In-House Validation of Four Duplex Droplet Digital PCR Assays to Quantify GM Soybean Events
Abstract
1. Introduction
2. Materials and Methods
2.1. Reference Material
2.2. DNA Extraction and Assessment of DNA Purity
2.3. Sample Preparation
2.4. Specificity
2.5. Droplet Digital PCR and Analysis
2.6. Calculation of GM Content
2.7. In-House Validation
2.8. Measurement Uncertainty
- -
- Uncertainty of the lec concentration of the DNA solution extracted from the CRM (uc,CRM): estimated as the standard error of the mean of cCRM.
- -
- Uncertainty of the lec concentration of the DNA solution from the non-GM material (uc,NGM): estimated as the standard error of the mean of cNGM. Uncertainty associated with the certified value of the CRM used (uw,CRM): the standard uncertainty of the certified value taken.
- -
- Uncertainty associated with the volume of the taken GM DNA solution (uv,CRM): estimated as the standard error of the pipette volume taken from the technical specification of the pipette.
- -
- Uncertainty associated with the volume of the taken NGM DNA solution (uv,NGM): estimated as the standard error of the pipette volume taken from the technical specification of the pipette.
- u: combined standard uncertainty;
- u(wGM,mGM): standard uncertainty in the function of wGM and the weighed GM material;
- u(wGM,pGM): standard uncertainty in the function of wGM and the purity of GM material;
- u(wGM,mNGM): standard uncertainty in the function of wGM and the weighed non-GM material;
- u(wGM,pNGM): standard uncertainty in the function of wGM and the impurity of non-GM material.
3. Results and Discussion
3.1. Specificity Evaluation
3.2. Cross-Talk
3.3. Robustness
3.4. Dynamic Range
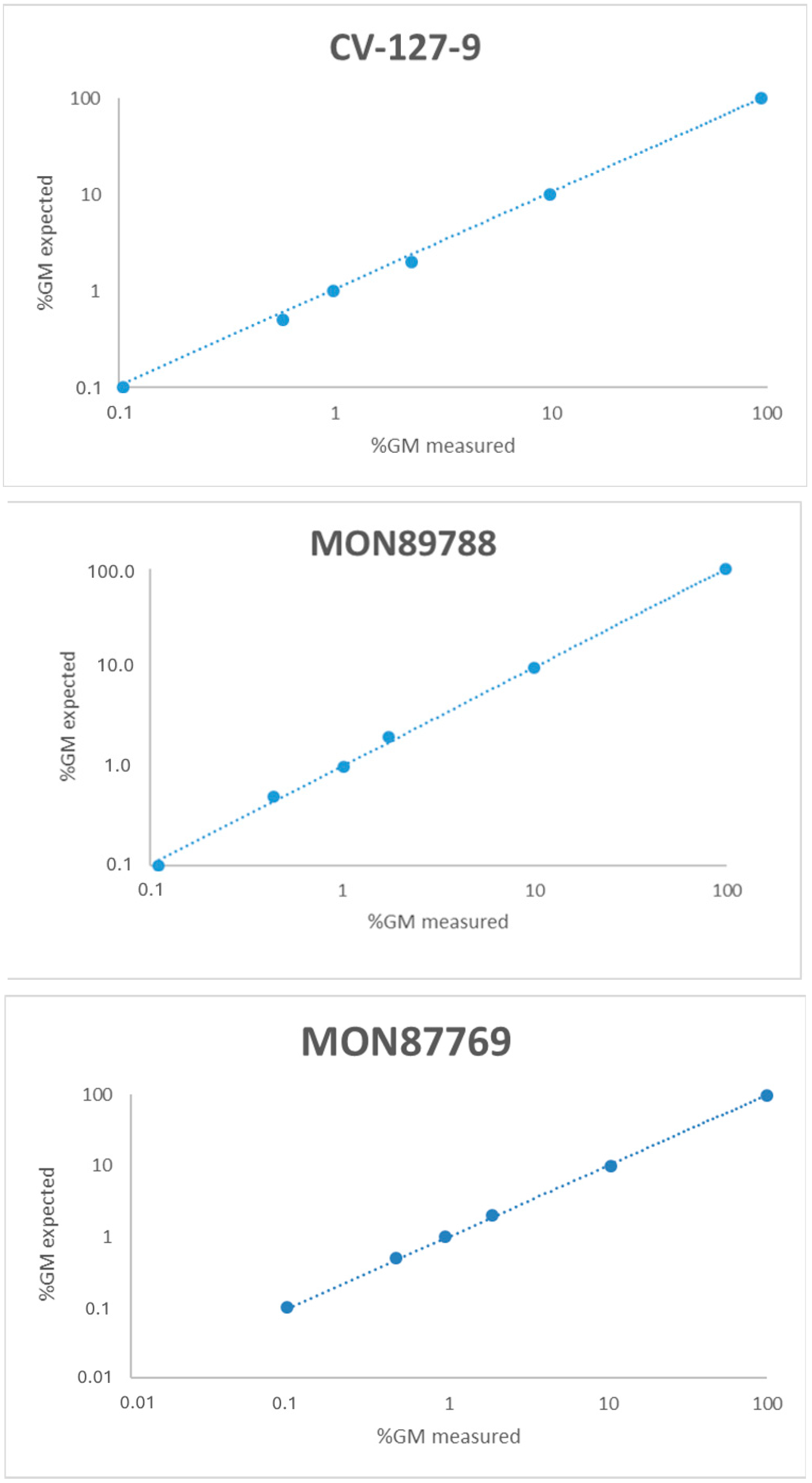
3.5. Linearity
3.6. Trueness
3.7. Precision
3.8. Asymmetric Limit of Quantification (LOQasym)
3.9. Measurement Uncertainty (MU)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Parliament, Council of the European Union. Regulation (EC) No 1829/2003 of the European Parliament and of the Council of 22 September 2003 on Genetically Modified Food and Feed; Publications Office of the European Union: Luxembourg, 2003. [Google Scholar]
- European Parliament, Council of the European Union. Regulation (EC) No 1830/2003 of the European Parliament and of the Council of 22 September 2003 Concerning the Traceability and Labelling of Genetically Modified Organisms and the Traceability of Food and Feed Products Produced from Genetically Modified Organisms and Amending Directive 2001/18/ECl; Publications Office of the European Union: Luxembourg, 2003. [Google Scholar]
- Biosafety Clearing House (BCH). Available online: https://bch.cbd.int/en/ (accessed on 5 February 2024).
- Luque-Perez, E.; Mazzara, M.; Weber, T.P.; Foti, N.; Grazioli, E.; Munaro, B.; Pinski, G.; Bellocchi, G.; Van Den Eede, G.; Savini, C. Testing the Robustness of Validated Methods for Quantitative Detection of GMOs Across qPCR Instruments. Food Anal. Methods 2012, 6, 343. [Google Scholar] [CrossRef][Green Version]
- Angers-Loustau, A.; Petrillo, M.; Bonfini, L.; Gatto, F.; Rosa, S.; Patak, A.; Kreysa, J. JRC GMO-Matrix: A web application to support Genetically Modified Organisms detection strategies. BMC Bioinform. 2014, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, S.; Berben, G.; Burns, M.; Corbisier, P.; De Giacomo, M.; De Loose, M.; Dagand, E.; Dobnik, D.; Eriksson, R.; Holst-Jensen, A.; et al. Overview and Recommendations for the Application of Digital PCR European Network of GMO Laboratories (ENGL); Publications Office of the European Union: Luxembourg, 2019. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.J.; Burrell, A.M.; Foy, C.A. The applicability of digital PCR for the assessment of detection limits in GMO analysis. Eur. Food Res. Technol. 2010, 231, 353. [Google Scholar] [CrossRef]
- Iwobi, A.; Gerdes, L.; Busch, U.; Pecoraro, S. Droplet digital PCR for routine analysis of genetically modified foods (GMO)—A comparison with real-time quantitative PCR. Food Control. 2020, 69, 205. [Google Scholar] [CrossRef]
- Corbisier, P.; Buttinger, G.; Savini, C.; Sacco, M.G.; Gatto, F.; Emons, H. Expression of GM content in mass fraction from digital PCR data. Food Control 2021, 133, 108626. [Google Scholar] [CrossRef] [PubMed]
- UNI CEI EN ISO/IEC 17025:2018; General Requirements for the Competence of Testing and Calibration Laboratories. ISO: Geneva, Switzerland, 2018.
- Mazzara, M.; Delobel, C.; Savini, C. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. European Network of GMO Laboratories (ENGL); Publications Office of the European Union: Luxembourg, 2008. [Google Scholar] [CrossRef]
- Grohmann, L.; Barbante, A.; Eriksson, R.; Gatto, F.; Georgieva, T.; Huber, I.; Hulin, J.; Köppel, R.; Marchesi, U.; Marmin, L.; et al. Guidance Document on Multiplex Real-Time PCR Methods; EUR 30708 EN; Publications Office of the European Union: Luxembourg, 2021; ISBN 978-92-76-37820-4. [Google Scholar]
- Mazzara, M.; Savini, J.; Delobel, J.; Broll, H.; Damant, A.; Paoletti, J.; Van den Eede, G. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing Part 2 European Network of GMO Laboratories European Union Reference Laboratory for Genetically Modified Food and Feed—Part 2; Publications Office of the European Union: Luxembourg, 2023. [Google Scholar] [CrossRef]
- Long, L.; Yan, W.; He, Y.; Dong, L.; Xing, Z.; Li, C.; Xia, W.; Li, F. Development of a Duplex Digital PCR Method to Quantify Five Genetically Modified Soybean Events. Food Anal. Methods 2021, 15, 294. [Google Scholar] [CrossRef]
- Demeke, T.; Lee, S.; Eng, M. Increasing the Efficiency of Canola and Soybean GMO Detection and Quantification Using Multiplex Droplet Digital PCR. Biology 2022, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Dobnik, D.; Štebih, D.; Blejec, A.; Morisset, D.; Žel, J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci. Rep. 2016, 6, 35451. [Google Scholar] [CrossRef] [PubMed]
- Nijland, J.A.B.; Hokken, M.W.J.; Prins, T.W. Set-up and in-house validation of two multiplex event-specific PCR methods to simplify screening for GMOs without common screening elements. J. Food Compos. Anal. 2023, 125, 105722. [Google Scholar] [CrossRef]
- Gatto, F.; Savini, C.; Sacco, M.G.; Vinciguerra, D.; Buttinger, G.; Corbisier, P.; Mazzara, M.; Emons, H. Single and multi-laboratory validation of a droplet digital PCR method. Food Control 2022, 140, 109117. [Google Scholar] [CrossRef] [PubMed]
- Košir, A.B.; Spilsberg, B.; Holst-Jensen, A.; Žel, J.; Dobnik, D. Development and inter-laboratory assessment of droplet digital PCR assays for multiplex quantification of 15 genetically modified soybean lines. Sci. Rep. 2017, 7, 8601. [Google Scholar] [CrossRef] [PubMed]
- Charles Delobel, C.; Bonfini, L.; Querci, M.; Mazzara, M.; Cordeil, S.; Van Den Eede, G. Event-Specific Method for the Quantification of Soybean MON 87701 Using Real-Time PCR—Validation Report and Protocol. 2011. Available online: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed on 5 February 2024).
- Savini, C.; Bonfini, L.; Querci, M.; Mazzara, M.; Cordeil, S.; Van den Eede, G. Event-specific Method for the Quantification of Soybean MON87769 Using Real-Time PCR—Validation Report and Protocol. 2012. Available online: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed on 5 February 2024).
- Delobel, C.; Bogni, A.; Pinski, G.; Mazzara, M.; Van Den Eede, G. Event-Specific Method for the Quantification of Soybean Line MON 89788 Using Real-Time PCR V. 1.01—Validation Report and Protocol; EUR 26153; Publications Office of the European Union: Luxembourg, 2013. [Google Scholar] [CrossRef]
- Savini, C.; Querci, M.; Bonfini, L.; Mazzara, M.; Cordeil, S.; Van den Eede, G. Event-Specific Method for the Quantification of Soybean CV127 Using Real-Time PCR—Validation Report and Protocol. 2011. Available online: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed on 5 February 2024).
- Trapmann, S.; Burns, M.; Corbisier, P.; Gatto, F.; Robouch, P.; Sowa, S.; Emons, H. Guidance Document on Measurement Uncertainty for GMO Testing Laboratories, 3rd ed.; Publications Office of the European Union: Luxembourg, 2020. [Google Scholar] [CrossRef]
- Hougs, L.; Gatto, F.; Goerlich, O.; Grohmann, L.; Lieske, K.; Mazzara, M.; Narendja, F.; Ovesna, J.; Papazova, N.; Scholtens, I. Verification of Analytical Methods for GMO Testing When Implementing Interlaboratory Validated Methods: Version 2; Publications Office of the European Union: Luxembourg, 2017. [Google Scholar] [CrossRef]
- ISO 21571:2013 Amd.1-2013; Foodstuffs—Methods of Analysis for the Detection of Genetically Modified Organisms and Derived Products—Nucleic Acid Extraction—Amendment 1. ISO: Geneva, Switzerland, 2013.
- Sinha, M.; Mack, H.; Coleman, T.P.; Fraley, S.I. A High-Resolution Digital DNA Melting Platform for Robust Sequence Profiling and Enhanced Genotype Discrimination. SLAS Technol. 2018, 23, 580. [Google Scholar] [CrossRef] [PubMed]
- BioRad. Droplet Digital™ PCR Droplet Digital™ PCR Applications Guide; BioRad: Hercules, CA, USA, 2018. [Google Scholar]
- EURL GMFF: European Union Reference Laboratories for Genetically Modified Food and Feed. Use of the Harmonised Conversion Factors to Transform PCR Results from the DNA Copy Number Ratio Domain into the Mass Fraction Domain: Application Note V1.0; European Union: Brussels, Belgium, 2019. [Google Scholar]
- Gudnason, H.; Dufva, M.; Bang, D.D.; Wolff, A. Comparison of multiple DNA dyes for real-time PCR: Effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007, 35, e127. [Google Scholar] [CrossRef] [PubMed]
- Trapmann, S.; Delobel, C.; Corbisier, P.; Emons, H.; Hougs, L.; Philipp, P.; Sandberg, M.; Schulze, M. European Technical Guidance Document for the Flexible Scope Accreditation of Laboratories Quantifying GMOs; Publications Office of the European Union: Luxembourg, 2013; ISBN 978-92-79-26176-3. [Google Scholar] [CrossRef]
- Rychlik, W. Selection of Primers for Polymerase Chain Reaction. PCR Protoc. 1993, 15, 31–40. [Google Scholar] [CrossRef]
- Breslauert, K.J.; Franks, R.; Blockers, H.; Markyt, L.A. Predicting DNA duplex stability from the base sequence (nearest-neighbor interactions/DNA structure/thermodynamics). Biochemistry 1986, 83, 3746–3750. [Google Scholar]
- Gunson, R.N.; Bennett, S.; Maclean, A.; Carman, W.F. Using multiplex real time PCR in order to streamline a routine diagnostic service. J. Clin. Virol. 2008, 43, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). Guidelines for the Verification of Methods Using Digital PCR. 2024. Available online: https://www.bvl.bund.de/SharedDocs/Downloads/07_Untersuchungen/Guidelines_for_the_verification_of_methods_using_digital_PCR.html (accessed on 31 October 2024).
- Montgomery, D.C. Design and Analysis of Experiments; John Wiley & Sons: Hoboken, NJ, USA, 1991. [Google Scholar]
- Corbisier, P.; Barbante, A.; Berben, G.; Broothaerts, W.; De Loose, M.; Emons, H.; Georgieva, T.; Lievens, A.; Mazzara, M.; Papazova, N.; et al. Recommendation for the Unit of Measurement and the Measuring System to Report Traceable and Comparable Results Expressing GM Content in Accordance with EU Legislation; Publications Office of the European Union: Luxembourg, 2017. [Google Scholar] [CrossRef]
- UNI CEI EN ISO/IEC 5725-2:2019; Accuracy (Trueness and Precision) of Measurement Methods and Results, Part 2: Basic Method for the Determination of Repeatability and Reproducibility of a Standard Measurement Method. ISO: Geneva, Switzerland, 2019.

| EXPERIMENTAL CONDITIONS | DATA | ACCEPTANCE CRITERIA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| INVESTIGATED PARAMETERS | Starting Material | Practical Approach | PCR Assay Design | Copies/Reaction per PCR Mix | Collecting Output and Analysis | Accepted Reference Value | Ref Source | ||||
| PCR Replicates | PCR Run | N. Operator | |||||||||
| Specificity (In silico theoretical test) | All oligonucleotides sequences combination for each target | Each duplex system was investigated through alignment with appropriate softwares to assess the generation of no specific PCR products e.g., cross-dimer or self-dimers | N.A. | N.A. | N.A. | N.A. | ∆G evaluation and secondary structure generation | Absence of unexpected/unwanted non-target amplification: ∆G ≤9 Kcal/mol | [14] | ||
| Experimental specificity (melting curve analysis of the PCR products) | DNA extracted from CRMs: AOCS 0809-A2, AOCS 0809-B2, AOCS 0906-B2, AOCS 0911-C2, AOCS 0906-A, AOCS 0911-A | DNA melting curve analysis using EvaGreen chemistry. qPCR run was performed using pair of oliognucleotides to detect GM event specific and endogeneous gene | 2 | 1 | 1 | N.A. | T melting analysis | No additional peak should be observed | [31] | ||
| Cross-talk | DNA extracted from CRMs non-modified soybean: AOCS 0906-A, AOCS 0911-A | Measured the fluorescence signal in presence of an excess of lectin gene and in absence of each GM soybean event | 3 | 1 | 1 | GM gene: 0 copies. Taxon-specific target:—48270 lectin copies determined for AOCS 0906-A.—33696 lectin copies determined for AOCS 0911-A | The fluorescence signals generated during the amplification of different targets evaluation | No cross-talk should be detected, but minimal cross-talk if below the fluorescence threshold is acceptable | [13] | ||
| Robustness | DNA extracted from: AOCS 0809-A2, AOCS 0809-B2, AOCS 0906-B2, AOCS 0911-C2 | Soybean DNA 0.1% GM (LOQasym value) prepared considering the practical dilution factor by measuring the content of Lectin reference gene for the GM positive and a GM negative DNA according to the formula described by Hougs et al., 2017, JRC [26] | 6 replicates per condition | 4 conditions tested, changing of: Temp ramp rate; Annealing temp; Oligont conc; Master mix vol | 1 | MON87701 0.1% (34.2 cp/rxn), MON87769 0.1% (24.6 cp/rxn), MON89788 0.1% (29 cp/rxn), CV-127-9 0.1% (22.4 cp/rxn) | The copy number for both GM event and lectin gene evaluation | Precision and trueness should not exceed 30% for all combinations | [14] | ||
| Dynamic range of the lectin gene | DNA extracted from CRMs non-modified soybean: AOCS 0906-A | A dilution series of conventional soybean at five different lectin copy number (10525, 5262, 2630, 263, 43) | 16 replicates per dilution level | 5 | 1 | 10525 cp/rxn (2100 cp/uL); 5262 cp/rxn (1050 cp/uL); 2630 cp/rxn (526 cp/uL); 263 cp/rxn (53 cp/uL); 43 cp/rxn (9 cp/uL) | R2 and slope evaluation between the theoretical and observed values of copy number for lectin gene. Outlier evaluation performed by Grubb’s test | The R2 should be ≥0.98% while slope should be 1.00 ± 0.25 | [13] | ||
| Dynamic range (duplex system for each event) | DNA extracted from: AOCS 0809-A2, AOCS 0809-B2, AOCS 0906-B2, AOCS 0911-C2 | GM soybean 0.1%, 0.5%, 1%, 2%, 10% prepared considering the practical dilution factor as described by Hougs et al., 2017, JRC [26] | 6 replicates per GM level tested | 5 runs 5 days | 1 | Six content level from 100% to 0.1% GM (0.1%, 0.5%, 1%, 2%, 10%, 100%) | R2 and slope evaluation between the theoretical and observed values of GM %. Outlier evaluation performed by Grubb’s test | The R2 should be ≥0.98% while slope should be 1.00 ± 0.25 | [13] | ||
| Linearity | GM target range; 0.1–100% GM | Regression analysis. Outlier evaluation performed by Grubb’s test | The slope of a plot observed vs expected value should be 1.00 ± 0.25 R2 ≥ 0.98% | [14] | |||||||
| Trueness (bias) | Six content level from 100% to 0.1% GM (0.1%, 0.5%, 1%, 2%, 10%, 100%) per each level 30 values are considered | Anova one-way and outliers evaluation by Grubb’s test | Bias ≤ 25% | [14] | |||||||
| Precision (RSDr) | RSDr ≤ 25% | [14] | |||||||||
| LOQasym | For each event 0.1% GM prepared considering the practical dilution factor as described by Hougs et al., 2017, JRC [26] | ≥60 replicates per module | 1 | 1 | GM target | Lec | Outlier evaluation performed by Grubb’s test | RSDr ≤ 25% Trueness within a bias of ±25% of the reference value (theorical value of the 0.1% GM prepared in laboratory with 100% GM and non-modified soybean) | [13] | ||
| cp/uL | cp/uL | ||||||||||
| MON87701 | 1.71 | 1776 | |||||||||
| MON87769 | 1.23 | 1316 | |||||||||
| MON89788 | 1.45 | 1411 | |||||||||
| CV-127-9 | 1.12 | 1165 | |||||||||
| Protocol | MON87701 | MON87769 | MON89788 | CV-127-9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (%) | RSDr (%) | Bias (%) | Mean (%) | RSDr (%) | Bias (%) | Mean (%) | RSDr (%) | Bias (%) | Mean (%) | RSDr (%) | Bias (%) | ||
| Ramp rate change and annealing temp unchanged | Original | 0.08 | 19.1 | 15.92 | 0.08 | 20.15 | 15.3 | 0.09 | 23.4 | 13.67 | 0.09 | 4.7 | 6.77 |
| −10% Primer Probe −10% Master Mix | 0.09 | 25.06 | 11.82 | 0.09 | 20.83 | 7.72 | 0.08 | 7.35 | 23.43 | 0.1 | 10.19 | 2.42 | |
| −10% Primer Probe | 0.1 | 16.92 | 2.48 | 0.09 | 11.15 | 7.12 | 0.08 | 21.06 | 17.91 | 0.09 | 14.02 | 7.04 | |
| −10% Master Mix | 0.1 | 21.4 | 0.28 | 0.1 | 11.82 | 0.38 | 0.07 | 20.9 | 25.31 | 0.09 | 3.03 | 6.42 | |
| Annealing temp +1 °C | No Change PCR reagents | 0.09 | 13.26 | 12.62 | 0.1 | 26.81 | 3.49 | 0.08 | 7.57 | 18.27 | 0.1 | 22.92 | 4.56 |
| −10% Primer Probe −10% Master Mix | 0.11 | 17.7 | 5.7 | 0.1 | 4.38 | 4.29 | 0.09 | 13.99 | 14.54 | 0.09 | 22.08 | 13.59 | |
| −10% Primer Probe | 0.08 | 10.23 | 16.3 | 0.1 | 24.54 | 0.28 | 0.08 | 7.83 | 20.29 | 0.1 | 26.61 | 0.24 | |
| −10% Master Mix | 0.09 | 15.88 | 10.99 | 0.11 | 7.37 | 14.57 | 0.08 | 9 | 21.87 | 0.11 | 5.35 | 11.64 | |
| Annealing temp +1 °C and ramp rate + 0.5 °C/s | No Change PCR Reagents | 0.08 | 8.47 | 15.42 | 0.09 | 23.12 | 12.36 | 0.08 | 14.02 | 22.24 | 0.11 | 18.47 | 14.44 |
| −10% Primer Probe −10% Master Mix | 0.09 | 2.28 | 9.97 | 0.11 | 5.86 | 6.52 | 0.09 | 21.28 | 12.33 | 0.09 | 15.78 | 8.63 | |
| −10% Primer Probe | 0.08 | 26.07 | 16.72 | 0.08 | 19.44 | 16.88 | 0.08 | 11.65 | 23.07 | 0.10 | 15.37 | 0.94 | |
| −10% Master Mix | 0.07 | 16.23 | 26.55 | 0.08 | 12.73 | 17.12 | 0.08 | 16.81 | 24.81 | 0.09 | 15.69 | 8.86 | |
| Ramp rate + 0.5 °C/s | No Change PCR Reagents | 0.08 | 6.66 | 23.13 | 0.08 | 21.42 | 18.85 | 0.09 | 16.92 | 14.66 | 0.11 | 24.38 | 11.89 |
| −10% Primer Probe −10% Master Mix | 0.08 | 21.44 | 15.23 | 0.08 | 12.64 | 15.27 | 0.08 | 12.93 | 21.6 | 0.08 | 24.78 | 16.01 | |
| −10% Primer Probe | 0.12 | 24.48 | 24.53 | 0.11 | 15.35 | 12.62 | 0.08 | 19.05 | 19.19 | 0.1 | 13.76 | 0.21 | |
| −10% Master Mix | 0.1 | 5.56 | 2.78 | 0.08 | 10.27 | 19.45 | 0.08 | 26.92 | 23.69 | 0.12 | 11.28 | 20.95 | |
| GM EVENT | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MON87701 | MON87769 | MON89788 | CV-127-9 | |||||||||
| GM Level (%) | Measured Mean (%) | Bias (%) | RSDr (%) | Measured Mean (%) | Bias (%) | RSDr (%) | Measured Mean (%) | Bias (%) | RSDr (%) | Measured Mean (%) | Bias (%) | RSDr (%) |
| 0.1 | 0.1 | 0.48 | 17.82 | 0.1 | 3.31 | 19.1 | 0.11 | 11.25 | 24.27 | 0.1 | 4.72 | 23.39 |
| 0.5 | 0.47 | 6.44 | 10.92 | 0.46 | 6.5 | 8.63 | 0.44 | 12.19 | 14.51 | 0.57 | 14.53 | 13.9 |
| 1 | 1 | 0.41 | 7.64 | 0.95 | 4.67 | 8.29 | 1.01 | 1.3 | 8.74 | 0.98 | 2.01 | 8.83 |
| 2 | 1.92 | 4.08 | 4.91 | 1.87 | 6.29 | 3.67 | 1.74 | 13.16 | 12.72 | 2.25 | 12.54 | 11.49 |
| 10 | 10.38 | 3.77 | 3.05 | 10.49 | 4.87 | 3.13 | 9.96 | 0.38 | 3.31 | 9.9 | 0.99 | 1.81 |
| 100 1 | 100.22 | 1.85 | 2.04 | 99.53 | 0.07 | 1.66 | 98.6 | 1 | 1.97 | 93.63 | 2.77 | 2.37 |
| MON87701 | MON89788 | MON87769 | CV-127-9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | R2 | Slope | Intercept | R2 | Slope | Intercept | R2 | Slope | Intercept | R2 | |
| R 1 | 0.98 | −0.01 | 1.00 | 1.01 | −0.01 | 1.00 | 1.02 | −0.07 | 1.00 | 1.05 | −0.16 | 1.00 |
| R 2 | 0.98 | 0.21 | 1.00 | 1.01 | 0.05 | 1.00 | 1.00 | −0.06 | 1.00 | 1.06 | −0.24 | 1.00 |
| R 3 | 0.99 | −0.01 | 1.00 | 1.01 | 0.09 | 1.00 | 1.00 | −0.07 | 1.00 | 1.07 | −0.21 | 1.00 |
| R 4 | 0.99 | −0.03 | 1.00 | 1.02 | 0.03 | 1.00 | 1.00 | −0.05 | 1.00 | 1.06 | −0.21 | 1.00 |
| R 5 | 0.97 | 0.05 | 1.00 | 1.00 | 0.05 | 1.00 | 0.99 | 0.004 | 1.00 | 1.07 | −0.22 | 1.00 |
| Mean | 0.98 | 0.04 | 1.00 | 1.01 | 0.04 | 1.00 | 1.00 | −0.05 | 1.00 | 1.06 | −0.21 | 1.00 |
| GM Level (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Performance Parameters | MON87701 | MON87769 | ||||||||||
| 0.1 | 0.5 | 1 | 2 | 10 | 98.40 | 0.1 | 0.5 | 1 | 2 | 10 | 99.60 | |
| N replicates | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 |
| N outliers | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Statistical outlier evaluation | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| Measured GM content (%) | 0.1 | 0.47 | 1 | 1.92 | 10.38 | 100.22 | 0.1 | 0.46 | 0.95 | 1.87 | 10.49 | 99.53 |
| Sr | 0.018 | 0.051 | 0.076 | 0.094 | 0.317 | 2.04 | 0.018 | 0.04 | 0.079 | 0.12 | 0.328 | 1.65 |
| RSDr % | 17.82 | 10.92 | 7.64 | 4.91 | 3.053 | 2.04 | 19.1 | 8.63 | 8.29 | 6.34 | 3.13 | 1.66 |
| Ubias | 0.31 | 0.48 | 1.52 | 3.62 | 7.52 | 8.58 | 0.32 | 0.38 | 1.56 | 5.60 | 7.35 | 5.92 |
| Bias | 0.0005 | 0.032 | 0.004 | 0.082 | 0.377 | 1.82 | 0.003 | 0.03 | 0.05 | 0.13 | 0.49 | 0.07 |
| Bias % | 0.48 | 6.44 | 0.41 | 4.08 | 3.77 | 1.85 | 3.31 | 6.5 | 4.67 | 6.29 | 4.87 | 0.07 |
| Difference with certified value 2 | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| Expanded uncertainty (%, with k = 2) | 0.03 | 0.05 | 0.15 | 0.36 | 0.75 | 0.91 | 0.03 | 0.05 | 0.16 | 0.56 | 0.74 | 0.81 |
| GM level (%) | ||||||||||||
| Performance parameters | MON89788 | CV-127-9 | ||||||||||
| 0.1 | 0.5 | 1 | 2 | 10 | 99.60 | 0.1 | 0.5 | 1 | 2 | 10 | 96.30 | |
| N replicates | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 |
| N outliers | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Statistical outlier evaluation | G 1 | N.A. | N.A. | N.A. | G 1 | G 1 | N.A. | N.A. | N.A. | N.A. | G 1 | N.A. |
| Measured GM content (%) | 0.11 | 0.44 | 1.01 | 1.74 | 9.96 | 98.6 | 0.1 | 0.57 | 0.98 | 2.25 | 9.9 | 93.63 |
| Sr | 0.027 | 0.06 | 0.089 | 0.22 | 0.33 | 1.94 | 0.02 | 0.08 | 0.09 | 0.26 | 0.18 | 2.22 |
| RSDr % | 24.27 | 14.51 | 8.74 | 12.72 | 3.31 | 1.97 | 23.39 | 13.9 | 8.83 | 11.49 | 1.81 | 2.37 |
| Ubias | 0.38 | 0.23 | 1.89 | 2.66 | 6.92 | 3.98 | 0.6 | 0.51 | 3.11 | 6 | 16.07 | 20.15 |
| Bias | 0.01 | 0.06 | 0.01 | 0.26 | 0.04 | 1 | 0.004 | 0.07 | 0.02 | 0.25 | 0.1 | 2.67 |
| Bias % | 11.25 | 12.19 | 1.3 | 13.16 | 0.38 | 1 | 4.72 | 14.53 | 2.01 | 12.54 | 0.99 | 2.77 |
| Difference with certified value 2 | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| Expanded uncertainty (%, with k = 2) | 0.04 | 0.05 | 0.19 | 0.34 | 0.7 | 0.53 | 0.06 | 0.05 | 0.31 | 0.6 | 1.61 | 2.22 |
| MON87701 | MON87769 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Performance Parameters | qPCR | ddPCR | qPCR | ddPCR | ||||||||||||||||||
| 0.085 | 0.26 | 0.9 | 2.7 | 8.1 | 0.1 | 0.5 | 1.00 | 2.00 | 10.00 | 98.4 ± 0.8 | 0.1 | 0.5 | 0.9 | 5.0 | 9.0 | 0.1 | 0.5 | 1.00 | 2.00 | 10.00 | 99.6 ± 0.2 | |
| N replicates | 48 | 48 | 48 | 48 | 48 | 30 | 30 | 30 | 30 | 30 | 30 | 48 | 48 | 48 | 48 | 48 | 30 | 30 | 30 | 30 | 30 | 30 |
| N outliers | 1 | 1 | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Statistical outlier evaluation | 1C | 1G | 1C 1G | 1C 2DG | 2C | - | - | - | - | - | - | 1C | - | 2C | 1C | - | - | - | - | - | - | - |
| Measured GM content (%) | 0.09 | 0.28 | 0.95 | 2.85 | 8.11 | 0.1 | 0.47 | 1 | 1.92 | 10.38 | 100 | 0.09 | 0.47 | 0.88 | 5.2 | 9.2 | 0.1 | 0.46 | 0.95 | 1.87 | 10.49 | 99.5 |
| Sr | 0.02 | 0.06 | 0.15 | 0.4 | 0.82 | 0.018 | 0.05 | 0.076 | 0.094 | 0.317 | 2.04 | 0.01 | 0.07 | 0.09 | 0.37 | 1.25 | 0.02 | 0.04 | 0.08 | 0.07 | 0.328 | 1.65 |
| RSDr % | 18 | 21 | 15 | 14 | 10 | 17.82 | 10.9 | 7.64 | 4.91 | 3.053 | 2.04 | 13 | 14 | 9.7 | 7 | 14 | 19.1 | 8.63 | 8.29 | 3.67 | 3.13 | 1.66 |
| Bias | 0.007 | 0.02 | 0.05 | 0.15 | 0.01 | 0.001 | 0.03 | 0.004 | 0.082 | 0.377 | 1.82 | −0.01 | −0.03 | −0.02 | 0.23 | 0.16 | 0 | 0.003 | 0.05 | 0.13 | 0.49 | 0.07 |
| Bias % | 8.6 | 6.4 | 5.2 | 5.6 | 0.1 | 0.48 | 6.44 | 0.41 | 4.08 | 3.77 | 1.85 | −5.8 | −5.2 | −2.2 | 4.6 | 1.8 | 3.31 | 6.5 | 4.67 | 6.29 | 4.87 | 0.07 |
| MON89788 | CV-127-9 | |||||||||||||||||||||
| Performance parameters | qPCR | ddPCR | qPCR | ddPCR | ||||||||||||||||||
| 0.1 | 0.4 | 0.9 | 4.00 | 8.00 | 0.1 | 0.5 | 1.00 | 2.00 | 10.00 | 99.6 ± 0.2 | 0.09 | 0.3 | 0.9 | 2.5 | 4.5 | 0.1 | 0.5 | 1.00 | 2.00 | 10.00 | 96.3 ± 1.8 | |
| N replicates | 48 | 48 | 48 | 48 | 48 | 30 | 30 | 30 | 30 | 30 | 30 | 48 | 48 | 48 | 48 | 48 | 30 | 30 | 30 | 30 | 30 | 30 |
| N outliers | 3 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Statistical outlier evaluation | 2C 1G | 1C | 1C | 1C | 1C | 1G | - | - | - | 1G | 1G | - | C | - | C | C | - | - | - | - | 1G | - |
| Measured GM content (%) | 0.09 | 0.38 | 0.89 | 4.42 | 8.22 | 0.11 | 0.44 | 1.01 | 1.74 | 9.96 | 98.6 | 0.1 | 0.32 | 0.92 | 2.68 | 4.76 | 0.1 | 0.57 | 0.98 | 2.25 | 9.9 | 93.6 |
| Sr | 0.01 | 0.08 | 0.13 | 0.57 | 0.99 | 0.027 | 0.06 | 0.089 | 0.22 | 0.33 | 1.94 | 0.01 | 0.036 | 0.144 | 0.19 | 0.44 | 0.02 | 0.08 | 0.09 | 0.26 | 0.18 | 2.22 |
| RSDr % | 16.2 | 21.7 | 14.9 | 12.9 | 12 | 24.27 | 14.5 | 8.74 | 12.72 | 3.31 | 1.97 | 11 | 11 | 16 | 7.1 | 9.2 | 23.4 | 13.9 | 8.83 | 11.5 | 1.81 | 2.37 |
| Bias | −0.01 | −0.02 | −0.01 | 0.42 | 0.22 | 0.01 | 0.06 | 0.01 | 0.26 | 0.04 | 1 | 0.01 | 0.02 | 0.02 | 0.18 | 0.26 | 0 | 0.07 | 0.02 | 0.25 | 0.1 | 2.67 |
| Bias % | −14.1 | −5 | −0.9 | 10.5 | 2.8 | 11.25 | 12.2 | 1.3 | 13.16 | 0.38 | 1 | 8.1 | 7.8 | 2.5 | 7.3 | 5.9 | 4.72 | 14.53 | 2.01 | 12.5 | 0.99 | 2.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verginelli, D.; Ciuffa, S.; Spinella, K.; La Rocca, D.; Misto, M.; Quarchioni, C.; Bonini, P.; Fusco, C.; Peroni, L.; Peddis, S.; et al. In-House Validation of Four Duplex Droplet Digital PCR Assays to Quantify GM Soybean Events. Foods 2024, 13, 4011. https://doi.org/10.3390/foods13244011
Verginelli D, Ciuffa S, Spinella K, La Rocca D, Misto M, Quarchioni C, Bonini P, Fusco C, Peroni L, Peddis S, et al. In-House Validation of Four Duplex Droplet Digital PCR Assays to Quantify GM Soybean Events. Foods. 2024; 13(24):4011. https://doi.org/10.3390/foods13244011
Chicago/Turabian StyleVerginelli, Daniela, Sara Ciuffa, Katia Spinella, Davide La Rocca, Marisa Misto, Cinzia Quarchioni, Pamela Bonini, Cristiana Fusco, Lorella Peroni, Stefania Peddis, and et al. 2024. "In-House Validation of Four Duplex Droplet Digital PCR Assays to Quantify GM Soybean Events" Foods 13, no. 24: 4011. https://doi.org/10.3390/foods13244011
APA StyleVerginelli, D., Ciuffa, S., Spinella, K., La Rocca, D., Misto, M., Quarchioni, C., Bonini, P., Fusco, C., Peroni, L., Peddis, S., & Marchesi, U. (2024). In-House Validation of Four Duplex Droplet Digital PCR Assays to Quantify GM Soybean Events. Foods, 13(24), 4011. https://doi.org/10.3390/foods13244011






