V.K.; Gajurel, P.R.; Gu, K.S.; Kumar, P. Ethnopharmacological Properties and Medicinal Uses of Litsea cubeba. Plants 2019, 8, 150. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, H.; Li, K. Litsea cubeba essential oil: Extraction, chemical composition, antioxidant and antimicrobial properties, and applications in the food industry. J. Food Sci. 2024, 89, 4583–4603. [Google Scholar] [CrossRef]
- Wu, J.; Liu, W.; Hou, S.; Wang, Y.; Fang, H.; Luo, S.; Yang, L.; Wen, C. Identification of Nrf2/Keap1 pathway and its transcriptional regulation of antioxidant genes after exposure to microcystins in freshwater mussel Cristaria plicata. Dev. Comp. Immunol. 2023, 141, 104629. [Google Scholar] [CrossRef]
- Borén, J.; Taskinen, M.-R.; Björnson, E.; Packard, C.J. Metabolism of triglyceride-rich lipoproteins in health and dyslipidaemia. Nat. Rev. Cardiol. 2022, 19, 577–592. [Google Scholar] [CrossRef]
- Li, J.; Ran, X.; Zhou, M.; Wang, K.; Wang, H.; Wang, Y. Oxidative stress and antioxidant mechanisms of obligate anaerobes involved in biological waste treatment processes: A review. Sci. Total Environ. 2022, 838, 156454. [Google Scholar] [CrossRef]
- Sadiq, I.Z. Free radicals and oxidative stress: Signaling mechanisms, redox basis for human diseases, and cell cycle regulation. Curr. Mol. Med. 2023, 23, 13–35. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Farag, M.A.; Cai, X.; Wang, S. Identification of novel antioxidant peptides from Lateolabrax japonicus and the underlying molecular mechanisms against oxidative stress injury in Caco-2 cells. Food Biosci. 2024, 62, 105538. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Sehrawat, A.; Mishra, J.; Sidhu, I.S.; Navik, U.; Khullar, N.; Kumar, S.; Bhatti, G.K.; Reddy, P.H. Oxidative stress in the pathophysiology of type 2 diabetes and related complications: Current therapeutics strategies and future perspectives. Free Radic. Biol. Med. 2022, 184, 114–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Botchway, B.O.A.; Zhang, Y.; Liu, X. Ellagic acid activates the Keap1-Nrf2-ARE signaling pathway in improving Parkinson’s disease: A review. Biomed. Pharmacother. 2022, 156, 113848. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhang, J.; Sun, J.; Min, T.; Bai, Y.; He, J.; Cao, H.; Che, Q.; Guo, J.; Su, Z. Oxidative stress in alcoholic liver disease, focusing on proteins, nucleic acids, and lipids: A review. Int. J. Biol. Macromol. 2024, 278, 134809. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.-M.; Zeng, X.-Y.; Hu, Y.; Zhang, J.; Wang, B.; Chen, S.-X. Bioactive proteins and antioxidant peptides from Litsea cubeba fruit meal: Preparation, characterization and ameliorating function on high-fat diet-induced NAFLD through regulating lipid metabolism, oxidative stress and inflammatory response. Int. J. Biol. Macromol. 2024, 280, 136186. [Google Scholar] [CrossRef]
- Alshehade, S.; Alshawsh, M.A.; Murugaiyah, V.; Asif, M.; Alshehade, O.; Almoustafa, H.; Al Zarzour, R.H. The role of protein kinases as key drivers of metabolic dysfunction-associated fatty liver disease progression: New insights and future directions. Life Sci. 2022, 305, 120732. [Google Scholar] [CrossRef]
- Renjuan, L.; Xiuli, Z.; Liping, S.; Yongliang, Z. Identification, in silico screening, and molecular docking of novel ACE inhibitory peptides isolated from the edible symbiot Boletus griseus-Hypomyces chrysospermus. LWT 2022, 169, 114008. [Google Scholar] [CrossRef]
- Tan, H.; Mi, N.; Tong, F.; Zhang, R.; Abudurexiti, A.; Lei, Y.; Zhong, Y.; Yan, J.; Yang, J.; Ma, X. Lactucopicrin promotes fatty acid β-oxidation and attenuates lipid accumulation through adenosine monophosphate-activated protein kinase activation in free fatty acid-induced human hepatoblastoma cancer cells. Food Sci. Nutr. 2024, 12, 5357–5372. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, S.; Li, Y.; Xie, D.; Li, J.-X. Residue profiles of peptides with cholesterol esterase and pancreatic lipase inhibitory activities through virtual screening and sequence analysis. Food Chem. 2024, 460, 140708. [Google Scholar] [CrossRef]
- Wei, K.; Wei, Y.; Xu, W.; Lu, F.; Ma, H. Corn peptides improved obesity-induced non-alcoholic fatty liver disease through relieving lipid metabolism, insulin resistance and oxidative stress. Food Funct. 2022, 13, 5782–5793. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.-L.; Zeng, H.-F.; Han, Z.-Y. Methionine alleviates heat stress-induced ferroptosis in bovine mammary epithelial cells through the Nrf2 pathway. Ecotoxicol. Environ. Saf. 2023, 256, 114889. [Google Scholar] [CrossRef] [PubMed]
- Dhaouafi, J.; Romdhani, M.; Deracinois, B.; Flahaut, C.; Nedjar, N.; Balti, R. Fractionation and identification of bioactive peptides from red macroalgae protein hydrolysates: In silico analysis and in vitro bioactivities. Biocatal. Agric. Biotechnol. 2024, 58, 103211. [Google Scholar] [CrossRef]
- Maseko, T.E.; Elkalaf, M.; Peterová, E.; Lotková, H.; Staňková, P.; Melek, J.; Dušek, J.; Žádníková, P.; Čížková, D.; Bezrouk, A.; et al. Comparison of HepaRG and HepG2 cell lines to model mitochondrial respiratory adaptations in non-alcoholic fatty liver disease. Int. J. Mol. Med. 2024, 53, 18. [Google Scholar] [CrossRef] [PubMed]
- Berscheid, A.; Straetener, J.; Schilling Nadine, A.; Ruppelt, D.; Konnerth Martin, C.; Schittek, B.; Krismer, B.; Peschel, A.; Steinem, C.; Grond, S.; et al. The microbiome-derived antibacterial lugdunin acts as a cation ionophore in synergy with host peptides. mBio 2024, 15, e00524–e00578. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Meng, Y.-Q.; Qiao, H.; Zhai, K.-R.; Li, Z.-Q.; Wei, S.-L.; Li, B. Melittin kills A549 cells by targeting mitochondria and blocking mitophagy flux. Redox Rep. 2023, 28, 2284517. [Google Scholar] [CrossRef]
- Ge, M.; Fontanesi, F.; Merscher, S.; Fornoni, A. The vicious cycle of renal lipotoxicity and mitochondrial dysfunction. Front. Physiol. 2020, 11, 732. [Google Scholar] [CrossRef]
- Javadov, S.; Jang, S.; Chapa-Dubocq, X.R.; Khuchua, Z.; Camara, A.K.S. Mitochondrial respiratory supercomplexes in mammalian cells: Structural versus functional role. J. Mol. Med. 2021, 99, 57–73. [Google Scholar] [CrossRef]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef]
- Canning, P.; Sorrell, F.J.; Bullock, A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015, 88, 101–107. [Google Scholar] [CrossRef]
- Crisman, E.; Duarte, P.; Dauden, E.; Cuadrado, A.; Rodríguez-Franco, M.I.; López, M.G.; León, R. Keap1-Nrf2 protein–protein interaction inhibitors: Design, pharmacological properties and therapeutic potential. Med. Res. Rev. 2023, 43, 237–287. [Google Scholar] [CrossRef]
- Zhao, Z.; Dong, R.; Cui, K.; You, Q.; Jiang, Z. An updated patent review of Nrf2 activators (2020-present). Expert Opin. Ther. Pat. 2023, 33, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.; Morales-González, J.A. Molecular recognition between potential natural inhibitors of the Keap1-Nrf2 complex. Int. J. Biol. Macromol. 2017, 105, 981–992. [Google Scholar] [CrossRef]
- Yurchenko, E.A.; Khmel, O.O.; Nesterenko, L.E.; Aminin, D.L. The Kelch/Nrf2 antioxidant system as a target for some marine fungal metabolites. Oxygen 2023, 3, 374–385. [Google Scholar] [CrossRef]
- Roy, A.; Paul, I.; Paul, T.; Hazarika, K.; Dihidar, A.; Ray, S. An in-silico receptor-pharmacophore based multistep molecular docking and simulation study to evaluate the inhibitory potentials against NS1 of DENV-2. J. Biomol. Struct. Dyn. 2024, 42, 6136–6164. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Gupta, P.S.S.; Rana, M.K. Potential targets of severe acute respiratory syndrome coronavirus 2 of clinical drug fluvoxamine: Docking and molecular dynamics studies to elucidate viral action. Cell Biochem. Funct. 2023, 41, 98–111. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef]
- Dullius, A.; Fassina, P.; Giroldi, M.; Goettert, M.I.; Volken de Souza, C.F. A biotechnological approach for the production of branched chain amino acid containing bioactive peptides to improve human health: A review. Food Res. Int. 2020, 131, 109002. [Google Scholar] [CrossRef]
- Hajieva, P.; Abrosimov, R.; Kunath, S.; Moosmann, B. Antioxidant and prooxidant modulation of lipid peroxidation by integral membrane proteins. Free Radic. Res. 2023, 57, 105–114. [Google Scholar] [CrossRef]
- Fan, L.; Mao, X.; Wu, Q. Purification, identification and molecular docking of novel antioxidant peptides from Walnut (Juglans regia L.) protein hydrolysates. Molecules 2022, 27, 8423. [Google Scholar] [CrossRef]
- Culletta, G.; Buttari, B.; Arese, M.; Brogi, S.; Almerico, A.M.; Saso, L.; Tutone, M. Natural products as non-covalent and covalent modulators of the Keap1/Nrf2 pathway exerting antioxidant effects. Eur. J. Med. Chem. 2024, 270, 116355. [Google Scholar] [CrossRef]
- Xiang, Z.; Xue, Q.; Gao, P.; Yu, H.; Wu, M.; Zhao, Z.; Li, Y.; Wang, S.; Zhang, J.; Dai, L. Antioxidant peptides from edible aquatic animals: Preparation method, mechanism of action, and structure-activity relationships. Food Chem. 2023, 404, 134701. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xiao, J.-H. The Keap1-Nrf2 System: A Mediator between Oxidative Stress and Aging. Oxidative Med. Cell. Longev. 2021, 2021, 6635460. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Pi, J.; Zhang, Q. Signal amplification in the Keap1-Nrf2-ARE antioxidant response pathway. Redox Biol. 2022, 54, 102389. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, F.; Liu, M.; Zhou, X.; Wang, M.; Cao, K.; Jin, S.; Shan, A.; Feng, X. Curcumin mitigates aflatoxin B1-induced liver injury via regulating the NLRP3 inflammasome and Nrf2 signaling pathway. Food Chem. Toxicol. 2022, 161, 112823. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Mandal, M.; Mandal, S. Discovery of multitarget-directed small molecule inhibitors from Andrographis paniculata for Nipah virus disease therapy: Molecular docking, molecular dynamics simulation and ADME-Tox profiling. Chem. Phys. Impact 2024, 8, 100493. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Vidal-Limon, A.; Liceaga, A.M. Multifunctional analysis of Chia Seed (Salvia hispanica L.) bioactive peptides using peptidomics and molecular dynamics simulations approaches. Int. J. Mol. Sci. 2022, 23, 7288. [Google Scholar] [CrossRef]
- Kryszczuk, M.; Kowalczuk, O. Significance of Nrf2 in physiological and pathological conditions an comprehensive review. Arch. Biochem. Biophys. 2022, 730, 109417. [Google Scholar] [CrossRef]
- Xiao, J.-L.; Liu, H.-Y.; Sun, C.-C.; Tang, C.-F. Regulation of Keap1-Nrf2 signaling in health and diseases. Mol. Biol. Rep. 2024, 51, 809. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Quan, Z.; Xiao, P.; Duan, J.-A. New insights into antioxidant peptides: An overview of efficient screening, evaluation models, molecular mechanisms, and applications. Antioxidants 2024, 13, 203. [Google Scholar] [CrossRef]
- Liu, W.; Liu, R.; Qin, Q.; Wang, H.; Zhang, X.; Meng, G. Molecular docking and molecular dynamics simulation of wheat gluten-derived antioxidant peptides acting through the Keap1–Nrf2 pathway. J. Sci. Food Agric. 2024, 104, 8150–8161. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Lu, A.; Sun, Y.; Liu, B.; Zhang, J.; Zhang, L.; Huang, P.; Yang, A.; Li, Z.; Cao, Y.; et al. Purification and identification of antioxidant and angiotensin converting enzyme-inhibitory peptides from Guangdong glutinous rice wine. LWT 2022, 169, 113953. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, K.; Jia, X.; Fu, C.; Yu, H.; Wang, Y. Antioxidant peptides, the guardian of life from oxidative stress. Med. Res. Rev. 2024, 44, 275–364. [Google Scholar] [CrossRef]
- Li, J.; Fu, A.; Zhang, L. An Overview of scoring functions used for protein–ligand interactions in molecular docking. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef]
- Akash, S.; Abdelkrim, G.; Bayil, I.; Hosen, M.E.; Mukerjee, N.; Shater, A.F.; Saleh, F.M.; Albadrani, G.M.; Al-Ghadi, M.Q.; Abdel-Daim, M.M.; et al. Antimalarial drug discovery against malaria parasites through haplopine modification: An advanced computational approach. J. Cell. Mol. Med. 2023, 27, 3168–3188. [Google Scholar] [CrossRef]
- Jena, S.; Dutta, J.; Tulsiyan, K.D.; Sahu, A.K.; Choudhury, S.S.; Biswal, H.S. Noncovalent interactions in proteins and nucleic acids: Beyond hydrogen bonding and π-stacking. Chem. Soc. Rev. 2022, 51, 4261–4286. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.E.; Fanourakis, A.; Phipps, R.J. Strategies That Utilize Ion Pairing Interactions to Exert Selectivity Control in the Functionalization of C–H Bonds. J. Am. Chem. Soc. 2022, 144, 18195–18211. [Google Scholar] [CrossRef]



| Gene | Forward Sequence (5′–3′) | Reverse Sequence (3′–5′) |
|---|---|---|
| Nrf2 | TGTCTTAATACCGAAAACAAGCAGC | GACCACAGTTGCCCACTTCTTTT |
| HO-1 | GCTAAGACCGCCTTCCTGCT | ACGAAGTGACGCCATCTGTGA |
| NQO1 | TATCACCACTGGGGGTAGCG | GGAGTGTGGCCAATGCTGTAA |
| MCJ | AAGTAATCACGGCAACAGGG | AATAAAAGCCTGGCAGCCTTGC |
| PPAR-α | TTTCACAAGTGCCTGTCTGTCG | TCTTCAGGTAGGCTTCGTGGAT |
| CPT-1a | GCCATGAAGCCCTCAAACAG | CACCCACCACCACGATAAGC |
| GAPDH | CCTCGTCCCGTAGACAAAATG | TGAGGTCAATGAAGGGGTCGT |
| Sequence | Predicted Score | Percentage of Hydrophobic Amino Acids (%) |
|---|---|---|
| LSPRDAVGH | 0.325905 | 44.44 |
| NAQRDAGIV | 0.176496 | 44.44 |
| EQRPTYSN | 0.131206 | 12.5 |
| EGDVFAVPR | 0.457817 | 55.56 |
| NAQRDAGLL | 0.467791 | 44.44 |
| HQEGPFVR | 0.472375 | 37.5 |
| QEGPFVRV | 0.452377 | 50 |
| DAGIVEGPK | 0.222096 | 44.44 |
| ALVVDGK | 0.129537 | 57.14 |
| ESEREVQ | 0.0394552 | 14.29 |
| LAGRDNIY | 0.211819 | 37.5 |
| QEGPFVR | 0.513535 | 42.68 |
| NAQRDAGIVEGPK | 0.185251 | 38.46 |
| AQRDAGLL | 0.581992 | 50 |
| AQRDAGIVEGPKGS | 0.167792 | 35.71 |
| RDAGIVEGPKGS | 0.142415 | 33.33 |
| EEVVHPY | 0.0867931 | 42.86 |
| GFDERVL | 0.36642 | 42.86 |
| DVPPPRGPL | 0.837006 | 66.67 |
| AGIVEGPKG | 0.298716 | 44.44 |
| Residues | Gbind | Coulomb | Solv | vdW | H-bond | Lipo | Pi–Pi |
|---|---|---|---|---|---|---|---|
| PeptideAA_4 (Pro) | −4.16 | −1.2 | −0.6 | −4.5 | −0.35 | −0.63 | 0 |
| PeptideAA_5 (Phe) | −5.97 | −5.12 | 7.08 | −6.69 | −0.15 | −3.4 | −1.13 |
| PeptideAA_6 (Val) | −4.98 | −7.54 | 8.71 | −6.27 | −0.38 | −1.41 | 0 |
| PeptideAA_7 (Arg) | −4.73 | 0.8 | 3.04 | −8.59 | −0.74 | −2.95 | −0.45 |
| Keap1_334 (Tyr) | −7.29 | −1.94 | 2.26 | −3.43 | −0.08 | −3.19 | −0.91 |
| Keap1_363 (Ser) | −2.53 | −1.35 | 0.05 | −1.06 | 0 | −0.18 | 0 |
| Keap1_380 (Arg) | −4.62 | −12.7 | 11.58 | −2.71 | −0.27 | −0.39 | −0.14 |
| Keap1_382 (Asn) | −5.42 | −2.76 | 1.44 | −2.95 | −0.36 | −0.47 | −0.31 |
| Keap1_387 (Asn) | −1.73 | −1.7 | 1.82 | −1.51 | −0.14 | −0.16 | −0.05 |
| Keap1_415 (Arg) | −2.26 | −3.62 | 4.3 | −2.1 | −0.02 | −0.82 | −0.01 |
| Keap1_555 (Ser) | −2.65 | −2.42 | 0.8 | −0.57 | −0.4 | −0.05 | 0 |
| Keap1_572 (Tyr) | −4.94 | −1.23 | 1 | −2.66 | −0.07 | −1.59 | −0.4 |
| Keap1_577 (Phe) | −1.87 | −0.24 | 0.18 | −1.04 | 0 | −0.77 | 0 |
| Keap1_602 (Ser) | −5.32 | −4.88 | 1.47 | −1.39 | −0.25 | −0.28 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Hu, Y.; Wang, Y.-M.; Wu, X.-X.; Lin, S.-T.; Li, H.; Zhang, J.; Fan, G.-R.; Wang, Z.-D.; Wang, B.; et al. Antioxidant Peptides from the Fruit Source of the Oil Crop Litsea cubeba Ameliorate FFA-Induced Oxidative Stress Injury: Based on Nrf2/Keap1 Pathway and Molecular Dynamics Simulations. Foods 2025, 14, 1707. https://doi.org/10.3390/foods14101707
Li L, Hu Y, Wang Y-M, Wu X-X, Lin S-T, Li H, Zhang J, Fan G-R, Wang Z-D, Wang B, et al. Antioxidant Peptides from the Fruit Source of the Oil Crop Litsea cubeba Ameliorate FFA-Induced Oxidative Stress Injury: Based on Nrf2/Keap1 Pathway and Molecular Dynamics Simulations. Foods. 2025; 14(10):1707. https://doi.org/10.3390/foods14101707
Chicago/Turabian StyleLi, Li, Ying Hu, Yu-Mei Wang, Xiao-Xue Wu, Si-Tong Lin, Hang Li, Ji Zhang, Guo-Rong Fan, Zong-De Wang, Bin Wang, and et al. 2025. "Antioxidant Peptides from the Fruit Source of the Oil Crop Litsea cubeba Ameliorate FFA-Induced Oxidative Stress Injury: Based on Nrf2/Keap1 Pathway and Molecular Dynamics Simulations" Foods 14, no. 10: 1707. https://doi.org/10.3390/foods14101707
APA StyleLi, L., Hu, Y., Wang, Y.-M., Wu, X.-X., Lin, S.-T., Li, H., Zhang, J., Fan, G.-R., Wang, Z.-D., Wang, B., & Chen, S.-X. (2025). Antioxidant Peptides from the Fruit Source of the Oil Crop Litsea cubeba Ameliorate FFA-Induced Oxidative Stress Injury: Based on Nrf2/Keap1 Pathway and Molecular Dynamics Simulations. Foods, 14(10), 1707. https://doi.org/10.3390/foods14101707







