Insights into the Paulownia Shan tong (Fortunei × Tomentosa) Essential Oil and In Silico Analysis of Potential Biological Targets of Its Compounds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Collection
2.2. PStEO Extraction
2.3. PStEO GC–MS Analysis
2.4. Molecular Docking
3. Results and Discussions
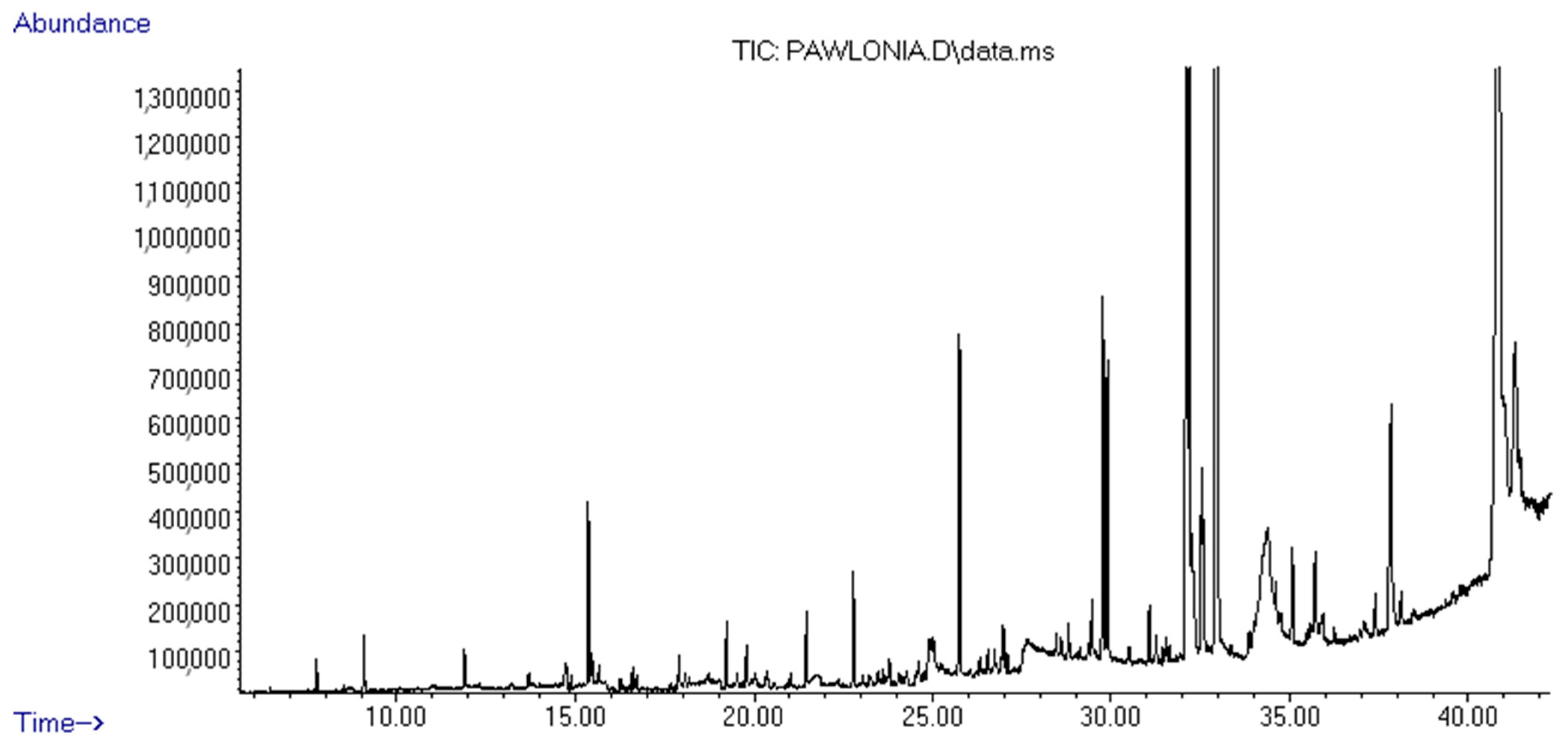
3.1. Chemical Composition of the PStEO
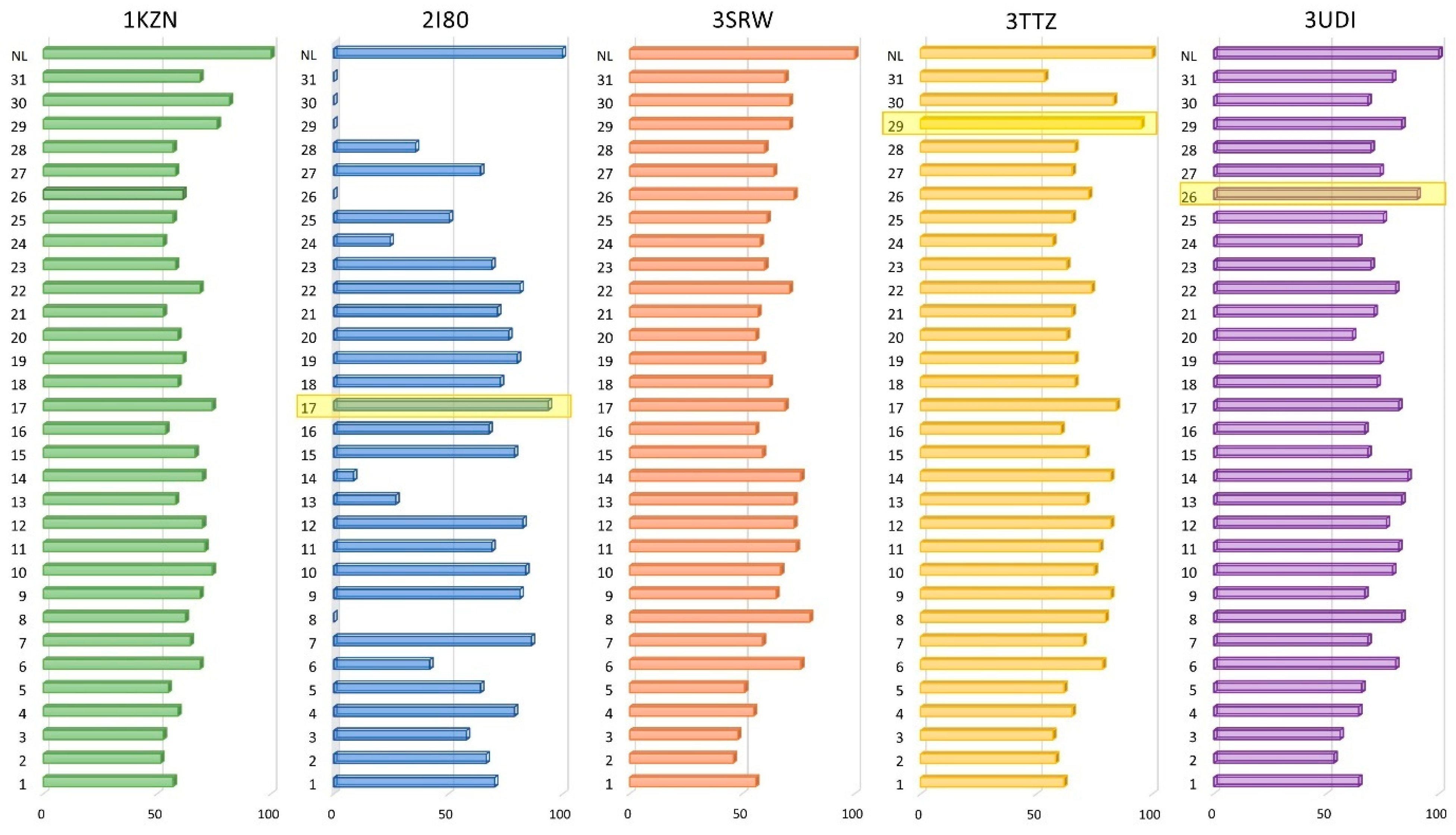
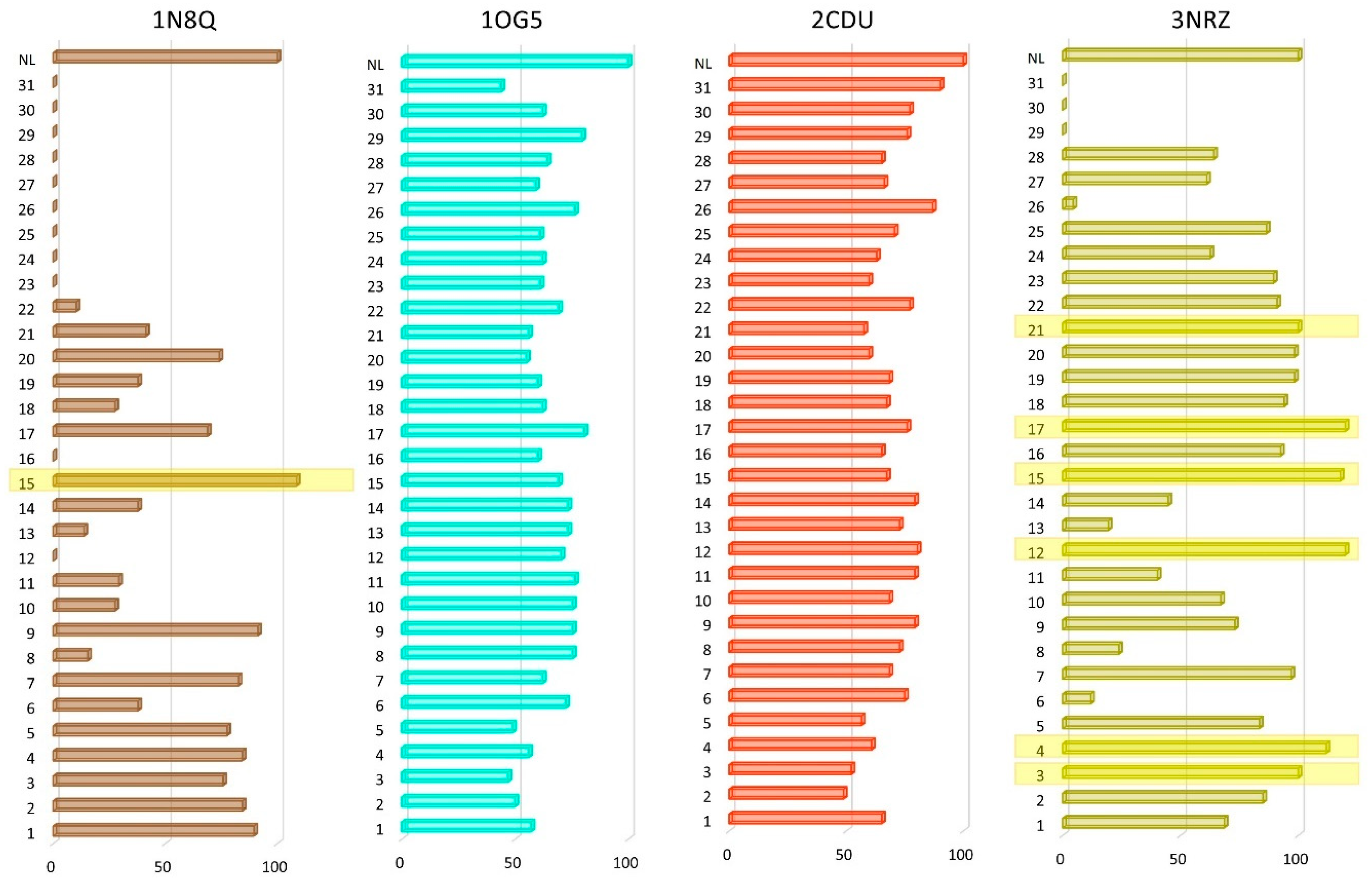
3.2. Molecular Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, X.; Zhou, X.; Wang, C.; Zhou, H. Environmental and human health impacts of volatile organic compounds: A perspective review. Chemosphere 2023, 313, 137489. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, J.; Xie, J. Progress on odor deterioration of aquatic products: Characteristic volatile compounds, analysis methods, and formation mechanisms. Food Biosci. 2023, 53, 102666. [Google Scholar] [CrossRef]
- Starowicz, M. Analysis of Volatiles in Food Products. Separations 2021, 8, 157. [Google Scholar] [CrossRef]
- Dhifi, W.; Bellili, S.; Jazi, S.; Bahloul, N.; Mnif, W. Essential Oils’ Chemical Characterization and Investigation of Some Biological Activities: A Critical Review. Medicines 2016, 3, 25. [Google Scholar] [CrossRef]
- de Oliveira, M.S.; Cruz, J.N.; Ferreira, O.O.; Pereira, D.S.; Pereira, N.S.; Oliveira, M.E.C.; Venturieri, G.C.; Guilhon, G.M.S.P.; Souza Filho, A.P.d.S.; Andrade, E.H.d.A. Chemical Composition of Volatile Compounds in Apis mellifera Propolis from the Northeast Region of Pará State, Brazil. Molecules 2021, 26, 3462. [Google Scholar] [CrossRef]
- Mesquita, K.d.S.M.; Feitosa, B.d.S.; Cruz, J.N.; Ferreira, O.O.; Franco, C.d.J.P.; Cascaes, M.M.; Oliveira, M.S.d.; Andrade, E.H.d.A. Chemical Composition and Preliminary Toxicity Evaluation of the Essential Oil from Peperomia circinnata Link var. circinnata. (Piperaceae) in Artemia salina Leach. Molecules 2021, 26, 7359. [Google Scholar] [CrossRef]
- Cascaes, M.M.; Silva, S.G.; Cruz, J.N.; Santana de Oliveira, M.; Oliveira, J.; Moraes, A.A.B.d.; Costa, F.A.M.d.; da Costa, K.S.; Diniz do Nascimento, L.; Helena de Aguiar Andrade, E. First report on the Annona exsucca DC. Essential oil and in silico identification of potential biological targets of its major compounds. Nat. Prod. Res. 2022, 36, 4009–4012. [Google Scholar] [CrossRef]
- Berg, E.C.; Zarnoch, S.J.; McNab, W.H. Survivorship, attained diameter, height and volume of three Paulownia species after 9 years in the southern Appalachians, USA. J. For. Res. 2020, 31, 2181–2191. [Google Scholar] [CrossRef]
- Franz, E. From ornamental to detrimental? The incipient invasion of Central Europe by Paulownia tomentosa. Preslia 2007, 79, 377–389. [Google Scholar]
- Nagata, T.; DuVal, A.; Schmull, M.; Tchernaja, T.A.; Crane, P.R. Paulownia tomentosa: A Chinese plant in Japan. Curtis’s Bot. Mag. 2013, 30, 261–274. [Google Scholar] [CrossRef]
- He, T.; Vaidya, B.; Perry, Z.; Parajuli, P.; Joshee, N. Paulownia as a medicinal tree: Traditional uses and current advances. Eur. J. Med. Plants 2016, 14, 1–15. [Google Scholar] [CrossRef]
- Guo, N.; Zhai, X.-Q.; Fan, G.-Q. Chemical composition, health benefits and future prospects of Paulownia flowers: A review. Food Chem. 2023, 412, 135496. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, M. Cultivation potential and uses of Paulownia wood: A review. Forests 2022, 13, 668. [Google Scholar] [CrossRef]
- Popescu, A.; Sabau, L. Paulownia Species growing for saplings in pots in romania: Technological aspects and comparative expenses, incomes and profit. Sci. Pap. Ser. Manag. Econ. Eng. Agric. Rural Dev. 2016, 16, 255–265. [Google Scholar]
- Fos, M.; Oliver-Villanueva, J.-V.; Vazquez, M. Radial variation in anatomical wood characteristics and physical properties of Paulownia elongata x Paulownia fortunei hybrid Cotevisa 2 from fast-growing plantations. Eur. J. Wood Wood Prod. 2023, 81, 819–831. [Google Scholar] [CrossRef]
- Akyildiz, M.H.; Kol Sahin, H. Some technological properties and uses of paulownia (Paulownia tomentosa Steud.) wood. J. Environ. Biol. 2010, 31, 351–355. [Google Scholar] [PubMed]
- Del Río, P.G.; Domínguez, V.D.; Domínguez, E.; Gullón, P.; Gullón, B.; Garrote, G.; Romaní, A. Comparative study of biorefinery processes for the valorization of fast-growing Paulownia wood. Bioresour. Technol. 2020, 314, 123722. [Google Scholar] [CrossRef] [PubMed]
- Alagawany, M.; Farag, M.R.; Sahfi, M.E.; Elnesr, S.S.; Alqaisi, O.; El-Kassas, S.; Al-Wajeeh, A.S.; Taha, A.E.; Abd E-Hack, M.E. Phytochemical characteristics of Paulownia trees wastes and its use as unconventional feedstuff in animal feed. Anim. Biotechnol. 2022, 33, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Seoane, P.; Díaz-Reinoso, B.; Moure, A.; Domínguez, H. Potential of Paulownia sp. for biorefinery. Ind. Crops Prod. 2020, 155, 112739. [Google Scholar] [CrossRef]
- Ibrahim, N.; El-Hawary, S.; Mohammed, M.; Faraid, M.; Nayera, A.; Refaat, E. Chemical composition, antimicrobial activity of the essential oil of the flowers of Paulownia tomentosa (Thunb.) Steud. growing in Egypt. J. Appl. Sci. 2013, 9, 3228–3232. [Google Scholar]
- Zhang, H.; Li, X.; Kang, H. Chitosan coatings incorporated with free or nano-encapsulated Paulownia Tomentosa essential oil to improve shelf-life of ready-to-cook pork chops. LWT 2019, 116, 108580. [Google Scholar] [CrossRef]
- Ferdosi, M.F.; Haider Khan, I.; Javaid, A.; Sattar, T.; Munir, A. Identification of antimicrobial constituents in essential oil from Paulownia fortunei flowers. Mycopath 2021, 18, 53–57. [Google Scholar]
- Jianu, C.; Goleț, I.; Stoin, D.; Cocan, I.; Bujancă, G.; Mișcă, C.; Mioc, M.; Mioc, A.; Șoica, C.; Lukinich-Gruia, A.T. Chemical profile of Ruta graveolens, evaluation of the antioxidant and antibacterial potential of its essential oil, and molecular docking simulations. Appl. Sci. 2021, 11, 11753. [Google Scholar] [CrossRef]
- Stahl, E. Das ätherische Öl aus Thymus praecox ssp. arcticus isländischer Herkunft. Planta Medica 1984, 50, 157–160. [Google Scholar] [CrossRef]
- Jianu, C.; Moleriu, R.; Stoin, D.; Cocan, I.; Bujancă, G.; Pop, G.; Lukinich-Gruia, A.T.; Muntean, D.; Rusu, L.-C.; Horhat, D.I. Antioxidant and antibacterial activity of Nepeta× faassenii bergmans ex stearn essential oil. Appl. Sci. 2021, 11, 442. [Google Scholar] [CrossRef]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry, 4th ed.; Allured Publishing Corporation: Carol Stream, IL, USA, 2007. [Google Scholar]
- Jianu, C.; Rusu, L.-C.; Muntean, I.; Cocan, I.; Lukinich-Gruia, A.T.; Goleț, I.; Horhat, D.; Mioc, M.; Mioc, A.; Șoica, C. In Vitro and In Silico Evaluation of the Antimicrobial and Antioxidant Potential of Thymus pulegioides Essential Oil. Antioxidants 2022, 11, 2472. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, C.; Liu, J.; Zheng, C. Chemical composition of the essential oil from Paulownia tomentosa flowers. Chem. Ind. For. Prod. 2005, 2, 99–102. [Google Scholar]
- Zhang, Y.; Sun, B.; Huang, M.; Chen, H. Analysis of the volatile compounds from the flower of Paulownia elongate. Chem. Ind. For. Prod. 2010, 30, 88–92. [Google Scholar]
- Linstrom, P.J.; Mallard, W.G.E. NIST Chemistry WebBook, NIST Standard Reference Database Number 69; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2003; p. 20899. [CrossRef]
- Thuy Hang, D.T.; Trang, D.T.; Dung, D.T.; Hai Yen, D.T.; Hoang, N.H.; Bang, N.A.; Cuc, N.T.; Nhiem, N.X.; Thanh Huong, P.T.; Tai, B.H.; et al. Guaianolide sesquiterpenes and benzoate esters from the aerial parts of Siegesbeckia orientalis L. and their xanthine oxidase inhibitory activity. Phytochemistry 2021, 190, 112889. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Pauff, J.M.; Hille, R. Substrate Orientation and Catalytic Specificity in the Action of Xanthine Oxidase: The sequential hydroxylation of hypoxanthine to uric acid. J. Biol. Chem. 2010, 285, 28044–28053. [Google Scholar] [CrossRef]

| Protein | PDB ID | Native Ligand Name/Chemical Structure | Grid Box Center Coordinates and Size (Å) |
|---|---|---|---|
| Escherichia coli DNA gyrase | 1KZN | Clorobiocin | Grid center coordinates: x = 18.0206 y = 30.6795 z = 35.3259 Grid size along the axes: x = 14.0835 Å y = 15.9180 Å z = 19.3350 Å |
| Staphylococcus aureus D-alanine: D-alanine ligase (DDl1) | 2I80 | 3-chloro-2,2-dimethyl-N-[4-(trifluoromethyl)phenyl]propenamide | Grid center coordinates: x = 35.8534 y = 3.9116 z = 25.8999 Grid size along the axes: x = 10.2217 Å y = 12.8046 Å z = 10.2436 Å |
| Staphylococcus aureus Dihydrofolate reductase (DHFR) | 3SRW | 7-(2-ethoxynaphthalen-1-yl)-6-methylquinazoline-2,4-diamine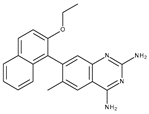 | Grid center coordinates: x = −4.5959 y = −29.9973 z = 6.2901 Grid size along the axes: x = 10.7001 Å y = 15.8621 Å z = 14.9382 Å |
| Staphylococcus aureus DNA gyrase subunit B | 3TTZ | 2-[(3S,4R)-4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}-3-fluoropiperidin-1-yl]-1,3-thiazole-5-carboxylic acid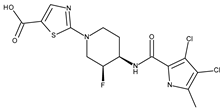 | Grid center coordinates: x = 16.5842 y = −18.6283 z = 6.6255 Grid size along the axes: x = 17.6122 Å y = 10.7091 Å z = 8.5457 Å |
| Acinetobacter baumannii Penicillin-binding protein 1a (PBP1a) | 3UDI | Penicillin G-open form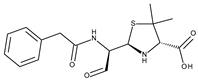 | Grid center coordinates: x = 34.3158 y = −1.1961 z = 12.2776 Grid size along the axes: x = 11.8683 Å y = 10.7091 Å z = 12.2516 Å |
| Lipoxygenase | 1N8Q | Protocatechuic acid | Grid center coordinates: x = 20.8705 y = 0.9784 z = 19.1589 Grid size along the axes: x = 9.4722 Å y = 6.7976 Å z = 10.6056 Å |
| CYP2C9 | 1OG5 | S-Warfarin | Grid center coordinates: x = −19.7808 y = 86.6496 z = 38.0890 Grid size along the axes: x = 11.6826 Å y = 11.6610 Å z = 10.9857 Å |
| NADPH-oxidase | 2CDU | Adenosine-5’-diphosphate | Grid center coordinates: x = 18.5709 y = −5.1763 z = −0.3019 Grid size along the axes: x = 17.8103 Å y = 15.7471 Å z = 15.5473 Å |
| Xanthine oxidase | 3NRZ | Hypoxanthine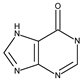 | Grid center coordinates: x = 38.1287 y = 19.4742 z = 18.4338 Grid size along the axes: x = 10.6615 Å y = 10.8740 Å z = 13.6877 Å |
| Common Name | RIcalc a | RIlit b | Area % |
|---|---|---|---|
| Terpenes | |||
| 3-Carene | 1029 | 1009 | 0.06 |
| Cedrane | 1444 | 1435 | 0.16 |
| α-Bergamotene | 1512 | - | 0.15 |
| β-Bisabolene | 1530 | 1547 | 0.25 |
| β-Copaene | 1535 | - | 0.15 |
| Guaiazulene | 1687 | 1772 | 0.47 |
| Sub-totals (Class compounds %) | 1.24 | ||
| Terpenoids | |||
| Globulol | 1473 | 1584 | 0.39 |
| Geranylgeraniol | 1593 | - | 0.35 |
| Caryophyllene oxide | 1602 | 1578 | 0.12 |
| Cadinol | 1659 | 1679 | 0.69 |
| Geranyl linalool | 1981 | 2034 | 3.93 |
| Sub-totals (Class compounds %) | 5.48 | ||
| Alcohols | |||
| 2,9-Octadecenyloxyethanol | 1715 | - | 0.36 |
| 2-Hexadecanol | 1885 | 1702 | 0.42 |
| 13-Heptadecyn-1-ol | 1898 | - | 0.16 |
| 3-Acetoxy-7, 8-epoxylanostan-11-ol | 2726 | - | 38.16 |
| Sub-totals (Class compounds %) | 39.1 | ||
| Esters | |||
| Ascabiol | 1770 | 1760 | 0.82 |
| Glycidol stearate | 2179 | - | 4.51 |
| Adipic acid, dioctyl ester | 2244 | - | 0.71 |
| 9,12-Octadecadienoic acid, 2-phenyl-1,3-dioxan-5-yl ester | 2295 | - | 9.21 |
| Ethyl iso-allocholate | 2577 | - | 4.71 |
| Sub-totals (Class compounds %) | 19.96 | ||
| Others | |||
| Nonanal | 1092 | 1089 | 0.33 |
| Quinol dimethyl ether | 1158 | 1163 | 0.53 |
| Anethole | 1297 | 1301 | 0.41 |
| 1,2,4-trimethoxybenzene | 1386 | 1377 | 0.21 |
| Geranyl acetone | 1468 | 1454 | 1.83 |
| Phytone | 1835 | 1838 | 1.25 |
| 9-Hexadecenoic acid | 1869 | 1942 | 0.16 |
| Heptacosane | 2186 | 429 | 3.71 |
| β-Monoolein | 2299 | - | 14.4 |
| 3-Ethyl-5-(2-ethylbutyl)-octadecane | 2441 | - | 1.16 |
| Lycopene, 1,2-dihydro-1-hydroxy- | 2749 | 4025 | 10.21 |
| Sub-totals (Class compounds %) | 34.2 | ||
| Total (Class compounds %) | 99.98 | ||
| Target PDB ID | 1KZN | 2I80 | 3SRW | 3TTZ | 3UDI |
|---|---|---|---|---|---|
| Docked EO component ID | Docking score. ∆G (kcal/mol) | ||||
| Native ligand | −9.3 | −8.1 | −10 | −8.4 | −7.3 |
| 1 | −5.3 | −5.7 | −5.6 | −5.2 | −4.7 |
| 2 | −4.8 | −5.4 | −4.6 | −4.9 | −3.9 |
| 3 | −4.9 | −4.7 | −4.8 | −4.8 | −4.1 |
| 4 | −5.5 | −6.4 | −5.5 | −5.5 | −4.7 |
| 5 | −5.1 | −5.2 | −5.1 | −5.2 | −4.8 |
| 6 | −6.4 | −3.4 | −7.6 | −6.6 | −5.9 |
| 7 | −6 | −7 | −5.9 | −5.9 | −5 |
| 8 | −5.8 | 1.20 | −8 | −6.7 | −6.1 |
| 9 | −6.4 | −6.6 | −6.5 | −6.9 | −4.9 |
| 10 | −6.9 | −6.8 | −6.7 | −6.3 | −5.8 |
| 11 | −6.6 | −5.6 | −7.4 | −6.5 | −6 |
| 12 | −6.5 | −6.7 | −7.3 | −6.9 | −5.6 |
| 13 | −5.4 | −2.2 | −7.3 | −6 | −6.1 |
| 14 | −6.5 | −0.7 | −7.6 | −6.9 | −6.3 |
| 15 | −6.2 | −6.4 | −5.9 | −6 | −5 |
| 16 | −5 | −5.5 | −5.6 | −5.1 | −4.9 |
| 17 | −6.9 | −7.6 | −6.9 | −7.1 | −6 |
| 18 | −5.5 | −5.9 | −6.2 | −5.6 | −5.3 |
| 19 | −5.7 | −6.5 | −5.9 | −5.6 | −5.4 |
| 20 | −5.5 | −6.2 | −5.6 | −5.3 | −4.5 |
| 21 | −4.9 | −5.8 | −5.7 | −5.5 | −5.2 |
| 22 | −6.4 | −6.6 | −7.1 | −6.2 | −5.9 |
| 23 | −5.4 | −5.6 | −6 | −5.3 | −5.1 |
| 24 | −4.9 | −2 | −5.8 | −4.8 | −4.7 |
| 25 | −5.3 | −4.1 | −6.1 | −5.5 | −5.5 |
| 26 | −5.7 | 0.9 | −7.3 | −6.1 | −6.6 |
| 27 | −5.4 | −5.2 | −6.4 | −5.5 | −5.4 |
| 28 | −5.3 | −2.9 | −6 | −5.6 | −5.1 |
| 29 | −7.1 | 3.40 | −7.1 | −8 | −6.1 |
| 30 | −7.6 | 16.40 | −7.1 | −7 | −5 |
| 31 | −6.4 | 26.10 | −6.9 | −4.5 | −5.8 |
| Target PDB ID | 1N8Q | 1OG5 | 2CDU | 3NRZ |
|---|---|---|---|---|
| Docked EO component ID | Docking score, ∆G (kcal/mol) | |||
| Native ligand | −5.8 | −9.8 | −9.2 | −6.7 |
| 1 | −5.2 | −5.6 | −6 | −4.6 |
| 2 | −4.9 | −4.9 | −4.5 | −5.7 |
| 3 | −4.4 | −4.6 | −4.8 | −6.7 |
| 4 | −4.9 | −5.5 | −5.6 | −7.5 |
| 5 | −4.5 | −4.8 | −5.2 | −5.6 |
| 6 | −2.2 | −7.1 | −6.9 | −0.8 |
| 7 | −4.8 | −6.1 | −6.3 | −6.5 |
| 8 | −0.9 | −7.4 | −6.7 | −1.6 |
| 9 | −5.3 | −7.4 | −7.3 | −4.9 |
| 10 | −1.6 | −7.4 | −6.3 | −4.5 |
| 11 | −1.7 | −7.5 | −7.3 | −2.7 |
| 12 | 0.3 | −6.9 | −7.4 | −8.1 |
| 13 | −0.8 | −7.2 | −6.7 | −1.3 |
| 14 | −2.2 | −7.2 | −7.3 | −3 |
| 15 | −6.3 | −6.8 | −6.2 | −7.9 |
| 16 | 1 | −5.9 | −6 | −6.2 |
| 17 | −4 | −7.9 | −7 | −8.2 |
| 18 | −1.6 | −6.1 | −6.2 | −6.3 |
| 19 | −2.2 | −5.9 | −6.3 | −6.6 |
| 20 | −4.3 | −5.4 | −5.5 | −6.6 |
| 21 | −2.4 | −5.5 | −5.3 | −6.7 |
| 22 | −0.6 | −6.8 | −7.1 | −6.1 |
| 23 | 4 | −6 | −5.5 | −6 |
| 24 | 10.40 | −6.1 | −5.8 | −4.2 |
| 25 | 4.80 | −6 | −6.5 | −5.8 |
| 26 | 17.40 | −7.5 | −8 | −0.3 |
| 27 | 4.40 | −5.8 | −6.1 | −4.1 |
| 28 | 5.90 | −6.3 | −6 | −4.3 |
| 29 | 35.3 | −7.8 | −7 | 8.70 |
| 30 | 71.5 | −6.1 | −7.1 | 19.60 |
| 31 | 76.9 | −4.3 | −8.3 | 28.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jianu, C.; Mioc, M.; Mioc, A.; Șoica, C.; Lukinich-Gruia, A.T.; Bujancă, G.; Rădulescu, M. Insights into the Paulownia Shan tong (Fortunei × Tomentosa) Essential Oil and In Silico Analysis of Potential Biological Targets of Its Compounds. Foods 2024, 13, 1007. https://doi.org/10.3390/foods13071007
Jianu C, Mioc M, Mioc A, Șoica C, Lukinich-Gruia AT, Bujancă G, Rădulescu M. Insights into the Paulownia Shan tong (Fortunei × Tomentosa) Essential Oil and In Silico Analysis of Potential Biological Targets of Its Compounds. Foods. 2024; 13(7):1007. https://doi.org/10.3390/foods13071007
Chicago/Turabian StyleJianu, Călin, Marius Mioc, Alexandra Mioc, Codruța Șoica, Alexandra Teodora Lukinich-Gruia, Gabriel Bujancă, and Matilda Rădulescu. 2024. "Insights into the Paulownia Shan tong (Fortunei × Tomentosa) Essential Oil and In Silico Analysis of Potential Biological Targets of Its Compounds" Foods 13, no. 7: 1007. https://doi.org/10.3390/foods13071007
APA StyleJianu, C., Mioc, M., Mioc, A., Șoica, C., Lukinich-Gruia, A. T., Bujancă, G., & Rădulescu, M. (2024). Insights into the Paulownia Shan tong (Fortunei × Tomentosa) Essential Oil and In Silico Analysis of Potential Biological Targets of Its Compounds. Foods, 13(7), 1007. https://doi.org/10.3390/foods13071007









