Lipidomic Profiling and Storage-Induced Changes in Cassava Flour Using LC-MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Sample Preparation and Extraction
2.3. HPLC Conditions
2.4. ESI-MS/MS Conditions
2.5. Lipid Qualitative and Quantitative Principle
2.6. Statistics
3. Results and Discussion
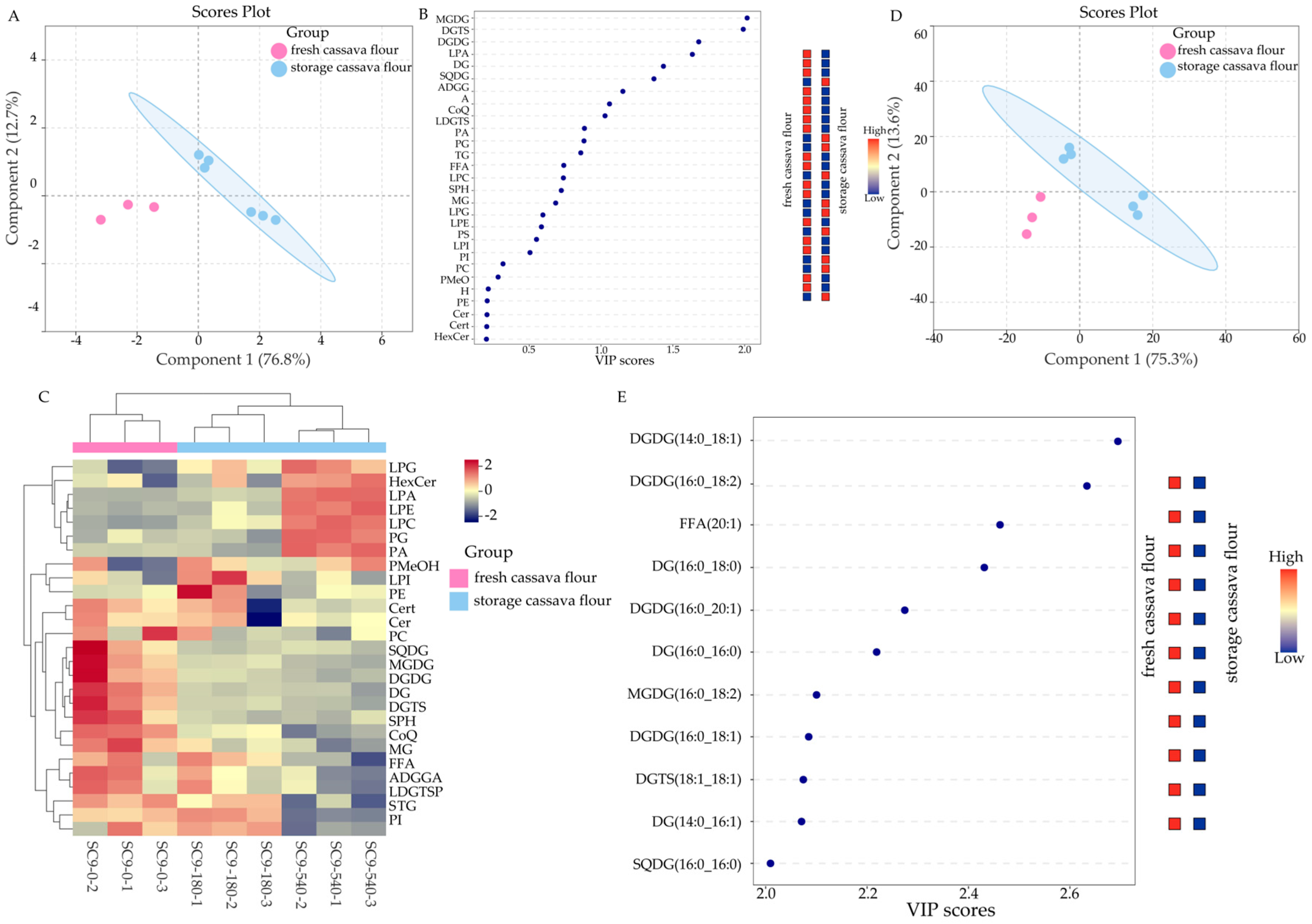
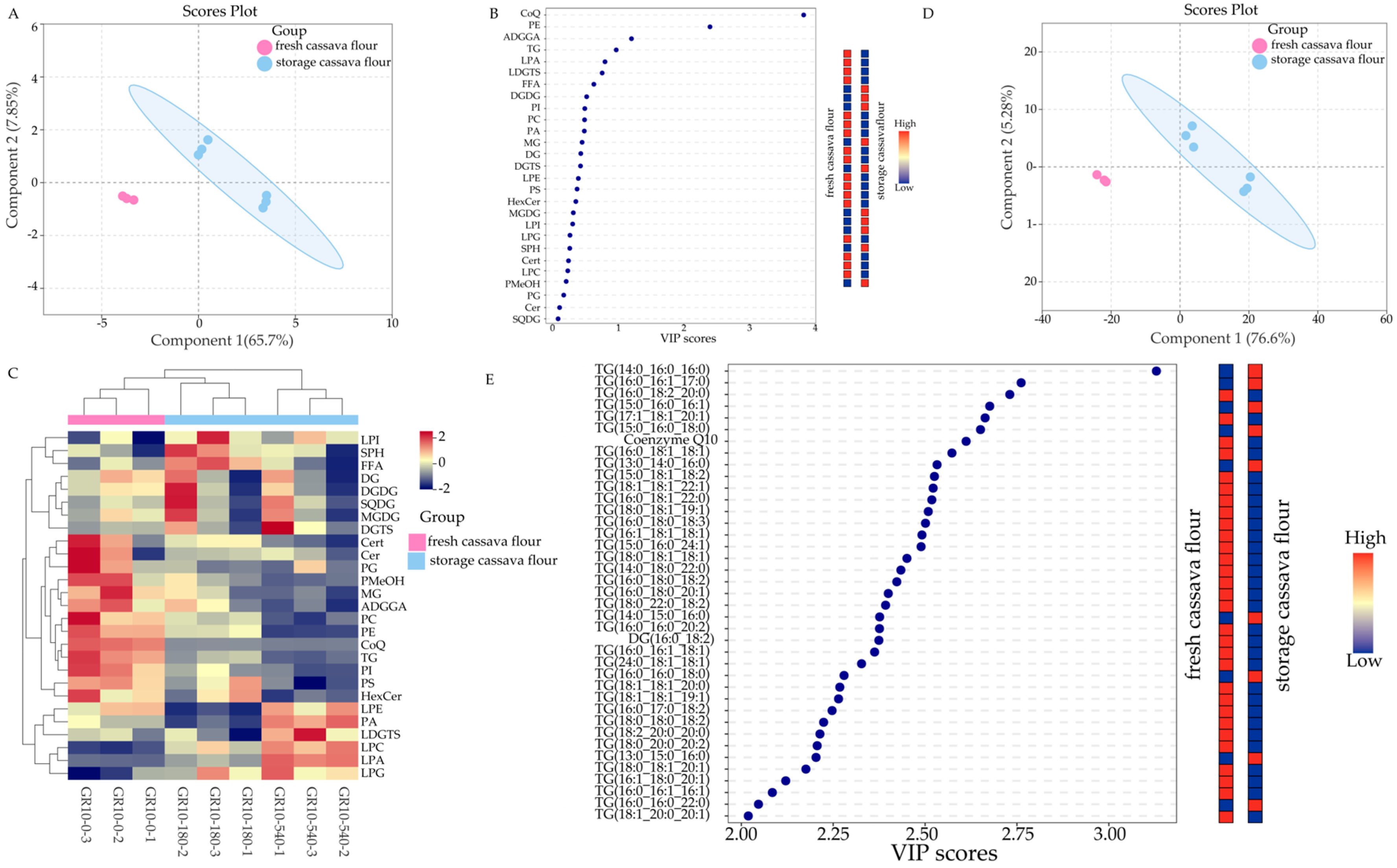
3.1. Qualitative and Quantitative Analysis and Differences in Cassava Flour Lipids during Storage
3.2. Significant Results of Differences in Lipids in Fresh and Stored Cassava Flour
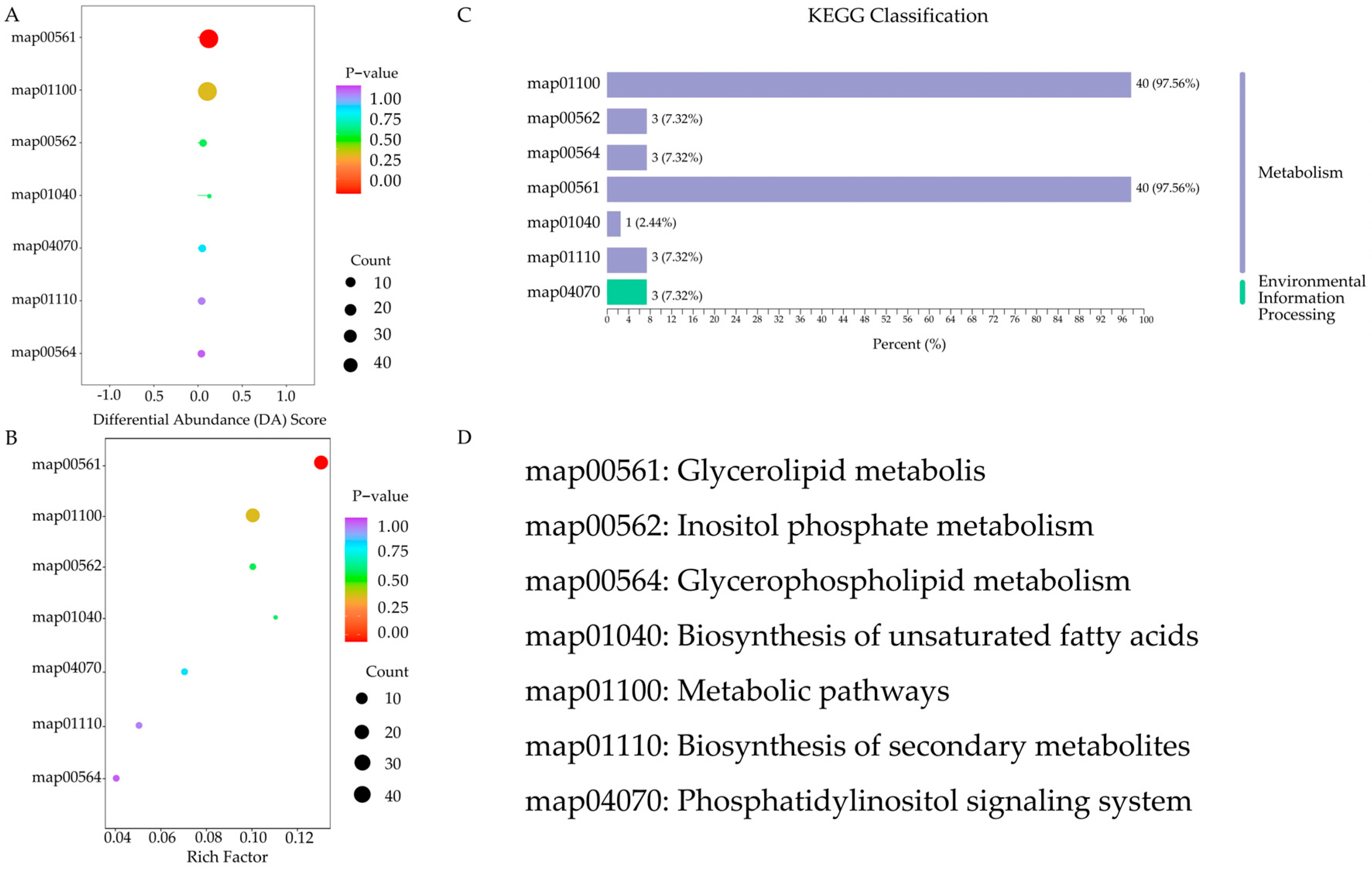
3.3. Lipid Metabolism Pathways during Cassava Flour Storage
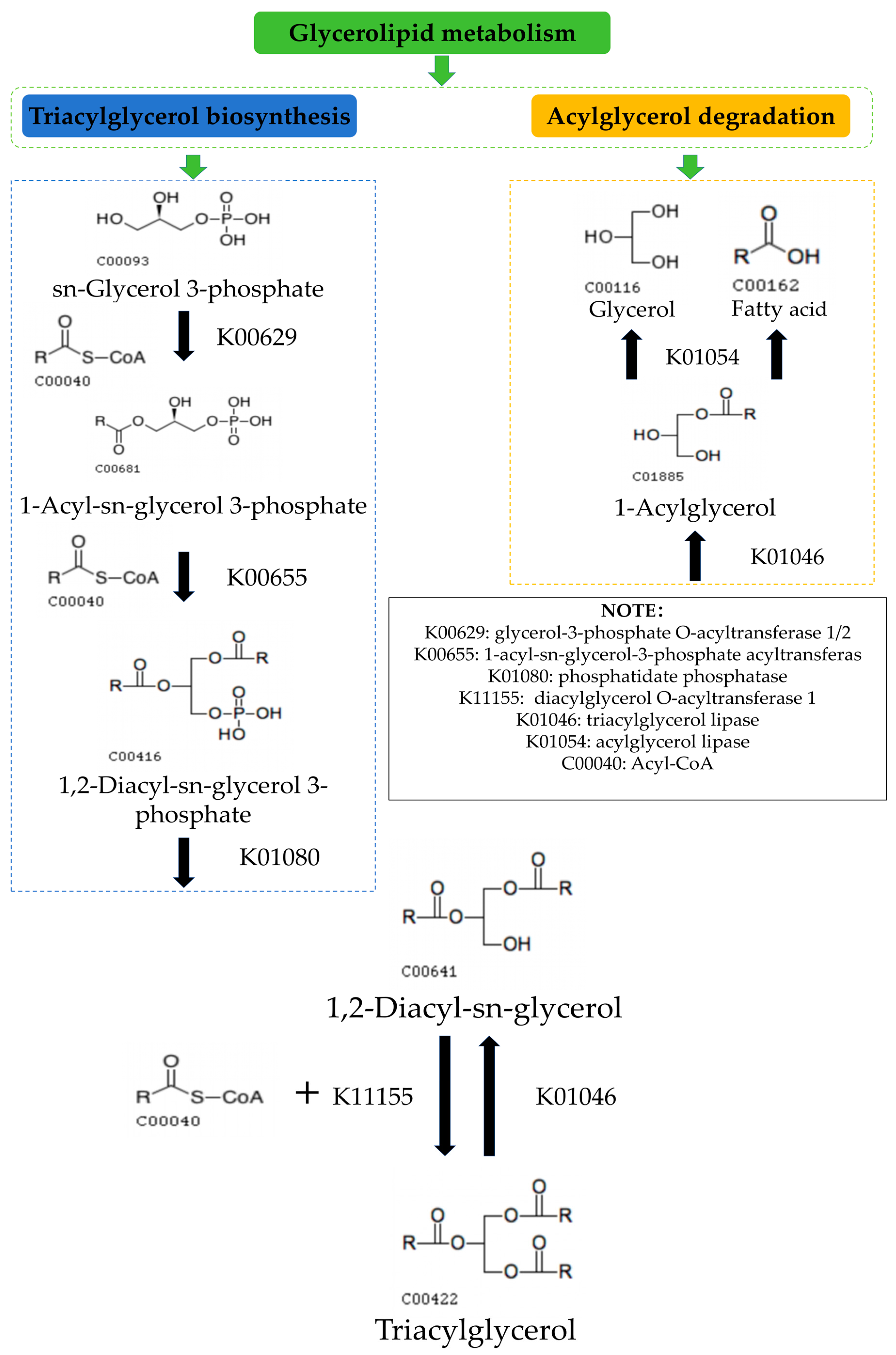
3.4. Possible Mechanisms of the Glyceride Metabolic Pathway in Cassava Flour during Storage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- McCallum, E.J.; Anjanappa, R.B.; Gruissem, W. Tackling Agriculturally Relevant Diseases in the Staple Crop Cassava (Manihot esculenta). Curr. Opin. Plant Biol. 2017, 38, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Mtunguja, M.K.; Laswai, H.S.; Kanju, E.; Ndunguru, J.; Muzanila, Y.C. Effect of Genotype and Genotype by Environment Interaction on Total Cyanide Content, Fresh Root, and Starch Yield in Farmer-Preferred Cassava Landraces in Tanzania. Food Sci. Nutr. 2016, 4, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Chisenga, S.M.; Workneh, T.S.; Bultosa, G.; Alimi, B.A. Progress in Research and Applications of Cassava Flour and Starch: A Review. J. Food Sci. Technol. 2019, 56, 2799–2813. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Guo, L.; Zhang, L.; Xie, C.; Li, W.; Gu, B.; Li, K. Study on Quality Characteristics of Cassava Flour and Cassava Flour Short Biscuits. Food Sci. Nutr. 2020, 8, 521–533. [Google Scholar] [CrossRef]
- Sangpueak, R.; Saengchan, C.; Laemchiab, K.; Kiddeejing, D.; Siriwong, S.; Thumanu, K.; Hoang, N.H.; Phansak, P.; Buensanteai, K. Flour on Gluten-Free Muffins from Different Edible Cassava Varieties in Thailand. Foods 2022, 11, 4053. [Google Scholar] [CrossRef]
- Lin, L.; Xu, H.; Wu, Q.; Wang, Q.; Yu, H.; Zhang, Z. Analysis and Evaluations of Sweet Cassava Flour during Storage. Food Sci. Technol. 2020, 45, 187–191. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, H.; Wu, R.; Lin, L.; Tan, W.; Zhang, Z.; Yang, X. Analysis of the Composition of Fatty Acids and the Effect of Storage Time on the Fatty Fcids in Edible Cassava Flour. Sci. Technol. Food Ind. 2023, 44, 229–237. [Google Scholar] [CrossRef]
- Hudson, B.J.F.; Ogunsua, A.O. Lipids of Cassava Tubers (Manihot Esculenta Crantz). J. Sci. Food Agric. 1974, 25, 1503–1508. [Google Scholar] [CrossRef]
- Lalaguna, F.; Agudo, M. Lipid Classes of Fresh Cassava Roots (Manihot Esculenta Crantz): Identification and Quantification. J. Am. Oil Chem. Soc. 1988, 65, 1808–1811. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A Comprehensive Classification System for Lipids. Eur. J. Lipid Sci. Technol. 2005, 107, 337–364. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS Comprehensive Classification System for Lipids1. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS Structure Database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Z.; OuYang, B.; Wang, W.; Lan, D.; Wang, Y. Lipidomics Analysis of Rice Bran during Storage Unveils Mechanisms behind Dynamic Changes in Functional Lipid Molecular Species. Food Chem. 2024, 447, 138946. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, S.; Ramaswamy, H.S.; Hu, F.; Yu, Y. Effect of High Pressure Processing on Rancidity of Brown Rice during Storage. LWT 2018, 93, 405–411. [Google Scholar] [CrossRef]
- Haman, N.; Romano, A.; Asaduzzaman, M.; Ferrentino, G.; Biasioli, F.; Scampicchio, M. A Microcalorimetry Study on the Oxidation of Linoleic Acid and the Control of Rancidity. Talanta 2017, 164, 407–412. [Google Scholar] [CrossRef]
- Drapal, M.; Ovalle Rivera, T.M.; Luna Meléndez, J.L.; Perez-Fons, L.; Tran, T.; Dufour, D.; Becerra Lopez-Lavalle, L.A.; Fraser, P.D. Biochemical Characterisation of a Cassava (Manihot esculenta Crantz) Diversity Panel for Post-Harvest Physiological Deterioration; Metabolite Involvement and Environmental Influence. J. Plant Physiol. 2024, 301, 154303. [Google Scholar] [CrossRef]
- Han, X. Global Analyses of Cellular Lipidomes Directly from Crude Extracts of Biological Samples by ESI Mass Spectrometry: A Bridge to Lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Han, X. Tutorial on Lipidomics. Anal. Chim. Acta 2019, 1061, 28–41. [Google Scholar] [CrossRef]
- Xuan, Q.; Hu, C.; Yu, D.; Wang, L.; Zhou, Y.; Zhao, X.; Li, Q.; Hou, X.; Xu, G. Development of a High Coverage Pseudotargeted Lipidomics Method Based on Ultra-High Performance Liquid Chromatography-Mass Spectrometry. Anal. Chem. 2018, 90, 7608–7616. [Google Scholar] [CrossRef]
- Hein, E.-M.; Hayen, H. Comparative Lipidomic Profiling of S. Cerevisiae and Four Other Hemiascomycetous Yeasts. Metabolites 2012, 2, 254–267. [Google Scholar] [CrossRef]
- Wenk, M.R. The Emerging Field of Lipidomics. Nat. Rev. Drug. Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Guo, J.; Tian, J.; Ma, K.; Liu, Y. Research progress on degradation methods and product properties of plant polysaccharides. J. Light Ind. 2023, 38, 55–62. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid Extraction by Methyl-Tert-Butyl Ether for High-Throughput Lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Hummel, J.; Segu, S.; Li, Y.; Irgang, S.; Jueppner, J.; Giavalisco, P. Ultra Performance Liquid Chromatography and High Resolution Mass Spectrometry for the Analysis of Plant Lipids. Front. Plant Sci. 2011, 2, 54. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Butka, E.; Wang, X. Comprehensive Quantification of Triacylglycerols in Soybean Seeds by Electrospray Ionization Mass Spectrometry with Multiple Neutral Loss Scans. Sci. Rep. 2014, 4, 6581. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Lu, S.; Liu, H.; Jin, C.; Li, Q.; Guo, L. An Efficient and Comprehensive Plant Glycerolipids Analysis Approach Based on High-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometer. Plant Direct 2019, 3, e00183. [Google Scholar] [CrossRef]
- Sun, C.; Li, T.; Song, X.; Huang, L.; Zang, Q.; Xu, J.; Bi, N.; Jiao, G.; Hao, Y.; Chen, Y.; et al. Spatially Resolved Metabolomics to Discover Tumor-Associated Metabolic Alterations. Proc. Natl. Acad. Sci. USA 2019, 116, 52–57. [Google Scholar] [CrossRef]
- Sun, J.; Hu, P.; Lyu, C.; Tian, J.; Meng, X.; Tan, H.; Dong, W. Comprehensive Lipidomics Analysis of the Lipids in Hazelnut Oil during Storage. Food Chem. 2022, 378, 132050. [Google Scholar] [CrossRef]
- Zhang, D.; Duan, X.; Shang, B.; Hong, Y.; Sun, H. Analysis of Lipidomics Profile of Rice and Changes during Storage by UPLC-Q-Extractive Orbitrap Mass Spectrometry. Food Res. Int. 2021, 142, 110214. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Qiu, J.; Li, Z. Effects of Superheated Steam Processing on Common Buckwheat Grains: Lipase Inactivation and Its Association with Lipidomics Profile during Storage. J. Cereal Sci. 2020, 95, 103057. [Google Scholar] [CrossRef]
- Kinney, A.J. Phospholipid Head Groups. In Lipid Metabolism in Plants; CRC Press: Boca Raton, FL, USA, 1993; ISBN 978-1-351-07407-0. [Google Scholar]
- Isidorov, V.A.; Rusak, M.; Szczepaniak, L.; Witkowski, S. Gas Chromatographic Retention Indices of Trimethylsilyl Derivatives of Mono- and Diglycerides on Capillary Columns with Non-Polar Stationary Phases. J. Chromatogr. A 2007, 1166, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Ba, Y.; Li, R.; Zhang, J.; Zou, L.; Wu, D.-T.; Hu, Y. Evaluation of Lipidomics Profile of Quinoa Flour and Changes during Storage Based on Ultra Performance Liquid Chromatography Coupled with Quadrupole Exactive Orbitrap Mass Spectrometry. Foods 2023, 12, 4434. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.S.; Proctor, A. Milled Rice Oxidation Volatiles and Odor Development. J. Food Sci. 2003, 68, 2676–2681. [Google Scholar] [CrossRef]
- Lam, H.S.; Proctor, A. Hydrolysis of Acylglycerols and Phospholipids of Milled Rice Surface Lipids during Storage. J. Am. Oil Chem. Soc. 2004, 81, 385–388. [Google Scholar] [CrossRef]
- Shadyro, O.; Samovich, S.; Edimecheva, I. Free-Radical and Biochemical Reactions Involving Polar Part of Glycerophospholipids. Free Radic. Biol. Med. 2019, 144, 6–15. [Google Scholar] [CrossRef]
- Baschiera, E.; Sorrentino, U.; Calderan, C.; Desbats, M.A.; Salviati, L. The Multiple Roles of Coenzyme Q in Cellular Homeostasis and Their Relevance for the Pathogenesis of Coenzyme Q Deficiency. Free Radic. Biol. Med. 2021, 166, 277–286. [Google Scholar] [CrossRef]
- Xu, J.-J.; Hu, M.; Yang, L.; Chen, X.-Y. How Plants Synthesize Coenzyme Q. Plant Commun. 2022, 3, 100341. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Hardré-Liénard, H.; Miras, S.; Miège, C.; Block, M.A.; Revah, F.; Joyard, J.; Maréchal, E. Refolding from Denatured Inclusion Bodies, Purification to Homogeneity and Simplified Assay of MGDG Synthases from Land Plants. Protein Expr. Purif. 2003, 31, 79–87. [Google Scholar] [CrossRef]
- Hernández-Corbacho, M.J.; Salama, M.F.; Canals, D.; Senkal, C.E.; Obeid, L.M. Sphingolipids in Mitochondria. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 56–68. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr. Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Pant, D.C.; Aguilera-Albesa, S.; Pujol, A. Ceramide Signalling in Inherited and Multifactorial Brain Metabolic Diseases. Neurobiol. Dis. 2020, 143, 105014. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Shang, K.; Li, X.; Zhang, C.-H.; Liu, J.-Q.; Huang, D.-Q. Characterization and Discrimination of Selected China’s Domestic Pork Using an LC-MS-Based Lipidomics Approach. Food Control 2019, 100, 305–314. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, Z.; Wang, Y.; Liu, P.; Hu, K. Taraxacum Officinale-Derived Exosome-like Nanovesicles Modulate Gut Metabolites to Prevent Intermittent Hypoxia-Induced Hypertension. Biomed. Pharmacother. 2023, 161, 114572. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, D.; Wang, Q.; Shang, B.; Liu, J.; Xing, X.; Hong, Y.; Duan, X.; Sun, H. Shotgun Lipidomics Reveals the Changes in Phospholipids of Brown Rice during Accelerated Aging. Food Res. Int. 2023, 171, 113073. [Google Scholar] [CrossRef]
- Moran, T.; Pace, J.; McDERMOTT, E.E. The Lipids in Flour: Oxidative Changes Induced by Storage and Improver Treatment. Nature 1954, 174, 449–452. [Google Scholar] [CrossRef]
- Clayton, T.A.; Morrison, W.R. Changes in Flour Lipids during the Storage of Wheat Flour. J. Sci. Food Agric. 1972, 23, 721–736. [Google Scholar] [CrossRef]
- Martin, J.J.J.; Wu, Q.; Feng, M.; Li, R.; Zhou, L.; Zhang, S.; Yang, C.; Cao, H. Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development. Metabolites 2023, 13, 727. [Google Scholar] [CrossRef]
- Mateos, P.S.; Navas, M.B.; Morcelle, S.R.; Ruscitti, C.; Matkovic, S.R.; Briand, L.E. Insights in the Biocatalyzed Hydrolysis, Esterification and Transesterification of Waste Cooking Oil with a Vegetable Lipase. Catal. Today 2021, 372, 211–219. [Google Scholar] [CrossRef]
- Murakami, C.; Sakane, F. Sphingomyelin Synthase-Related Protein Generates Diacylglycerol via the Hydrolysis of Glycerophospholipids in the Absence of Ceramide. J. Biol. Chem. 2021, 296, 100454. [Google Scholar] [CrossRef]
- Robertson, F.; Passaro, L. Hydrolysis of Triglycerides in Cream Using Porcine Pancreatic Lipase. J. Chem. Educ. 2020, 97, 4438–4444. [Google Scholar] [CrossRef]
- Xue, L.-L.; Chen, H.-H.; Jiang, J.-G. Implications of Glycerol Metabolism for Lipid Production. Prog. Lipid Res. 2017, 68, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.W.; Byerrum, R.U.; Gee, R.W.; Tolbert, N.E. Glycerol Concentrations in Crop Plants. Plant Sci. 1988, 56, 31–38. [Google Scholar] [CrossRef]
- Mao, Y.; Lee, Y.-Y.; Xie, X.; Wang, Y.; Zhang, Z. Preparation, Acyl Migration and Applications of the Acylglycerols and Their Isomers: A Review. J. Funct. Foods 2023, 106, 105616. [Google Scholar] [CrossRef]
- Bhunia, R.K.; Sinha, K.; Kaur, R.; Kaur, S.; Chawla, K. A Holistic View of the Genetic Factors Involved in Triggering Hydrolytic and Oxidative Rancidity of Rice Bran Lipids. Food Rev. Int. 2023, 39, 441–466. [Google Scholar] [CrossRef]

| Lipid | Content of Lipids (μg/g) | ||||||
|---|---|---|---|---|---|---|---|
| Class I | Class II | SC9 (Day 0) | SC9 (Day 180) | SC9 (Day 540) | GR10 (Day 0) | GR10 (Day 180) | GR10 (Day 540) |
| GL | ADGGA | 32.37 ± 9.06 abc | 25.05 ± 6.57 ab | 15.59 ± 5.69 a | 50.25 ± 13.31 c | 35.69 ± 13.80 bc | 14.26 ± 5.97 a |
| DG | 34.37 ± 5.26 ab | 17.89 ± 0.56 a | 16.31 ± 2.28 a | 48.06 ± 7.51 b | 39.08 ± 21.97 ab | 36.07 ± 19.18 ab | |
| DGDG | 42.90 ± 13.11 ab | 20.35 ± 1.16 ab | 15.98 ± 1.51 a | 66.24 ± 8.92 b | 62.91 ± 50.17 ab | 45.52 ± 29.34 ab | |
| DGTS | 5.07 ± 1.32 ab | 2.10 ± 0.11 ab | 1.62 ± 0.26 a | 3.73 ± 0.47 ab | 4.31 ± 3.37 ab | 6.62 ± 4.69 b | |
| LDGTS | 749.41 ± 197.12 c | 629.13 ± 175.44 abc | 402.61 ± 135.00 ab | 414.60 ± 75.37 ab | 339.79 ± 147.57 a | 645.23 ± 160.55 bc | |
| MG | 361.47 ± 38.28 b | 278.51 ± 26.11 a | 233.50 ± 27.64 a | 467.43 ± 59.14 c | 358.59 ± 59.74 b | 292.58 ± 24.36 ab | |
| MGDG | 14.94 ± 5.18 a | 6.32 ± 0.47 a | 4.22 ± 0.47 a | 23.14 ± 5.80 a | 23.22 ± 17.75 a | 20.37 ± 14.53 a | |
| DGTS | 6.83 ± 1.77 ab | 2.84 ± 0.15 ab | 2.18 ± 0.36 a | 5.02 ± 0.63 ab | 5.81 ± 4.55 ab | 8.94 ± 6.35 b | |
| SQDG | 25.15 ± 8.42 a | 12.35 ± 0.57 a | 12.98 ± 1.45 a | 25.04 ± 4.71 a | 31.60 ± 26.57 a | 29.50 ± 19.13 a | |
| TG | 308.88 ± 11.10 b | 343.69 ± 17.17 b | 177.34 ± 10.00 a | 626.65 ± 74.23 c | 343.81 ± 39.52 b | 235.01 ± 15.19 a | |
| SP | Cer | 75.77 ± 3.38 a | 69.03 ± 12.58 a | 71.22 ± 1.43 a | 76.89 ± 13.74 a | 70.20 ± 0.64 a | 68.79 ± 4.57 a |
| Cert | 111.11 ± 5.23 ab | 102.24 ± 18.18 ab | 99.61 ± 1.25 ab | 116.19 ± 20.79 b | 108.67 ± 2.80 ab | 89.66 ± 5.21 a | |
| HexCer | 5.94 ± 0.60 ab | 6.06 ± 0.61 ab | 7.03 ± 0.16 b | 7.42 ± 1.15 b | 6.60 ± 1.38 b | 4.89 ± 0.36 a | |
| SPH | 1.13 ± 0.15 b | 0.80 ± 0.03 a | 0.80 ± 0.04 a | 1.02 ± 0.07 b | 1.17 ± 0.09 b | 1.04 ± 0.09 b | |
| PR | CoQ | 0.18 ± 0.01 ab | 0.12 ± 0.01 ab | 0.09 ± 0.01 ab | 2.60 ± 0.17 c | 0.21 ± 0.01 b | 0.08 ± 0.01 a |
| FA | FFA | 468.01 ± 88.12 b | 484.91 ± 44.58 b | 323.03 ± 54.81 a | 525.16 ± 59.36 b | 717.51 ± 46.61 c | 491.69 ± 87.51 b |
| GP | LPA | 7.97 ± 0.04 a | 10.59 ± 0.50 b | 28.01 ± 0.61 d | 7.71 ± 0.13 a | 10.09 ± 0.78 b | 18.99 ± 0.64 c |
| LPC | 3.84 ± 0.11 bc | 4.65 ± 0.33 d | 6.65 ± 0.16 e | 3.15 ± 0.05 a | 3.65 ± 0.16 b | 4.00 ± 0.11 c | |
| LPE | 5.87 ± 0.15 b | 6.30 ± 0.56 b | 9.10 ± 0.26 c | 6.05 ± 0.26 b | 4.91 ± 0.11 a | 6.19 ± 0.33 b | |
| LPG | 2.18 ± 0.25 a | 2.71 ± 0.16 bc | 3.05 ± 0.18 c | 2.15 ± 0.20 a | 2.59 ± 0.23 b | 2.71 ± 0.21 bc | |
| LPI | 9.46 ± 0.96 a | 11.23 ± 0.84 ab | 9.39 ± 0.62 a | 9.89 ± 1.34 a | 12.10 ± 1.46 b | 11.24 ± 0.88 ab | |
| PA | 16.39 ± 0.25 bc | 14.65 ± 1.42 a | 28.25 ± 0.97 e | 17.97 ± 0.88 c | 14.94 ± 0.71 ab | 21.86 ± 0.80 d | |
| PC | 11.73 ± 1.62 b | 10.57 ± 1.17 ab | 9.83 ± 0.83 ab | 15.12 ± 2.42 c | 11.62 ± 0.40 b | 8.78 ± 0.75 a | |
| PE | 0.31 ± 0.01 a | 0.34 ± 0.06 a | 0.31 ± 0.01 a | 1.66 ± 0.27 c | 0.95 ± 0.09 b | 0.12 ± 0.02 a | |
| PG | 3.75 ± 0.48 ab | 3.47 ± 0.47 a | 6.54 ± 0.23 d | 4.67 ± 0.43 c | 4.11 ± 0.11 abc | 4.19 ± 0.32 bc | |
| PI | 58.62 ± 7.66 ab | 62.49 ± 0.95 b | 46.80 ± 2.45 a | 103.63 ± 10.90 d | 76.25 ± 8.60 c | 62.74 ± 4.51 b | |
| PMeOH | 0.02 ± 0.003 a | 0.03 ± 0.001 ab | 0.03 ± 0.001 ab | 0.03 ± 0.003 b | 0.03 ± 0.002 a | 0.02 ± 0.003 a | |
| PS | 49.90 ± 2.25 c | 46.06 ± 2.86 bc | 32.85 ± 5.32 a | 60.89 ± 3.08 d | 53.80 ± 8.14 cd | 39.93 ± 5.27 ab | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; Wang, Q.; He, Y.; Yu, H.; Lin, L.; Zhang, Z. Lipidomic Profiling and Storage-Induced Changes in Cassava Flour Using LC-MS/MS. Foods 2024, 13, 3039. https://doi.org/10.3390/foods13193039
Du P, Wang Q, He Y, Yu H, Lin L, Zhang Z. Lipidomic Profiling and Storage-Induced Changes in Cassava Flour Using LC-MS/MS. Foods. 2024; 13(19):3039. https://doi.org/10.3390/foods13193039
Chicago/Turabian StyleDu, Peixu, Qinfei Wang, Yi He, Houmei Yu, Liming Lin, and Zhenwen Zhang. 2024. "Lipidomic Profiling and Storage-Induced Changes in Cassava Flour Using LC-MS/MS" Foods 13, no. 19: 3039. https://doi.org/10.3390/foods13193039
APA StyleDu, P., Wang, Q., He, Y., Yu, H., Lin, L., & Zhang, Z. (2024). Lipidomic Profiling and Storage-Induced Changes in Cassava Flour Using LC-MS/MS. Foods, 13(19), 3039. https://doi.org/10.3390/foods13193039







