Abstract
This study aimed to investigate the probiogenomic features of artisanal bacteriocin-producing Enterococcus faecium BGPAS1-3 and the use of the improved pMALc5HisEk expression vector for overexpressing class II bacteriocins and the application of purified bacteriocin 31 in a milk model as a preservative against L. monocytogenes. The BGPAS1-3 strain was isolated from traditional fresh soft cheese manufactured in households on a small scale in rural locations surrounding Pale Mountain City in Bosnia and Herzegovina. The whole-genome sequencing approach and bioinformatics analyses revealed that the strain BGPAS1-3 was non-pathogenic to humans. The presence of bacteriocin operons suggested the ability of the isolate to suppress the growth of pathogens. Coding regions for three maturated bacteriocins (bacteriocin 31, bacteriocin 32, and enterocin P) produced by BGPAS1-3 were amplified and expressed in Escherichia coli ER2523 using the pMALc5HisEk system. All three bacteriocins were successfully overexpressed and purified after enterokinase cleavage but showed different antimicrobial activity. Bacteriocin 31 showed significantly stronger antimicrobial activity compared with bacteriocin 32. It was the only one that proved to be suitable for use as a food preservative against L. monocytogenes in a milk model.
1. Introduction
The genus Listeria consists of Gram-positive bacilli that are ubiquitous in various environments and can infect humans or warm-blooded animals through contaminated food []. Within this genus, in addition to non-pathogenic species, there are only two pathogenic species for humans and animals, Listeria monocytogenes and Listeria ivanovii []. The Listeria outbreaks have a significant economic impact on the food industry []. Many cases of listeriosis are linked to outbreaks. At the same time, numerous sporadic cases can also arise from persistent L. monocytogenes, underscoring the severe public health and economic consequences of L. monocytogenes’ persistence in the food system. This persistence can lead to the contamination of finished products, resulting in numerous recalls, product redesigns, and internal rejections, all of which amplify the economic impact []. Moreover, extensive testing programs, which are designed to identify growth niches and environmental persistence, add to the financial burden []. Although precisely quantifying the economic impact of L. monocytogenes persistence is challenging, it is evident that a significant portion of the overall costs related to L. monocytogenes contamination in the food industry stems from the persistent presence of this major foodborne pathogen in processing environments []. In addition to that, infections caused by pathogenic species are widespread globally and have significant health importance. Listeriosis is a serious zoonotic and systemic disease affecting human health after the consumption of contaminated foods []. Listeria monocytogenes is an important foodborne pathogen in humans and a cause of foodborne illness []. After being ingested through contaminated food, L. monocytogenes infects the intestinal epithelium, passes through the intestinal barrier, and causes systemic infections in other body organs []. By producing a soluble cytolysin, listeriolysin O (LLO), and intracellular replication, L. monocytogenes leads to a high hospitalization rate and potential mortality in immunocompromised individuals []. The ability to produce and form biofilms plays a role in the persistence of L. monocytogenes in the environment for long periods []. Biofilm formation affects resistance to various antibiotics and organic sanitizers []. The biofilm increases cell density, which provides an optimal environment for the exchange of eDNA via conjugation, some of which may code for antibiotic resistance. Cells in biofilms are about 1000 times more resistant to antibiotics than the planktonic form, and one of the reasons is the presence of extracellular polymeric substance (EPS) matrices. Additionally, the EPS matrix protects bacterial cells in deeper layers from antimicrobial agents by limiting diffusion [,]. The multiple drug-resistant L. monocytogenes strains isolated from environmental and animal sources, as well as various foods, have been extensively reported. These strains are associated with the excessive use of antibiotics in clinical environments and farms []. In recent years, the application of alternative approaches in the control of L. monocytogenes has been a priority [,,]. Live probiotic strains of lactic acid bacteria (LAB) and their bioactive compounds have become important in controlling L. monocytogenes in the food industry []. Increased attention has been focused on the ability of LAB to produce bacteriocins, specific proteinaceous substances, which inhibit the growth of pathogens such as Listeria and extend the shelf life of food products [,]. Many bacteriocins of LAB are effective and safe natural inhibitors of pathogenic and food spoilage bacteria and have been suggested for use as food “preservatives” [,]. The enterococci produce bacteriocins, small molecular weight peptides, usually characterized by specific antimicrobial activity against Gram-positive pathogens and spoilage bacteria. Enterococcal bacteriocins or enterocins are classified into four classes: class I, lantibiotic; class II, non-lantibiotic; class III, cyclic enterocin; and class IV, enterocin with a high molecular weight []. Most enterocins produced by Enterococcus sp. belong to class II and have strong inhibitory effects against L. monocytogenes. Class II enterocins are heat-stable, ribosomally synthesized peptides that do not undergo extensive post-translational modifications besides cleavage of a leader peptide during their transport out of the cell []. In the food industry, bacteriocins can be applied either as added food additives or through in situ production by starters. The use of bacteriocin-producing enterococci as starter cultures in the production of certain Mediterranean cheeses may prevent spoilage and growth of L. monocytogenes []. Considering that enterococci can also be pathogenic, using them in the food industry presents challenges []. A whole-genome sequencing (WGS)-based method is the only effective method to fully characterize the safety status of Enterococcus strains [] and is in line with the statement of the European Food Safety Authority (EFSA) on the requirements for WGS analysis of strains intended for use in the food industry []. Based on whole-genome information, the prediction of potential harmful determinants or genetic features associated with beneficial effects provides a comprehensive insight into potential probiotic strains. Combining the results of WGS and phenotypic tests provides insights into the potential of the selected strain for further application []. Although the use of bacteriocins is considered safe [], in most cases the use of bacteriocins has not been proven to be successful. The reasons for the lower antimicrobial activity are most likely either the degradation or the adsorption of bacteriocins into complex food matrices []. The only bacteriocin approved for use in the food industry is Nisin []; however, its application is limited due to its low activity in non-acidic environments. Bacteriocins are used in the preservation of various foods, either alone or combination with other conservation methods, most often pasteurization [].
To our knowledge, this is the first study to report the use of probiogenomics to assess the safety and probiotic properties and use of the improved pMALc5HisEk expression vector for overexpressing class II bacteriocins and application of purified bacteriocin 31 in milk model as a preservative against L. monocytogenes.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Enterococcus faecium BGPAS1-3 from the laboratory collection of the Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Serbia, was grown in M17 medium (Merck GmbH, Darmstadt, Germany) supplemented with D-glucose (0.5% w/v) (GM17) at 37 °C. Escherichia coli ER2523 (New England Biolabs, Ltd., Hitchin, UK) was grown in Luria Bertani (LB) medium containing 0.5% NaCl, 0.5% yeast extract, and 1% tryptone (Torlak, Belgrade, Serbia) at 37 °C with aeration (180 rpm). Solid medium and soft agar were prepared by adding 1.5% and 0.7% (w/v) agar (Torlak, Belgrade, Serbia) to the liquid media, respectively. Ampicillin (100 μg/mL) was used for selecting and maintaining transformants. Isopropyl-β-D-1-thiogalactopyranoside (IPTG; Serva, Heidelberg, Germany) was used at suitable concentrations to induce protein expression. For the count of L. monocytogenes BG322, IMS1, IMS2, IMS3, and IMS4 strains in the milk model, Difco Oxford Medium Base (Becton, Dickinson and Company, Sparks, MD, USA) was used. Indicator strains and growth conditions are listed in Table 1.

Table 1.
Indicator strains used in this study.
2.2. Whole-Genome Sequencing, Standard Analysis, and Data Deposition
The total DNA of En. faecium BGPAS1-3 was extracted by the method described previously []. Total DNA quality and concentration were evaluated by agarose gel electrophoresis and by a Bio-Spec nano (Shimadzu, Tokyo, Japan) spectrophotometer. The whole-genome sequencing (WGS) of the En. faecium BGPAS1-3 genome was performed on the Illumina HiSeq 2500 platform by the MicrobesNG service (MicrobesNG, IMI-School of Biosciences, University of Birmingham, Birmingham, UK) using a 2 × 250 bp paired-end strategy. Library preparation was carried out using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), with minor modifications to the manufacturer’s protocol. Specifically, 2 ng of input DNA was used, and the PCR elongation time was extended to 1 min. The process was executed on a Hamilton Microlab STAR (Hamilton, Reno, NV, USA). Pooled libraries were quantified with a Roche LightCycler 96 qPCR machine (Roche, Indianapolis, IN, USA) using the Kapa Biosystems Library Quantification Kit for Illumina. Trimmomatic v0.30 [] with a sliding window quality cutoff of Q15 was used for adapter trimming. De novo assembly was performed using SPAdes v3.7 [], and contigs were annotated using Prokka 1.11 []. Kraken software [] was used to identify the closest available reference genome, and BWA MEM [] was used for mapping the reads to the reference genome. Draft genome sequences of BGPAS1-3 have been deposited in GenBank under accession number JAUKVL000000000.
2.3. Genotypic Characterization for Safety and Probiotic-Related Traits
The En. faecium BGPAS1-3 complete genome sequence was analyzed for safety assessment using the online bioinformatics tools accessible at the Center for Genomic Epidemiology: ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/; accessed on 26 April 2024). [] following the thresholds of 60% identity over a length of 60% coverage, respectively. Further, VirulenceFinder (https://cge.food.dtu.dk/services/VirulenceFinder/; accessed on 26 April 2024) [] and the Virulence Factor of Bacterial Pathogens Database (VFDB) (http://www.mgc.ac.cn/VFs/main.htm; accessed on 26 April 2024) [] were used to predict potential virulence genes. The identification threshold was set at 90% identity over a length of 60% for VirulenceFinder. Pathogen-Finder v1.1 (http://cge.cbs.dtu.dk/services/PathogenFinder/; accessed on 26 April 2024) [] was used to investigate acquired pathogenic and antibiotic resistance factors. PlasmidFinder v2.1.1 (http://cge.cbs.dtu.dk/services/PlasmidFinder; accessed on 26 April 2024) [] was used for plasmid identification against a Gram-positive database with a 60% minimum identity threshold and minimum 60% coverage. For beneficial traits, the BGPAS1-3 complete genome was mined for genes associated with biosynthetic pathways of essential amino acids using the KAAS-KEGG Automatic Annotation Server (http://www.genome.jp/kegg/kaas/; accessed on 26 April 2024) [] and BAGEL4 (http://bagel4.molgenrug.nl/; accessed on 26 April 2024) [] for the detection of potential antimicrobial compounds. The Proksee web platform (https://proksee.ca/; accessed on 26 April 2024) was used for circular genome visualization and comparison [].
2.4. DNA Manipulations
For plasmid isolation from E. coli ER2523 transformants, a Thermo Fisher Scientific GeneJET Plasmid Miniprep kit was used according to the manufacturer’s instructions (Thermo Scientific, Vilnius, Lithuania). Digestion with restriction enzymes was conducted according to the supplier’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). By the manufacturer’s recommendations, DNA was ligated using T4 DNA ligase (Agilent Technologies, Santa Clara, CA, USA). The standard heat shock transformation method was used for the transformation of E. coli with a plasmid [].
2.5. Overexpression of Bacteriocins from Enterococcus faecium BGPAS1-3 in Escherichia coli ER2523
Transformants of E. coli ER2523 (pMALc5HisEk_Bac31, pMALc5HisEk_Bac32, and pMALc5HisEk_EntP) were maintained overnight on LB Agar Petri dishes containing ampicillin (100 µg/mL) and glucose (1%) at 30 °C. The next day, new cultures (each 200 mL of LB with 1% glucose and 100 µg/mL ampicillin) were inoculated using a 2% overnight culture and incubated at 30 °C with aeration (180 rpm on a rotatory shaker). Expression of recombinant peptides was induced in the logarithmic growth phase (OD600 = 0.8–1.0) with the addition of 0.3 mmol/L (final) IPTG for 3 h. Bacterial cells were collected by centrifugation at 4500× g and frozen at −20 °C. Before purification, the level of induction was tested by comparing the amounts of total proteins from the same amount of induced and non-induced cells. Purification including cell lysis, affinity chromatography, and cleavage of the fusion protein with enterokinase was performed according to the manufacturer’s instructions (amylose resin purification, according to pMAL Protein Fusion & Purification System, New England Biolabs, Ltd., UK; Ni-NTA agarose resin purification, according to The QIAexpressionist, Qiagen Gmbh, Hilden, Germany; cleavage with recombinant bovine enterokinase in 1× enterokinase buffer (20 mmol/L Tris-HCl, pH 7.4, 50 mmol/L NaCl, 2 mmol/L CaCl2) using 10 U, according to GenScript, Piscataway, NJ, USA), with the addition of a cell lysis step that was performed in column buffer (CB) with 1 mg/mL lysozyme for 30 min on ice. The total protein concentrations were determined using the BSA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Total protein concentrations were measured using the BSA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Proteins from all stages of expression, purification, and proteolysis were analyzed by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For SDS-PAGE, samples were mixed with 2 × sample loading buffer (125 mmol/L Tris-HCl pH 6.8, 10 mmol/L EDTA, 4% SDS, 25% glycerol, 5% β-mercaptoethanol, 0.07% bromophenol blue) at a 1:1 ratio. Before loading onto the gel, samples were denatured by heating at 100 °C for 5 min.
2.6. Bacteriocin Activity Assay
To detect bacteriocin activity, an agar-well diffusion assay was conducted []. Cultures for the bacteriocin assay were grown without antibiotic selection. A spot-on-the-lawn inhibition assay [] was used to test the antimicrobial activity of recombinant antimicrobial peptides. The sensitive strain used in bacteriocin tests was L. monocytogenes BG322. Inhibition zones were examined after 24 h of incubation at 37 °C, with a clear zone indicating bacteriocin activity. The bacteriocin activity assay was performed in at least two independent experiments. Bacteriocin activity was quantified by performing two-fold serial dilutions of the purified bacteriocins, and activity was expressed in arbitrary units per milliliter (AU/mL). One arbitrary unit (AU) is defined as the reciprocal of the highest dilution of bacteriocin that still produces a clear zone of growth inhibition in the indicator strain [].
2.7. Amplified Fragments and Constructs Sequencing and Sequence Analysis
Amplified fragments and constructs were sequenced by the Macrogen sequencing service (Macrogen Europe, Amsterdam, The Netherlands). Sequence annotation and similarity searches were performed using the BLAST program of the National Center for Biotechnology Information (NCBI) []. Open reading frames (ORFs) and restriction enzyme sites were predicted using the DNA Strider3 program.
2.8. Effects of Purified Bacteriocin 31 on the Abundance of Listeria monocytogenes in a Milk Model
A milk model was used to investigate the effects of purified bacteriocin 31 on the abundance of L. monocytogenes BG322, IMS1, IMS2, IMS3, and IMS4 according to the previously described protocol [] with slight modifications. Autoclaved reconstituted skimmed milk (11% w/v RSM) was independently inoculated with 107 CFU/mL of L. monocytogenes BG322, IMS1, IMS2, IMS3, and IMS4. Different amounts of BGPAS1-3 purified bacteriocin 31 at final concentrations of 100 AU/mL, 500 AU/mL, and 1000 AU/mL were added. The milk samples were monitored during the 7-day storage at 4 °C. For analysis, serial dilutions of each sample (ranging from 10−1 to 10−7) were prepared, and 100 μL of each dilution was plated onto Difco Oxford Medium Base agar plates. The plates were then incubated for 48 h at 37 °C under aerobic conditions.
2.9. Statistical Analysis
All experiments were independently repeated at least three times, with each experiment performed in triplicate. The data are presented as mean values ± standard deviation across all experiments. For comparisons among multiple groups, two-way ANOVA followed by the Bonferroni post hoc test was applied. Statistical significance was defined as p < 0.05. Statistical analyses and graph preparations were carried out using GraphPad Prism 10 software.
3. Results
Previous studies have shown that En. faecium BGPAS1-3 has a strong direct antilisterial effect, which was observed at a similar level after treatment with heat-killed BGPAS1-3 cells []. Furthermore, our previous results show that live and heat-killed BGPAS1-3 cells can prevent tight junction disruption, enable IL-8 mRNA induction, and stimulate TGF-β mRNA expression in differentiated Caco-2 infected with L. monocytogenes []. Taking into account all these demonstrated features and that the tested strain can synthesize thermostable bacteriocins, in this study we analyzed the genome of BGPAS1-3 using a variety of bioinformatic tools to assess the strain’s safety status and analyzed beneficial features with particular emphasis on the presence of bacteriocins and to clone, overexpress, prove antimicrobial activity, and apply bacteriocins in a milk model for the L. monocytogenes control.
3.1. Enterococcus faecium BGPAS1-3 Was Predicted as a Non-Human Pathogen
Whole-genome sequencing (WGS) of Enterococcus faecium BGPAS1-3 revealed that the genome size was 2,745,343 bp (175 contigs), comprising 2.664 protein-coding sequences (CDS), 66 tRNAs, and 1 rRNA operon, with a guanine-cytosine (GC) content of 37.97% (Figure 1).
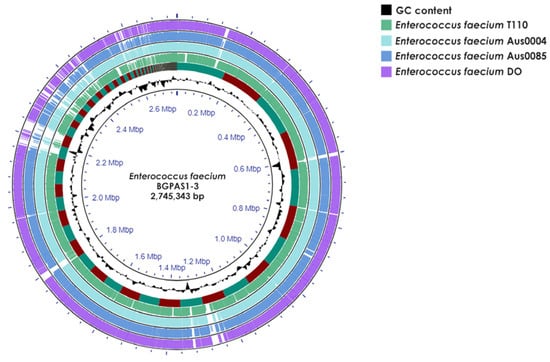
Figure 1.
Circular genome map of Enterococcus faecium BGPAS1-3 visualized and compared with selected pathogenic (Aus0004, Aus0085, and DO) and probiotic (T110) Enterococcus faecium strains using Proksee. Note that the map’s rings are arranged as follows from the inside out: the first ring shows scale marks in megabase pairs (Mbp) and GC content (black); the second ring depicts a comparative analysis of the BGPAS1-3 genome against the T110 genome using BLAST+ 2.12.0 software; and subsequent rings compare the BGPAS1-3 genome with those of strains Aus0004, Aus0085, and DO.
Antimicrobial resistance (AMR) analysis was performed using ResFinder (v4.1), which identified two genes involved in resistance to antibiotics. These two genes corresponded to a homolog of aac(6′)-Ii involved in aminoglycoside resistance (% identity, 99.82; query/HSP length, 549/549; accession number, L12710) and a homolog to msr(C) involved in MLS (Macrolide, Lincosamide and Streptogramin B) (% identity, 98.98; query/HSP length, 1479/1479; accession number, AY004350) (Supplementary Table S1).
Virulence factors screening was performed using VirulenceFinder (v2.0). The analysis discovered the presence of two virulence genes: acm (98.98%; 1866/2166; CP003351.1) and efaAfm (90.71%; 861/879; AF042288.1), which encode collagen-binding protein and cell wall adhesin, respectively. Moreover, the Virulence Factor of Bacterial Pathogens Database (VFDB) was used to search for virulence factors and showed that the BGPAS1-3 genome contained genes responsible for adhesion (ebpA, ebpB, ebpC, srtC, and efaA), antiphagocytosis (cpsA/uppS, cpsB/cdsA, cpsC, and cpsK), biofilm formation (bopD), and immune evasion (cps4I) (Supplementary Table S2).
The pathogenic potential of BGPAS1-3 was determined via PathogenFinder (v1.1.). Based on the results, BGPAS1-3 was predicted as a non-human pathogen (the probability of being a human pathogen was 0.262), with five not-pathogenic and two pathogenic families. PlasmidFinder (accessed on 26 April 2024) was used for plasmid identification against a Gram-positive database with a 60% minimum identity threshold and minimum 60% coverage. Seven plasmid sequences were discovered: rep18a (99.89%), rep29 (100%), repUS42 (87.29%), repUS53 (88.8%), repUS15 (99.52%), rep2 (99.93%), and rep1 (96.97%) (Supplementary Table S3).
3.2. Enterococcus faecium BGPAS1-3 Contains Genes Predicted to Be Involved in Biosynthetic Pathways of Essential Amino Acids
Desirable traits of the probiotic strains included the production of essential amino acids. Using the KAAS-KEGG Automatic Annotation Server to search the BGPAS1-3 genome, genes involved in biosynthetic pathways of essential amino acids were found. It was noticed that the BGPAS1-3 genome contained genes involved in the biosynthesis of the essential amino acids alanine, aspartate, arginine, cysteine, and valine (Supplementary Table S4).
3.3. Enterococcus faecium BGPAS1-3 Contains Operons/Gene Clusters Predicted to Encode the Production of Four Antimicrobial Peptides
The genome of En. faecium BGPAS1-3 was analyzed for genes encoding bacteriocins and non-bactericidal post-translationally modified peptides using the BAGEL4 online tool. The BAGEL4 analysis identified four candidate genes in En. faecium BGPAS1-3 that are associated with the production of antimicrobial peptides: enterolysin A, bacteriocin 31, bacteriocin 32, and enterocin P. Based on the analysis in BAGEL4, enterolysin A showed a similarity of 36.4% to those previously expressed where activity was obtained and was not the subject of further investigations (Table 2, Supplementary Figure S1).

Table 2.
Results of BAGEL4 search of Enterococcus faecium BGPAS1-3 genome sequence.
3.4. Overexpression of Bacteriocin Genes from Enterococcus faecium BGPAS1-3 in Escherichia coli ER2523
To test which of the thermostable bacteriocins produced by En. faecium BGPAS1-3 are naturally active on L. monocytogenes BG322, three bacteriocin genes (bacteriocin 31, bacteriocin 32, and enterocin P) were amplified by PCR using primers listed in Table 3. Amplified fragments of the expected sizes were purified, digested with corresponding restriction enzymes (bacteriocin 31 with BamHI, and bacteriocin 32 and enterocin P with HindIII), and ligated into pMALc5HisEk [] predigested with BsaBI-HindIII or BsaBI-BamHI. The ligation mixtures were transformed into ER2523 (New England Biolabs) competent cells. Ampicillin-resistant transformants were screened for the presence of corresponding fragments by plasmid isolation and restriction enzyme (SacI-HindIII) analysis. Promising constructs (three per cloning) were sequenced using the pMalE primer to confirm the presence of the corresponding DNA fragment in frame with the maltose binding protein gene. Two clones per bacteriocin were stored in LB with 15% glycerol at −80 °C until the overexpression experiments were performed (Figure 2).

Table 3.
List of primers used in this study.
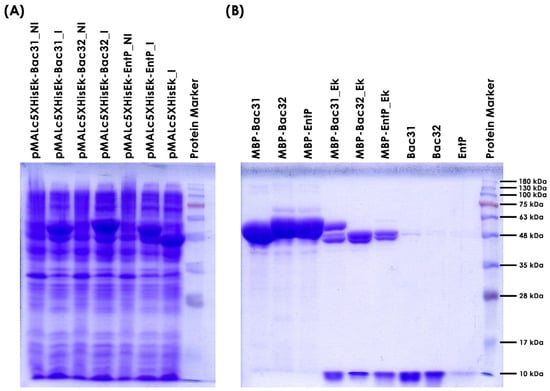
Figure 2.
SDS-PAGE analysis of three Enterococcus faecium BGPAS1-3 bacteriocins expression and purification from E. coli ER2523. (A) Analysis of the level of induction/expression of three enterococcal bacteriocins (tag MBP-bacteriocin hybrid protein) in E. coli ER2523. Total proteins of E. coli ER2523 carrying pMALc5HisEk_Bac31, pMALc5HisEk_Bac32, pMALc5HisEk_EntP, and empty vector pMALc5HisEk before (NI) and after induction (I) with 0.3 mmol/L of IPTG were loaded on 12.5% SDS PAGE gel. (B) Analysis of the level of tag-bacteriocin purification (MBP-), efficiency of enterokinase digestion (Ek), and tag release of three enterococcal bacteriocins on 15% SDS PAGE gel.
Furthermore, the sequences of the expressed enterocin P and bacteriocin 32 proteins from strain BGPAS1-3 were 100% identical to those previously expressed where activity was obtained. Bacteriocin 31 showed the greatest difference, characterized by changes in three amino acids (Supplementary Figure S2).
3.5. Purified Bacteriocins Are Active against Listeria sp. and Lactic Acid Bacteria
After cloning and purification of three thermostable bacteriocins, the antimicrobial potential against different species was tested. It was shown that the purified bacteriocin 31 and bacteriocin 32 were active against L. monocytogenes ATCC19111, L. monocytogenes BG322, L. monocytogenes IMS1, L. monocytogenes IMS2, L. monocytogenes IMS3, L. monocytogenes IMS4, L. ivanovii ATCC19119, and L. inocua ATCC33090, while bacteriocin 31 exhibited much larger zones of inhibition and showed additional antimicrobial activity against En. faecium DDE4, Lactobacillus plantarum BGGO7-29, Lb. plantarum BGVL2a-18, and Leuconostoc mesenteroides subsp. cremoris BGTRS1-2. Enterocin P did not show antimicrobial activity against any of the tested indicator strains (Figure 3).
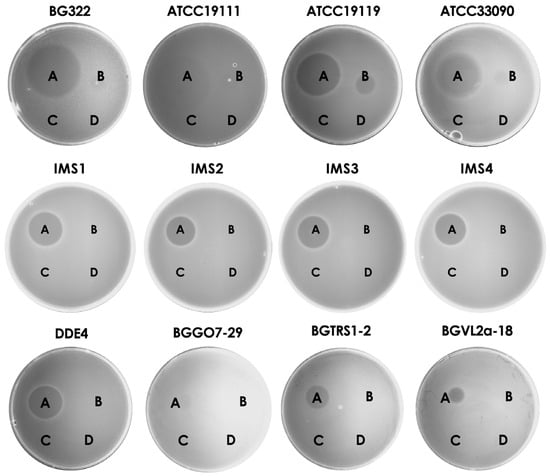
Figure 3.
Antimicrobial effect of purified bacteriocins on indicator strains. Note: A—bacteriocin 31; B—bacteriocin 32; C—enterocin P; D—Enterokinase buffer.
Further, the same volume used for testing individual bacteriocins was applied in combination to investigate the synergistic effect of purified bacteriocins. It was demonstrated that bacteriocins did not exhibit a synergistic effect, and the size of the antimicrobial zone on the indicator strain was the same in bacteriocin combination, as well as after the application of bacteriocin 31 alone (Figure 4).

Figure 4.
Synergistic effect of purified bacteriocins. Note: 1—bacteriocin 31; 2—bacteriocin 32; 3—enterocin P; 4—the same volume of bacteriocin 31 and bacteriocin 32; 5—the same volume of bacteriocin 31 and enterocin P; 6—the same volume of bacteriocin 32 and enterocin P; 7—the same volume of bacteriocin 31, bacteriocin 32, and enterocin P; 8—enterokinase buffer.
Since we demonstrated the strong antimicrobial activity of bacteriocin 31 and the weak antimicrobial activity of bacteriocin 32, the next goal was to determine arbitrary units per milliliter/mg of purified bacteriocins. We showed that bacteriocin 31 retained its antimicrobial activity after the eight two-fold dilutions against BG322, while bacteriocin 32 lost its activity after the second two-fold dilution. Since the concentrations of both bacteriocins after purification were similar (bacteriocin 31, 2.3 ± 0.3 μg/μL, and bacteriocin 32, 2.8 ± 0.2 μg/μL), it was concluded that bacteriocin 31 was more potent against all L. monocytogenes strains (Figure 5).
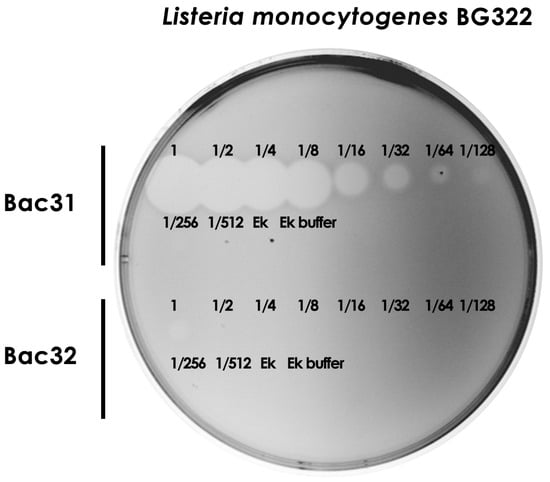
Figure 5.
Determination of arbitrary units per milliliter of purified bacteriocins. Note: Fractions represent a two-fold dilution of purified bacteriocin in Ek buffer. Ek—enterokinase; Ek buffer—enterokinase buffer; Bac31—bacteriocin 31; Bac32—bacteriocin 32.
3.6. Application of Purified Bacteriocin 31 Effectively Reduces Listeria monocytogenes in a Milk Model
Although the application of bacteriocin and/or bacteriocin-producing starters is considered a good way to protect a food product from pathogenic and food spoilage bacteria, the limiting factor is the weak activity of bacteriocins in complex food systems or the sensitivity of bacteriocins to various technological procedures []. Our study demonstrated that purified bacteriocin 31 (100, 500, and 1000 AU/mL) had a significant inhibitory effect after three days of storage on the growth of the L. monocytogenes BG322, L. monocytogenes IMS1, L. monocytogenes IMS2, L. monocytogenes IMS3, and L. monocytogenes IMS4 in the milk model. A decrease in the number of L. monocytogenes BG322 during three days of storage by applying different concentrations of the purified bacteriocin (100, 500, and 1000 AU/mL) was observed from 1.4 × 107 CFU/mL to 5 × 106 CFU/mL, 2.1 × 103 CFU/mL, and 1.6 × 103 CFU/mL, respectively. Also, during the additional four days of storage, the number of L. monocytogenes BG322 compared with the control milk was reduced, and the application of the highest concentration of bacteriocin 31 failed to eliminate L. monocytogenes BG322 from the milk in 5 days (Figure 6A). Similarly, the application of bacteriocin 31 in three concentrations reduced the presence of L. monocytogenes isolated from different types of cheese. The lowest applied concentration of bacteriocin 31 reduced the abundance of all four L. monocytogenes after the third day of storage, while concentrations of 500 and 1000 AU/mL reduced the abundance from about 108 to 27–57 CFU/mL. After the fifth and seventh days of storage, with the application of the highest concentration of 1000 AU/mL, the abundance of all four L. monocytogenes was significantly reduced compared with untreated milk samples. On the seventh day, the number of L. monocytogenes IMS1 compared with the control milk was reduced with the highest concentration from 2.67 × 108 to 6.00 × 104 CFU/mL (Figure 6B), L. monocytogenes IMS2 from 6.33 × 108 to 2.93 × 105 CFU/mL (Figure 6C), L. monocytogenes IMS3 from 4.33 × 108 to 2.67 × 104 CFU/mL (Figure 6D), and L. monocytogenes IMS4 from 1.43 × 108 to 5.00 × 104 CFU/mL (Figure 6E).
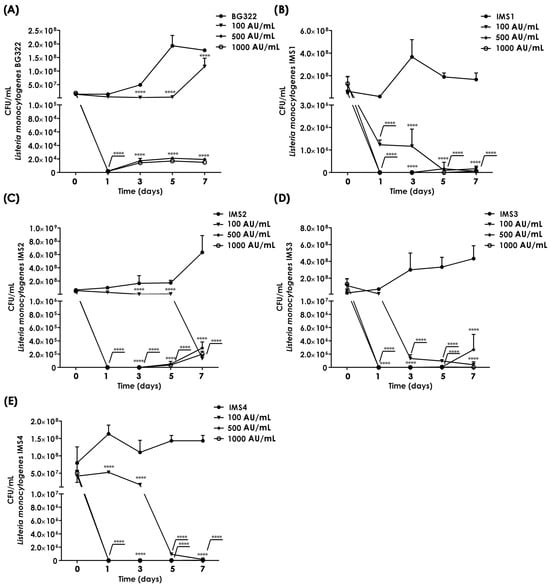
Figure 6.
Reduction in Listeria monocytogenes in a milk model with purified bacteriocin 31. L. monocytogenes BG322 (A), L. monocytogenes IMS1 (B), L. monocytogenes IMS2 (C), L. monocytogenes IMS3 (D), and L. monocytogenes IMS4 (E). Note: Statistical significance p < 0.05 ****.
4. Discussion
Nowadays, there is a tendency to consume ready-to-eat food without or with minimal food processing to reduce the nutrients lost []. On the other hand, eating ready-to-eat food might contaminate it with pathogens, which can cause illnesses in humans. One of the important foodborne pathogens is L. monocytogenes, which causes listeriosis. Listeriosis is a very serious foodborne disease among high-risk population groups such as the immunocompromised, which is associated with a high mortality rate []. Additionally, the consumption of raw milk and raw milk products or post-pasteurization contamination can cause listeriosis outbreaks. There has been a growing need for alternative, natural agents for food preservation and extending food’s shelf life in recent years. Bacteriocins represent safe and harmless alternatives in the control of pathogenic bacteria in food []. Because of bacteriocin production, many LABs have been successfully applied to inhibit foodborne pathogens [,]. Among LAB species, Enterococcus sp. is generally used as a nonstarter lactic acid bacteria (NSLAB) in a variety of cheeses, especially artisan cheeses produced in southern Europe []. Enterococcus species do not have a generally recognized as safe (GRAS) status, despite the knowledge that the pathogenesis of enterococci is a strain-specific property and is more common to clinical enterococci than to food-based enterococci []. Due to their controversial nature, varying from pathogens to probiotics [], one of the aims of this work was to apply a WGS-based approach for safety assessment and probiotic potential analysis of bacteriocin-producing En. faecium BGPAS1-3 originating from young cheese. The genome size of enterococci varies between 2.7 and 3.6 Mb, with a GC content ranging from 37% to 45%. According to Dublin and Palmer [], the commensal enterococcal strains have smaller genomes compared with the clinical isolates, where genomic variability is caused by the presence of foreign genetic material. The BGPAS1-3 genome size is 2.75 Mb with a total of 2664 protein-coding genes and contains seven plasmids, and when compared with the pathogenic Aus0004, Aus0085, and DO, as well as the probiotic strain T110, differences in sequences are observed. Similar to our study, a comparison of the genomes of probiotic and pathogenic enterococci revealed that the probiotic strain Enterococcus faecalis EF-2001 is significantly different from the pathogenic strains []. Analysis of the draft genome using bioinformatics programs showed that En. faecium BGPAS1-3 is not pathogenic. However, BGPAS1-3 carries the enterococcal intrinsic resistance gene msr(C), responsible for resistance to macrolides and aac(6′)-Ii, which encodes an aminoglycoside acetyltransferase responsible for low-level resistance to aminoglycoside. The previous study showed that BGPAS1-3 was sensitive to erythromycin and resistant to low-level gentamicin []. In addition, it was found that BGPAS1-3 did not harbor any AMR genes reported for pathogenic enterococci, such as vancomycin resistance genes [,]. Moreover, the identification of virulence genes is crucial before classifying enterococci strains as a probiotic. After screening for the presence of virulence genes, genes associated with adhesion, biofilm formation, and antiphagocytosis were found in the BGPAS1-3 genome. In previous studies, a correlation between the presence of these genes and colonization ability was shown. It is well known that adhesion is essential for commensal and probiotic enterococci to promote colonization, persist, and avoid elimination from the host []. For beneficial properties, the BGPAS1-3 genome was mined for genes associated with essential amino acid biosynthetic pathways. It has been shown that the BGPAS1-3 genome contains genes involved in the biosynthesis of five amino acids: alanine, aspartate, arginine, cysteine, and valine. A previous study showed that En. faecium 17OM39 has the potential to synthesize essential amino acids, and Ghattargi and coauthors [] assumed that these amino acids serve as precursors for the synthesis of short-chain fatty acids. Enterococcus sp. produces a wide diversity of bacteriocins, called enterocins, ribosomally synthesized antimicrobial peptides with specific antimicrobial activity to inhibit the growth of species closely related to the producing bacteria []. They have been widely studied because of their activities against some foodborne pathogens and spoilage bacteria []. Most enterocins are class II bacteriocins and are defined as unmodified and heat stable []. One of the goals of this study was the cloning and overexpression of the thermostable bacteriocins of the BGPAS1-3 strain, which was shown in a previous study to contribute to the reduction in L. monocytogenes ATCC19111 adhesion to epithelial cells [,]. Using the BAGEL4 software, genes for four bacteriocins (enterolysin A, bacteriocin 31, bacteriocin 32, and enterocin P) were found in the BGPAS1-3 genome. Enterocin P and bacteriocin 31 are classified as “Class II.1. enterocins of the pediocin family” according to Franz and co-authors [], whereas bacteriocin 32 is classified as “Class II.3. other linear nonpediocin-like enterocins”. Furthermore, three small thermostable bacteriocins (bacteriocin 31, bacteriocin 32, and enterocin P) of BGPAS1-3 were selected for cloning and overexpression. All three bacteriocins have been previously described [], but bacteriocins from strain BGPAS1-3 showed amino acid differences, especially in the leader peptide sequence. Our previous studies demonstrated that the enhanced E. coli pMAL expression system was highly effective for expressing lactococci-like peptides [,]. Consequently, we successfully used this system to express and purify three bacteriocins from strain BGPAS1-3. It was surprising to observe a significant variation in the antimicrobial activity of the bacteriocins expressed in E. coli: bacteriocin 31 exhibited exceptional specific antimicrobial activity, bacteriocin 32 displayed weak activity, and enterocin P showed no detectable activity. Similar results were obtained with expressed potential bacteriocins of L. lactis S50 []. For strain BGPAS1-3, the antimicrobial peptides responsible for its antimicrobial activity have not been previously purified or characterized. Therefore, it is unclear whether the analyzed bacteriocins exhibit antimicrobial activity in the natural strain or if the expression system in E. coli fails to produce the active form of these peptides. Further research will focus on determining whether additional processing is required for these bacteriocins’ proper synthesis and activity. Enterococcal bacteriocins have been successfully expressed in various heterologous systems, including E. coli []. Interestingly, no antimicrobial activity was obtained in this study, although the sequence of the expressed enterocin P protein from strain BGPAS1-3 was 100% identical to the previously expressed one where activity was obtained. The difference is that in the previous study, together with enterocin P, the authors expressed a potential protein responsible for immunity []. Bacteriocin 31, which showed the greatest difference in the amino acid sequence, showed the highest antimicrobial activity, but considering that we do not have both clones (bacteriocin 31 from BGPAS1-3 and other enterococci), we cannot make a comparison and claim whether it is a consequence of changed amino acids or all clones can exhibit high antimicrobial activity. Site-specific mutagenesis of the pMAL5cHisEk_Bac31 clone, which is planned in the future, will provide an answer about the contribution of individual amino acids to the antimicrobial activity and spectrum of action.
This study showed that purified bacteriocins (bacteriocin 31 and bacteriocin 32) had antimicrobial activities against bacteria from the genera Listeria, Enterococcus, Lactobacillus, and Leuconostoc. While previous studies have shown that enterocin P is active against LAB and foodborne pathogens like L. monocytogenes [], this study found no evidence of activity for enterocin P. One of the approaches to eliminating foodborne pathogens is a combination of bacteriocins that show a synergistic effect. The effect of Nisin with other antimicrobial compounds in the effective elimination of pathogens is known []. Therefore, we examined the synergistic effect of three bacteriocins in reducing the number of L. monocytogenes, and we showed that the use of bacteriocin 31, bacteriocin 32, and enterocin P does not have a synergistic effect. However, it remains an open question for future studies of the synergistic effect of these bacteriocins with technological processes of food production. Bearing in mind that the examined bacteriocins are thermostable, the effect of synergism should be examined with processes that require high temperatures. The application of bacteriocins in milk and the subsequent heat treatment of milk are possible means to eliminate L. monocytogenes. In this study, we showed that the application of the most potent purified bacteriocin 31 at different concentrations was able to reduce L. monocytogenes BG322, IMS1, IMS2, IMS3, and IMS4 strains in the milk model after three days of storage. However, during storage at 4 °C, even the highest concentration of bacteriocin failed to eliminate L. monocytogenes. Dal Bello and coauthors proposed the pretreatment of milk before cheese production as the most effective method in the biocontrol of L. monocytogenes [].
5. Conclusions
Probiogenomic analysis of En. faecium BGPAS1-3 allows us to better understand its safety status and probiotic potential. Also, these data helped us to investigate the potential application of live strains and holobiotics in the food industry. However, due to controlled application and efficiency, the purified thermostable bacteriocins provide a more practical solution for combating foodborne pathogens. The use of practical tools for the overexpression of such bacteriocins contributed to an efficient reduction in L. monocytogenes in milk. In this study, we demonstrated that some enterococcal bacteriocins could be successfully expressed in massive amounts in E. coli hosts and applied in food preservation. However, additional experiments are necessary to establish a correlation between the antimicrobial potential, the spectrum of activity, and the amino acid sequence of the bacteriocins.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/foods13162637/s1: Supplementary Figure S1: The presence of genes encoding bacteriocins and non-bactericidal post-translationally modified peptides using the BAGEL4 online tool; Supplementary Figure S2: Comparison of amino acid sequences of Enterococcus faecium BGPAS1-3 bacteriocin 31 and representative bacteriocin 31; Supplementary Table S1: Putative antimicrobial resistance genes in Enterococcus faecium BGPAS1-3; Supplementary Table S2: Putative virulence factors in Enterococcus faecium BGPAS1-3 and comparison with other Enterococcus faecium strains; Supplementary Table S3: Putative plasmids identification against a Gram-positive database using PlasmidFinder of Enterococcus faecium BGPAS1-3; Supplementary Table S4: Putative genes involved in the synthesis of essential amino acids in Enterococcus faecium BGPAS1-3.
Author Contributions
N.P. and M.K., conceptualization, formal analysis, investigation, data curation, writing—original draft, and writing—review and editing; D.R. and E.B., methodology, software, and writing—original draft; D.S., K.V. and M.Ž., investigation and formal analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Ministry of Science, Technological Development and Innovation of the Republic of Serbia under registration No. 451-03-66/2024-03/200042.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors. Draft genome sequences of BGPAS1-3 have been deposited in GenBank under accession number JAUKVL000000000. https://www.ncbi.nlm.nih.gov/nuccore/JAUKVL000000000 (accessed on 13 July 2023).
Acknowledgments
We acknowledge Branko Velebit, principal research fellow, head of the Department of Microbiology and Molecular Biology, Institute of Meat Hygiene and Technology (INMES), University of Belgrade, Serbia.
Conflicts of Interest
The strains Streptococcus thermophilus BGKMJ1-36, Lactobacillus bulgaricus BGVLJ1-21, Lactobacillus plantarum BGGO7-29, Leuconostoc mesenteroides subsp. cremoris BGTRS1-2, Lactobacillus plantarum BGVL2a-18, Lactococcus lactis subsp. lactis BGVL2-8, Lactococcus lactis subsp. l actisBGTRK4-21, Lactococcus lactis subsp. lactis biovar. diacetylactis BGTRK10-2, and Lactococcus lactis subsp. cremoris BGTRM1-22 were deposited in the BCCM/LMG Bacterial Culture Collection in Belgium under patent deposition and are governed by a license agreement between the Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Serbia, and Invetlab d.o.o. Belgrade, Serbia. The funders had no involvement in the study design, data collection, analysis, interpretation, manuscript writing, or decision to publish the results.
References
- Jibo, G.G.; Raji, Y.E.; Salawudeen, A.; Amin-Nordin, S.; Mansor, R.; Jamaluddin, T.Z.M.T. A Systematic Review and Meta-Analysis of the Prevalence of Listeria monocytogenes in South-East Asia; a One-Health Approach of Human-Animal-Food-Environment. One Health 2022, 15, 100417. [Google Scholar] [CrossRef] [PubMed]
- Hafner, L.; Pichon, M.; Burucoa, C.; Nusser, S.H.A.; Moura, A.; Garcia-Garcera, M.; Lecuit, M. Listeria monocytogenes Faecal Carriage Is Common and Depends on the Gut Microbiota. Nat. Commun. 2021, 12, 6826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Chen, X.; Qu, C. Review Controlling Listeria monocytogenes in Ready-to-Eat Meat and Poultry Products: An Overview of Outbreaks, Current Legislations, Challenges, and Future Prospects. Trends Food Sci. Technol. 2021, 116, 24–35. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Olanya, O.M.; Hoshide, A.K.; Ijabadeniyi, O.A.; Ukuku, D.O.; Mukhopadhyay, S.; Niemira, B.A.; Ayeni, O. Cost Estimation of Listeriosis (Listeria monocytogenes) Occurrence in South Africa in 2017 and Its Food Safety Implications. Food Control 2019, 102, 231–239. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Libera, K.; Konieczny, K.; Grabska, J.; Szopka, W.; Augustyniak, A.; Pomorska-Mól, M. Selected Livestock-Associated Zoonoses as a Growing Challenge for Public Health. Infect. Dis. Rep. 2022, 14, 63–81. [Google Scholar] [CrossRef]
- Thakur, M.; Asrani, R.K.; Patial, V. Listeria monocytogenes: A Food-Borne Pathogen. In Foodborne Diseases; Holban, A.M., Grumezescu, A.M., Eds.; Handbook of Food Bioengineering, Academic Press: Cambridge, MA, USA, 2018; pp. 157–192. ISBN 978-0-12-811444-5. [Google Scholar]
- Drolia, R.; Bhunia, A.K. Crossing the Intestinal Barrier via Listeria Adhesion Protein and Internalin A. Trends Microbiol. 2019, 27, 408–425. [Google Scholar] [CrossRef]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and Virulence of Listeria monocytogenes : A Trip from Environmental to Medical Microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes Biofilms in the Food Industry: Is the Current Hygiene Program Sufficient to Combat the Persistence of the Pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef]
- Skowron, K.; Hulisz, K.; Gryń, G.; Olszewska, H.; Wiktorczyk, N.; Paluszak, Z. Comparison of Selected Disinfectants Efficiency against Listeria monocytogenes Biofilm Formed on Various Surfaces. Int. Microbiol. 2018, 21, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Givskov, M. Quorum-Sensing Inhibitors as Anti-Pathogenic Drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic Characterization of Antibiotic Resistance in Listeria and Production of Antimicrobial and Virulence Factors. Int. J. Mol. Sci. 2021, 22, 8141. [Google Scholar] [CrossRef]
- Farber, J.M.; Zwietering, M.; Wiedmann, M.; Schaffner, D.; Hedberg, C.W.; Harrison, M.A.; Hartnett, E.; Chapman, B.; Donnelly, C.W.; Goodburn, K.E.; et al. Alternative Approaches to the Risk Management of Listeria monocytogenes in Low Risk Foods. Food Control 2021, 123, 107601. [Google Scholar] [CrossRef]
- Ceruso, M.; Liu, Y.; Gunther, N.W.; Pepe, T.; Anastasio, A.; Qi, P.X.; Tomasula, P.M.; Renye, J.A. Anti-Listerial Activity of Thermophilin 110 and Pediocin in Fermented Milk and Whey. Food Control 2021, 125, 107941. [Google Scholar] [CrossRef]
- Liu, Y.; Ceruso, M.; Jiang, Y.; Datta, A.R.; Carter, L.; Strain, E.; Pepe, T.; Anastasi, A.; Fratamico, P. Construction of Listeria monocytogenes Mutants with In-Frame Deletions in the Phosphotransferase Transport System (PTS) and Analysis of Their Growth under Stress Conditions. J. Food Sci. 2013, 78, M1392–M1398. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health Benefits of Lactic Acid Bacteria (LAB) Fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef]
- Kasimin, M.E.; Shamsuddin, S.; Molujin, A.M.; Sabullah, M.K.; Gansau, J.A.; Jawan, R. Enterocin: Promising Biopreservative Produced by Enterococcus sp. Microorganisms 2022, 10, 684. [Google Scholar] [CrossRef]
- Olatunde, O.O.; Benjakul, S. Natural Preservatives for Extending the Shelf-Life of Seafood: A Revisit. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1595–1612. [Google Scholar] [CrossRef]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, Synthesis, Mechanism of Action and Resistance Development in Food Spoilage Causing Bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef]
- Lozo, J.; Topisirovic, L.; Kojic, M. Natural Bacterial Isolates as an Inexhaustible Source of New Bacteriocins. Appl. Microbiol. Biotechnol. 2021, 105, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.A.P.; van Belkum, M.J.; Holzapfel, W.H.; Abriouel, H.; Gálvez, A. Diversity of Enterococcal Bacteriocins and Their Grouping in a New Classification Scheme. FEMS Microbiol. Rev. 2007, 31, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as Probiotics and Their Implications in Food Safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef]
- Favaro, L.; Barretto Penna, A.L.; Todorov, S.D. Bacteriocinogenic LAB from Cheeses—Application in Biopreservation? Trends Food Sci. Technol. 2015, 41, 37–48. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef]
- Peng, X.; Ed-Dra, A.; Yue, M. Whole Genome Sequencing for the Risk Assessment of Probiotic Lactic Acid Bacteria. Crit. Rev. Food Sci. Nutr. 2023, 63, 11244–11262. [Google Scholar] [CrossRef]
- EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain-2021—EFSA Journal—Wiley Online Library. Available online: https://efsa.onlinelibrary.wiley.com/doi/full/10.2903/j.efsa.2021.6506 (accessed on 3 July 2024).
- Haranahalli Nataraj, B.; Behare, P.V.; Yadav, H.; Srivastava, A.K. Emerging Pre-Clinical Safety Assessments for Potential Probiotic Strains: A Review. Crit. Rev. Food Sci. Nutr. 2023, 11, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Zhao, X.; Zhen, Z.; Wang, X.; Guo, N. Synergy of a Combination of Nisin and Citric Acid against Staphylococcus aureus and Listeria monocytogenes. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2017, 34, 2058–2068. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial Production of Bacteriocins: Latest Research Development and Applications. Biotechnol. Adv. 2018, 36, 2187–2200. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, A.; Abriouel, H.; López, R.L.; Ben Omar, N. Bacteriocin-Based Strategies for Food Biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [CrossRef]
- Parish, J.H. Genetic Manipulation of Streptomyces—A Laboratory Manual: By D A Hopwood, M J Bibb, K F Chater; T Kieser CJ Bruton, H M Kieser, D J Lydiate, C P Smith, J M Ward and H Schrempf. Pp 356. The John Innes Foundation, Norwich, UK and Cold Spring Harbour Laboratory. 1985. $25 ISBN 0-7084-0336-0. Biochem. Educ. 1986, 14, 196. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An Open Online Resource for Identification of Antimicrobial Resistance Genes in Next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder Web-Services for Easy Identification of Acquired Antibiotic Resistance and E. coli Virulence Genes in Bacteriophage and Prophage Nucleotide Sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Enuh, B.M.; Gedikli, S.; Aytar Çelik, P.; Çabuk, A. Genome Sequence and Probiotic Potential of Newly Isolated Enterococcus durans Strain MN187066. Lett. Appl. Microbiol. 2023, 76, ovad035. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on Transformation of Escherichia coli with Plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Kojic, M.; Svircevic, J.; Banina, A.; Topisirovic, L. Bacteriocin-Producing Strain of Lactococcus lactis subsp. diacitilactis S50. Appl. Environ. Microbiol. 1991, 57, 1835–1837. [Google Scholar] [CrossRef] [PubMed]
- Vukotic, G.; Polovic, N.; Mirkovic, N.; Jovcic, B.; Stanisavljevic, N.; Fira, D.; Kojic, M. Lactococcin B Is Inactivated by Intrinsic Proteinase PrtP Digestion in Lactococcus lactis subsp. lactis BGMN1-501. Front. Microbiol. 2019, 10, 874. [Google Scholar] [CrossRef]
- Ansari, A.; Zohra, R.R.; Tarar, O.M.; Qader, S.A.U.; Aman, A. Screening, Purification and Characterization of Thermostable, Protease Resistant Bacteriocin Active against Methicillin Resistant Staphylococcus aureus (MRSA). BMC Microbiol. 2018, 18, 192. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.L.; Gaya, P.; Medina, M.; Nuñez, M. Bactericidal Effect of Enterocin 4 on Listeria monocytogenes in a Model Dairy System. J. Food Prot. 1997, 60, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Popović, N.; Đokić, J.; Brdarić, E.; Dinić, M.; Terzić-Vidojević, A.; Golić, N.; Veljović, K. The Influence of Heat-Killed Enterococcus faecium BGPAS1-3 on the Tight Junction Protein Expression and Immune Function in Differentiated Caco-2 Cells Infected With Listeria monocytogenes ATCC 19111. Front. Microbiol. 2019, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Gardijan, L.; Miljkovic, M.; Obradovic, M.; Borovic, B.; Vukotic, G.; Jovanovic, G.; Kojic, M. Redesigned pMAL Expression Vector for Easy and Fast Purification of Active Native Antimicrobial Peptides. J. Appl. Microbiol. 2022, 133, 1001–1013. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, L. Encapsulated Natural Antimicrobials: A Promising Way to Reduce Microbial Growth in Different Food Systems. Food Control 2021, 123, 107678. [Google Scholar] [CrossRef]
- Archer, D.L. The Evolution of FDA’s Policy on Listeria monocytogenes in Ready-to-Eat Foods in the United States. Curr. Opin. Food Sci. 2018, 20, 64–68. [Google Scholar] [CrossRef]
- Sebastianski, M.; Bridger, N.A.; Featherstone, R.M.; Robinson, J.L. Disease Outbreaks Linked to Pasteurized and Unpasteurized Dairy Products in Canada and the United States: A Systematic Review. Can. J. Public. Health Rev. Can. Santé Publique 2022, 113, 569–578. [Google Scholar] [CrossRef]
- Verma, D.K.; Thakur, M.; Singh, S.; Tripathy, S.; Gupta, A.K.; Baranwal, D.; Patel, A.R.; Shah, N.; Utama, G.L.; Niamah, A.K.; et al. Bacteriocins as Antimicrobial and Preservative Agents in Food: Biosynthesis, Separation and Application. Food Biosci. 2022, 46, 101594. [Google Scholar] [CrossRef]
- Golić, N.; Veljović, K.; Popović, N.; Đokić, J.; Strahinić, I.; Mrvaljević, I.; Terzić-Vidojević, A. In vitro and in vivo Antagonistic Activity of New Probiotic Culture against Clostridium difficile and Clostridium perfringens. BMC Microbiol. 2017, 17, 108. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Giraffa, G. Functionality of Enterococci in Dairy Products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Terzić-Vidojević, A.; Veljović, K.; Popović, N.; Tolinački, M.; Golić, N. Enterococci from Raw-Milk Cheeses: Current Knowledge on Safety, Technological, and Probiotic Concerns. Foods 2021, 10, 2753. [Google Scholar] [CrossRef]
- Popović, N.; Dinić, M.; Tolinački, M.; Mihajlović, S.; Terzić-Vidojević, A.; Bojić, S.; Djokić, J.; Golić, N.; Veljović, K. New Insight into Biofilm Formation Ability, the Presence of Virulence Genes and Probiotic Potential of Enterococcus sp. Dairy Isolates. Front. Microbiol. 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectr. 2014, 5, 1110–1128. [Google Scholar] [CrossRef]
- Panthee, S.; Paudel, A.; Hamamoto, H.; Ogasawara, A.A.; Iwasa, T.; Blom, J.; Sekimizu, K. Complete Genome Sequence and Comparative Genomic Analysis of Enterococcus faecalis EF-2001, a Probiotic Bacterium. Genomics 2021, 113, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- Gorrie, C.; Higgs, C.; Carter, G.; Stinear, T.P.; Howden, B. Genomics of Vancomycin-Resistant Enterococcus faecium. Microb. Genom. 2019, 5, e000283. [Google Scholar] [CrossRef]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of Virulence in Enterococcus faecium, a Hospital-Adapted Opportunistic Pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Ghattargi, V.C.; Nimonkar, Y.S.; Burse, S.A.; Davray, D.; Kumbhare, S.V.; Shetty, S.A.; Gaikwad, M.A.; Suryavanshi, M.V.; Doijad, S.P.; Utage, B.; et al. Genomic and Physiological Analyses of an Indigenous Strain, Enterococcus faecium 17OM39. Funct. Integr. Genom. 2018, 18, 385–399. [Google Scholar] [CrossRef]
- Negash, A.W.; Tsehai, B.A. Current Applications of Bacteriocin. Int. J. Microbiol. 2020, 2020, 4374891. [Google Scholar] [CrossRef]
- Popović, N.; Stevanović, D.; Radojević, D.; Veljović, K.; Đokić, J.; Golić, N.; Terzić-Vidojević, A. Insight into the Postbiotic Potential of the Autochthonous Bacteriocin-Producing Enterococcus faecium BGZLM1-5 in the Reduction in the Abundance of Listeria monocytogenes ATCC19111 in a Milk Model. Microorganisms 2023, 11, 2844. [Google Scholar] [CrossRef]
- Malesevic, M.; Stanisavljevic, N.; Miljkovic, M.; Jovcic, B.; Filipic, B.; Studholme, D.J.; Kojic, M. TThe Large Plasmidome of Lactococcus lactis subsp. lactis Bv. diacetylactis S50 Confers Its Biotechnological Properties. Int. J. Food Microbiol. 2021, 337, 108935. [Google Scholar] [CrossRef]
- Fathizadeh, H.; Saffari, M.; Esmaeili, D.; Moniri, R.; Salimian, M. Evaluation of Antibacterial Activity of Enterocin A-Colicin E1 Fusion Peptide. Iran. J. Basic Med. Sci. 2020, 23, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.; Criado, R.; Citti, R.; Martín, M.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Cloning, Production and Functional Expression of Enterocin P, a Sec-Dependent Bacteriocin Produced by Enterococcus faecium P13, in Escherichia coli. Int. J. Food Microbiol. 2005, 103, 239–250. [Google Scholar] [CrossRef]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and Genetic Characterization of Enterocin P, a Novel Sec-Dependent Bacteriocin from Enterococcus faecium P13 with a Broad Antimicrobial Spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [CrossRef] [PubMed]
- Dal Bello, B.; Cocolin, L.; Zeppa, G.; Field, D.; Cotter, P.D.; Hill, C. Technological Characterization of Bacteriocin Producing Lactococcus lactis Strains Employed to Control Listeria monocytogenes in Cottage Cheese. Int. J. Food Microbiol. 2012, 153, 58–65. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).