Use of Transcriptomics to Reveal the Joint Immunotoxicity Mechanism Initiated by Difenoconazole and Chlorothalonil in the Human Jurkat T-Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Jurkat Cell Culture
2.3. Immunotoxicity Evaluation of Single Pesticide Exposure
2.4. Joint Immunotoxicity Evaluation
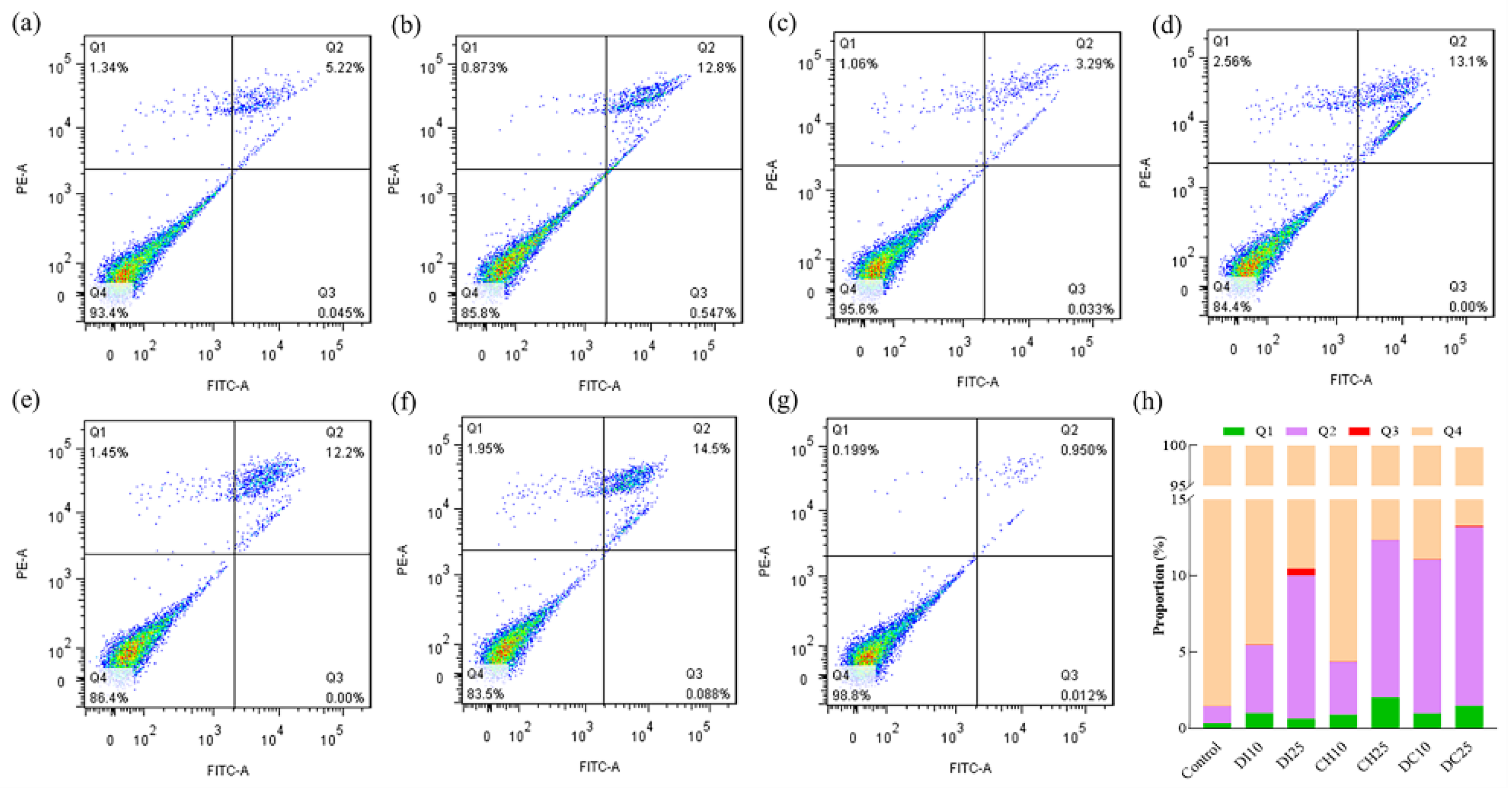
2.5. Cell Apoptosis Analysis
2.6. RNA Extraction and Sequencing
2.7. Read Mapping and Differentially Expressed Gene Analysis
2.8. Quantitative Real-Time PCR (qRT–PCR)
2.9. Statistical Analysis
3. Results and Discussion
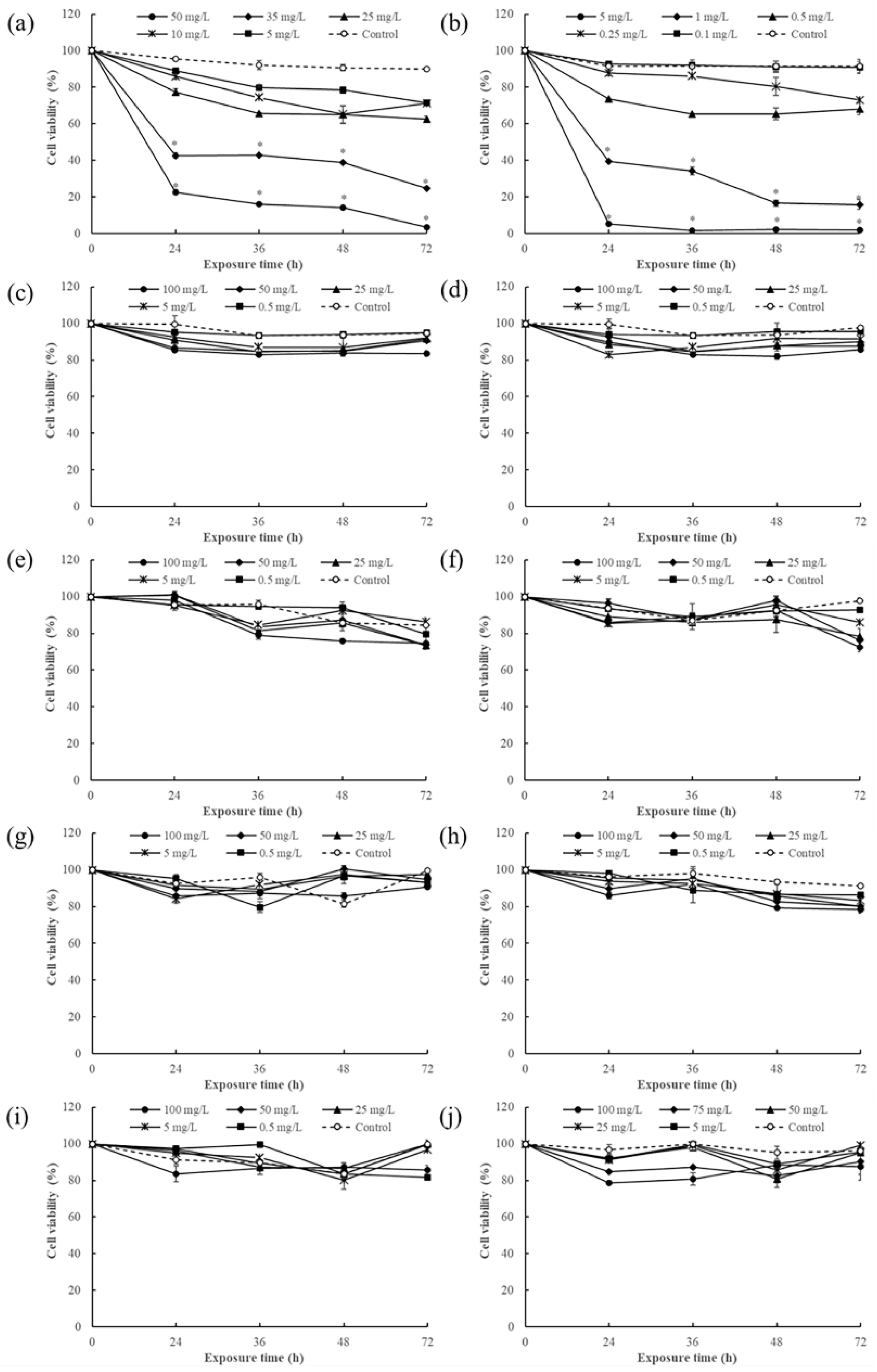
3.1. Immunotoxicity Screening of Pesticides Based on Human Jurkat Cells
3.2. Effect of Single Pesticide Exposure on Cell Viability
3.3. Effects of Combined Exposure to Difenoconazole and Chlorothalonil on Cell Viability
3.4. Effects of Pesticide Exposure on Cell Apoptosis
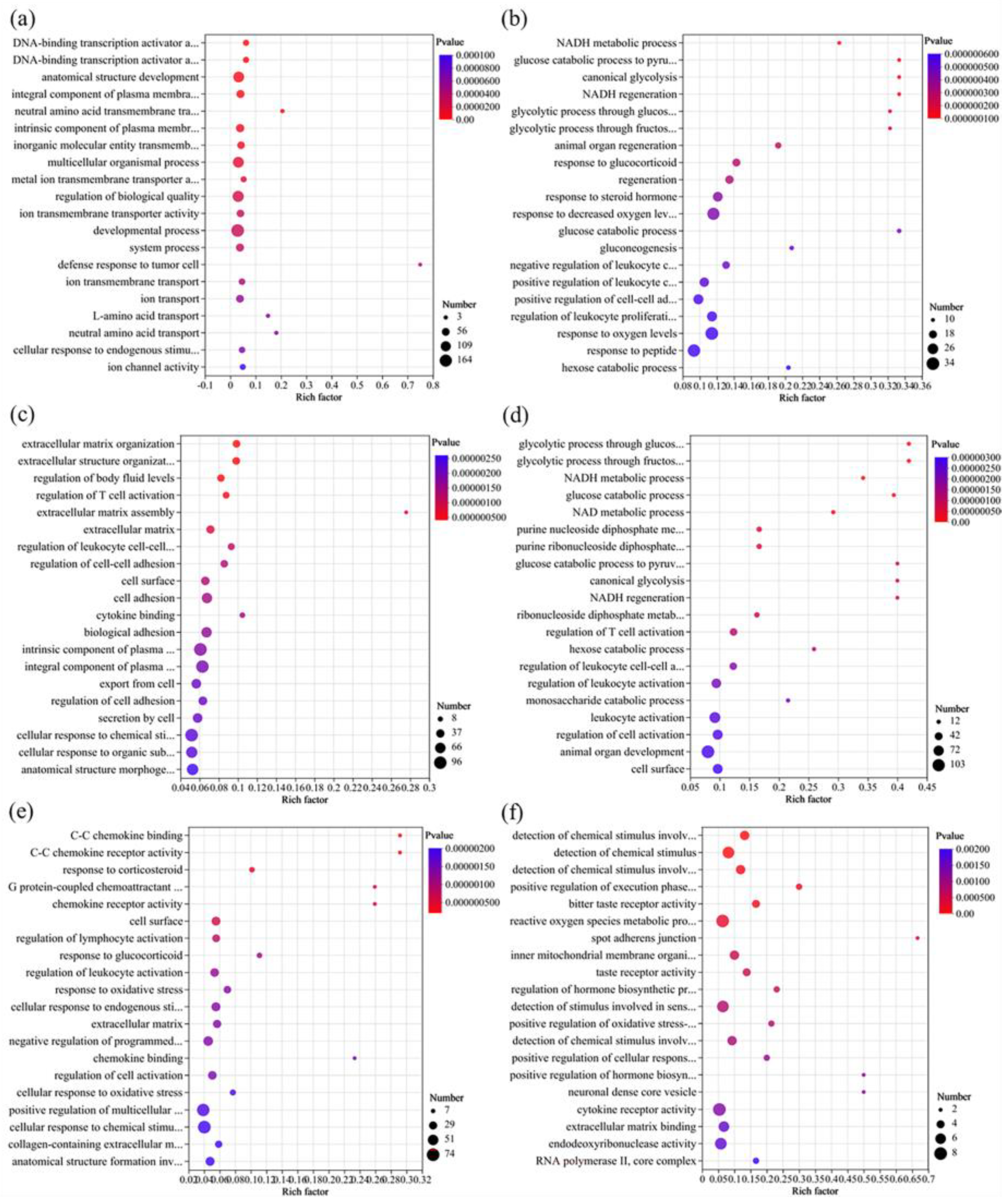
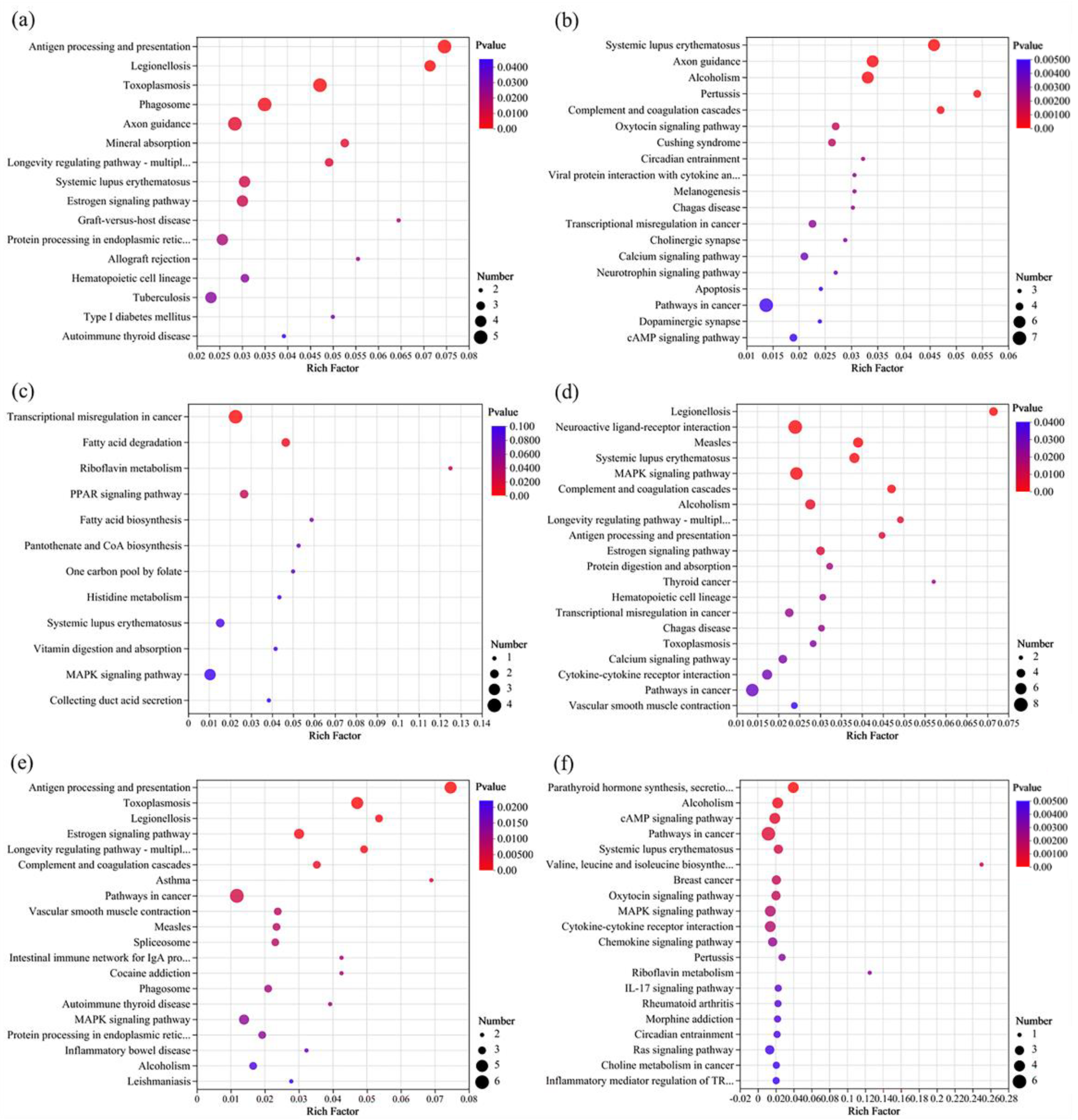
3.5. Transcriptomics Analysis
3.5.1. Overview
3.5.2. GO Enrichment Analysis of DEGs
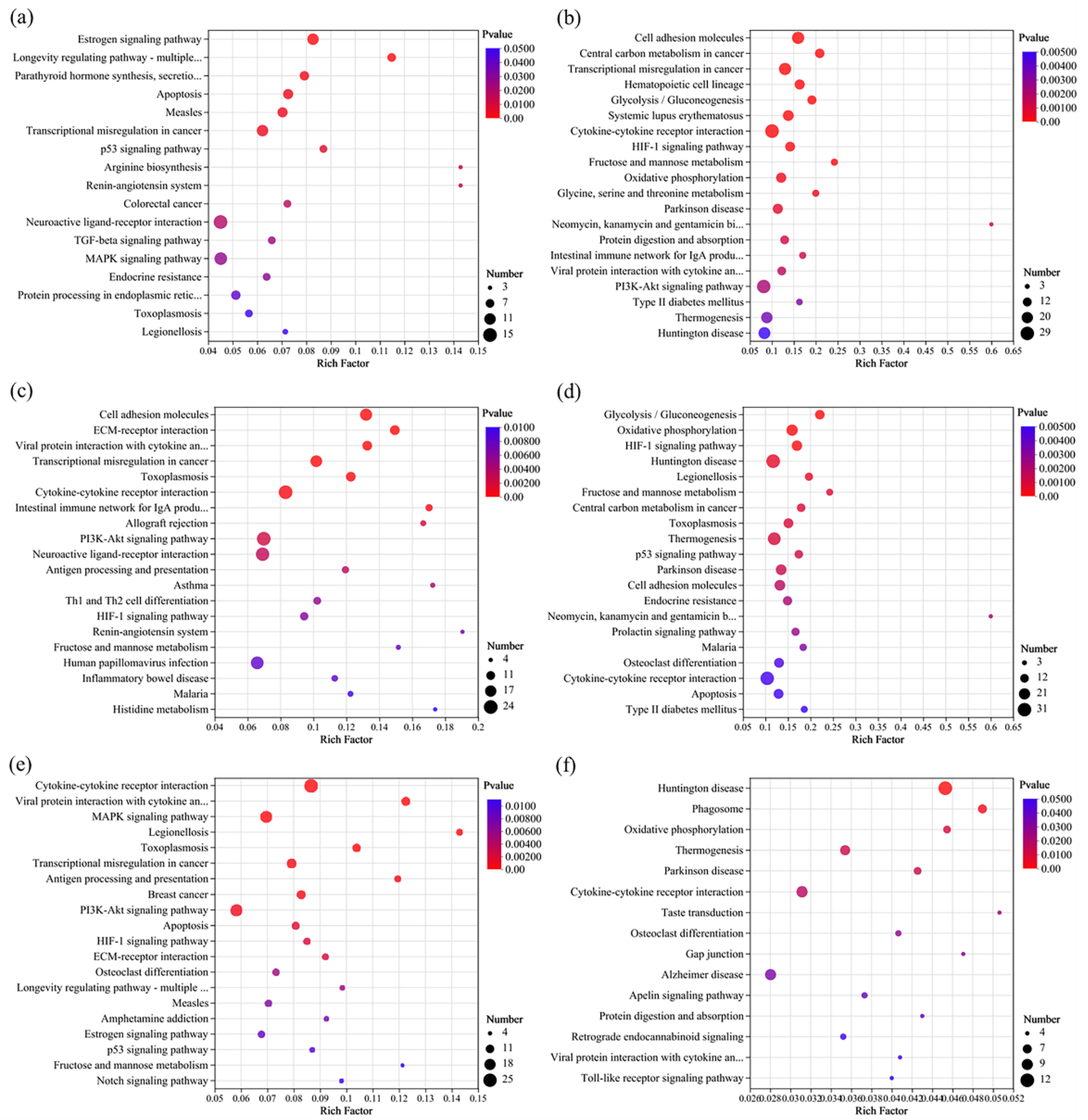
3.5.3. KEGG Enrichment Analysis of DEGs
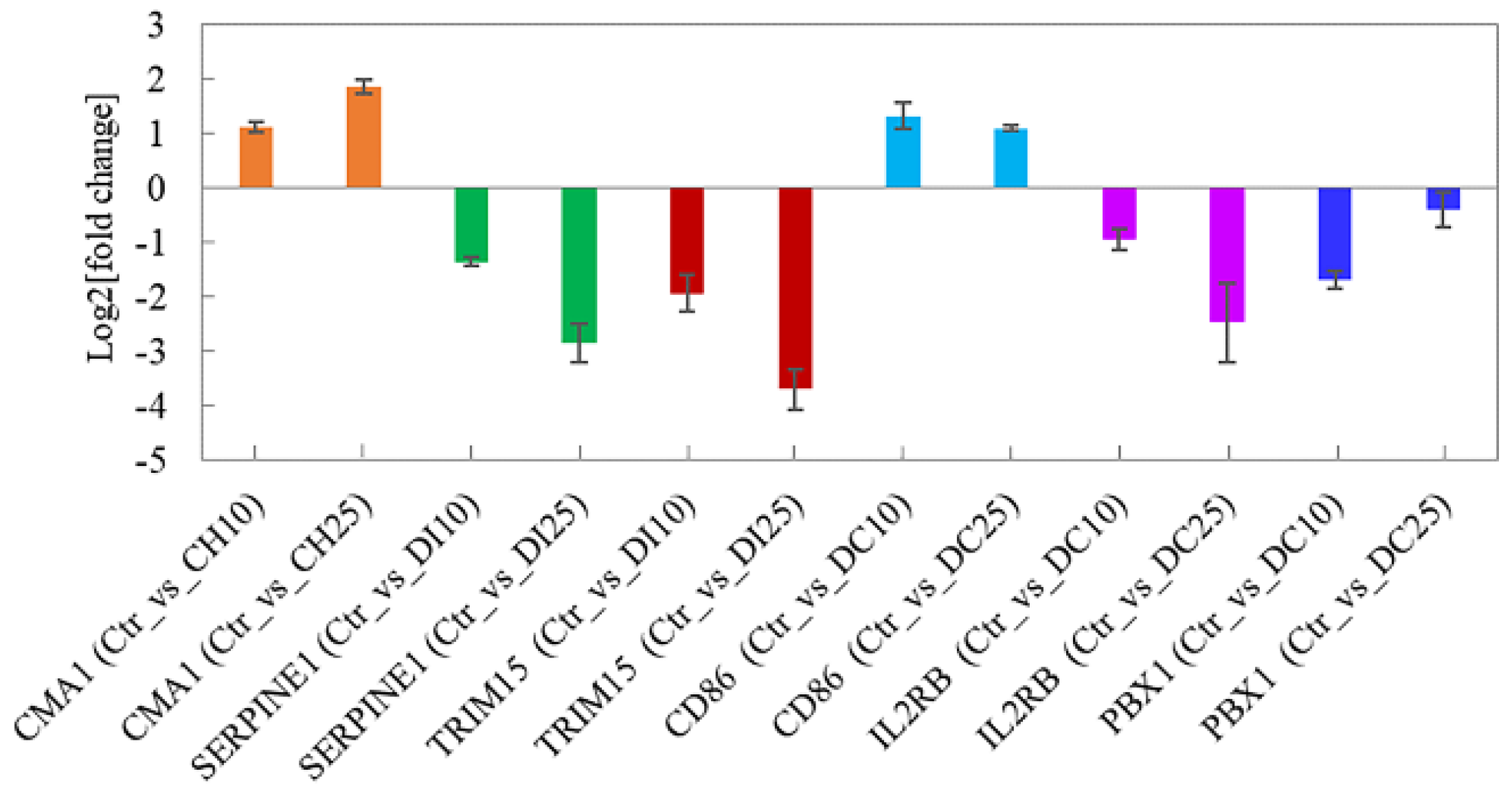
3.6. Quantitative Real-Time PCR Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nehra, M.; Dilbaghi, N.; Marrazza, G.; Kaushik, A.; Sonne, C.; Kim, K.H.; Kumar, S. Emerging nanobiotechnology in agriculture for the management of pesticide residues. J. Hazard. Mater. 2021, 401, 123369. [Google Scholar] [CrossRef] [PubMed]
- Samsidar, A.; Siddiquee, S.; Shaarani, S.M. A review of extraction, analytical and advanced methods for determination of pesticides in environment and foodstuffs. Trends Food Sci. Technol. 2018, 71, 188–201. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, R.; Luo, S.; Ying, L.; Jin, Y.; Li, Y.; Xiong, M.; Yuan, X.; Jia, L.; Chen, Y. Antagonistic effects and mechanisms of carbendazim and chlorpyrifos on the neurobehavior of larval zebrafish. Chemosphere 2022, 293, 133522. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Jiang, H.; Zhao, Y.; He, J.; Cheng, H.; Martyniuk, C.J. A comprehensive review of 1,2,4-triazole fungicide toxicity in zebrafish (Danio rerio): A mitochondrial and metabolic perspective. Sci. Total Environ. 2022, 809, 151177. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qiu, J.; Xu, Y.; Liao, G.; Jia, Q.; Pan, Y.; Wang, T.; Qian, Y. Integrated non-targeted lipidomics and metabolomics analyses for fluctuations of neonicotinoids imidacloprid and acetamiprid on Neuro-2a cells. Environ. Pollut. 2021, 284, 117327. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, M.; Chen, C.; Yang, X.; Qian, Y. Three widely used pesticides and their mixtures induced cytotoxicity and apoptosis through the ROS-related caspase pathway in HepG2 cells. Food Chem. Toxicol. 2021, 152, 112162. [Google Scholar] [CrossRef] [PubMed]
- Dhouib, I.; Jallouli, M.; Annabi, A.; Marzouki, S.; Gharbi, N.; Elfazaa, S.; Lasram, M.M. From immunotoxicity to carcinogenicity: The effects of carbamate pesticides on the immune system. Environ. Sci. Pollut. Res. Int. 2016, 23, 9448–9458. [Google Scholar] [CrossRef]
- Corsini, E.; Sokooti, M.; Galli, C.L.; Moretto, A.; Colosio, C. Pesticide induced immunotoxicity in humans: A comprehensive review of the existing evidence. Toxicology 2013, 307, 123–135. [Google Scholar] [CrossRef]
- Fukuyama, T.; Tajima, Y.; Ueda, H.; Hayashi, K.; Kosaka, T. Prior exposure to immunosuppressive organophosphorus or organochlorine compounds aggravates the TH1- and TH2-type allergy caused by topical sensitization to 2,4-dinitrochlorobenzene and trimellitic anhydride. J. Immunotoxicol. 2011, 8, 170–182. [Google Scholar] [CrossRef]
- Bini Dhouib, I.; Annabi, A.; Jallouli, M.; Marzouki, S.; Gharbi, N.; Elfazaa, S.; Lasram, M.M. Carbamates pesticides induced immunotoxicity and carcinogenicity in human: A review. J. Appl. Biomed. 2016, 14, 85–90. [Google Scholar] [CrossRef]
- Lee, G.H.; Choi, K.C. Adverse effects of pesticides on the functions of immune system. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2020, 235, 108789. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wang, D.; Li, H.; You, J. Joint toxicity of imidacloprid and azoxystrobin to Chironomus dilutus at organism, cell, and gene levels. Aquat. Toxicol. 2021, 233, 105783. [Google Scholar] [CrossRef]
- Cheng, L.; Lu, Y.; Zhao, Z.; Hoogenboom, R.; Zhang, Q.; Liu, X.; Song, W.; Guan, S.; Song, W.; Rao, Q. Assessing the combined toxicity effects of three neonicotinoid pesticide mixtures on human neuroblastoma SK-N-SH and lepidopteran Sf-9 cells. Food Chem. Toxicol. 2020, 145, 111632. [Google Scholar] [CrossRef] [PubMed]
- Benka-Coker, W.; Hoskovec, L.; Severson, R.; Balmes, J.; Wilson, A.; Magzamen, S. The joint effect of ambient air pollution and agricultural pesticide exposures on lung function among children with asthma. Environ. Res. 2020, 190, 109903. [Google Scholar] [CrossRef] [PubMed]
- Rizzati, V.; Briand, O.; Guillou, H.; Gamet-Payrastre, L. Effects of pesticide mixtures in human and animal models: An update of the recent literature. Chem. Biol. Interact. 2016, 254, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Boobis, A.; Budinsky, R.; Collie, S.; Crofton, K.; Embry, M.; Felter, S.; Hertzberg, R.; Kopp, D.; Mihlan, G.; Mumtaz, M.; et al. Critical analysis of literature on low-dose synergy for use in screening chemical mixtures for risk assessment. Crit. Rev. Toxicol. 2011, 41, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Huang, R.; Roberts, R.; Tong, W. Toxicogenomics: A 2020 Vision. Trends Pharmacol. Sci. 2019, 40, 92–103. [Google Scholar] [CrossRef]
- Schmitz-Spanke, S. Toxicogenomics—What added Value Do These Approaches Provide for Carcinogen Risk Assessment? Environ. Res. 2019, 173, 157–164. [Google Scholar] [CrossRef]
- Porreca, I.; D’Angelo, F.; De Franceschi, L.; Matte, A.; Ceccarelli, M.; Iolascon, A.; Zamo, A.; Russo, F.; Ravo, M.; Tarallo, R.; et al. Pesticide toxicogenomics across scales: In vitro transcriptome predicts mechanisms and outcomes of exposure in vivo. Sci. Rep. 2016, 6, 38131. [Google Scholar] [CrossRef]
- Lee, G.H.; Hwang, K.A.; Choi, K.C. Effects of fludioxonil on the cell growth and apoptosis in T and B lymphocytes. Biomolecules 2019, 9, 500. [Google Scholar] [CrossRef]
- Li, Y.C.; Liu, S.Y.; Meng, F.B.; Xu, S.H.; Qiu, J.; Qian, Y.Z.; Xu, Y.Y.; Li, Y. Comparative transcriptome analysis to investigate the immunotoxicity mechanism triggered by dimethomorph on human Jurkat T cell lines. Foods 2022, 11, 3848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.K.; Williams, T.D.; Abdallah, M.A.E.; Harrad, S.; Chipman, J.K.; Viant, M.R. Transcriptomic and metabolomic approaches to investigate the molecular responses of human cell lines exposed to the flame retardant hexabromocyclododecane (HBCD). Toxicol. Vitr. 2015, 29, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Verschoor, C.P.; Lelic, A.; Bramson, J.L.; Bowdish, D.M.E. An introduction to automated flow cytometry gating tools and their implementation. Front. Immunol. 2015, 6, 380. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Hu, Z.K.; Ye, N.; Yin, H.F. IsoSplitter: Identification and characterization of alternative splicing sites without a reference genome. RNA 2021, 27, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Zhang, H.; Qian, M.; Wang, J.; Yang, G.; Weng, Y.; Jin, C.; Li, Y.; Jin, Y. Insights into the effects of difenoconazole on the livers in male mice at the biochemical and transcriptomic levels. J. Hazard. Mater. 2022, 422, 126933. [Google Scholar] [CrossRef]
- Liu, F.; Li, X.; Bello, B.K.; Zhang, T.; Yang, H.; Wang, K.; Dong, J. Difenoconazole causes spleen tissue damage and immune dysfunction of carp through oxidative stress and apoptosis. Ecotoxicol. Environ. Saf. 2022, 237, 113563. [Google Scholar] [CrossRef]
- da Silveira Guerreiro, A.; Rola, R.C.; Rovani, M.T.; da Costa, S.R.; Sandrini, J.Z. Antifouling biocides: Impairment of bivalve immune system by chlorothalonil. Aquat. Toxicol. 2017, 189, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Naasri, S.; Helali, I.; Aouni, M.; Mastouri, M.; Harizi, H. N-acetylcysteine reduced the immunotoxicity effects induced in vitro by azoxystrobin and iprodione fungicides in mice. Environ. Toxicol. 2021, 36, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.-B.; Zhou, L.; Li, J.-J.; Li, Y.-C.; Wang, M.; Zou, L.-H.; Liu, D.-Y.; Chen, W.-J. The combined effect of protein hydrolysis and Lactobacillus plantarum fermentation on antioxidant activity and metabolomic profiles of quinoa beverage. Food Res. Int. 2022, 157, 111416. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Wang, J.; Bao, Z.; Jin, Y.; Xiao, Y. Acute chlorothalonil exposure had the potential to influence the intestinal barrier function and micro-environment in mice. Sci. Total Environ. 2023, 894, 165038. [Google Scholar] [CrossRef] [PubMed]
- Altevogt, P.; Sammar, M.; Hüser, L.; Kristiansen, G. Novel insights into the function of CD24: A driving force in cancer. Int. J. Cancer 2021, 148, 546–559. [Google Scholar] [CrossRef]
- Shi, S.; Ye, S.; Mao, J.; Ru, Y.; Lu, Y.; Wu, X.; Xu, M.; Zhu, T.; Wang, Y.; Chen, Y.; et al. CMA1 is potent prognostic marker and associates with immune infiltration in gastric cancer. Autoimmunity 2020, 53, 210–217. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, P.; Xing, D.; Liu, H.; Shi, F.; Hu, L.; Zou, X.; Nie, H.; Zuo, J.; Zhuang, Z.; et al. Developmental Neurotoxicity of Difenoconazole in Zebrafish Embryos. Toxics 2023, 11, 353. [Google Scholar] [CrossRef]
- Worthington, C.R.; Elliott, G.F. Muscle contraction: Energy rate equations in relation to efficiency and step-size distance. Int. J. Biol. Macromol. 2003, 32, 149–158. [Google Scholar] [CrossRef]
- Clegg, A.; Hassan-Smith, Z. Frailty and the endocrine system. Lancet Diabetes Endocrinol. 2018, 6, 743–752. [Google Scholar] [CrossRef]
- Wilkinson, H.N.; Hardman, M.J. The role of estrogen in cutaneous ageing and repair. Maturitas 2017, 103, 60–64. [Google Scholar] [CrossRef]
- Guan, Y.; Zhu, X.; Liang, J.; Wei, M.; Huang, S.; Pan, X. Upregulation of HSPA1A/HSPA1B/HSPA7 and downregulation of HSPA9 were related to poor survival in colon cancer. Front. Oncol. 2021, 11, 749673. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Shen, S.; Fu, S.; Fu, J. HSPA6 and its role in cancers and other diseases. Mol. Biol. Rep. 2022, 49, 10565–10577. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Hwa, J. Regulation of VWF expression, and secretion in health and disease. Curr. Opin. Hematol. 2016, 23, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Pang, L.; Liu, Z.; Meng, X. SERPINE1 associated with remodeling of the tumor microenvironment in colon cancer progression: A novel therapeutic target. BMC Cancer 2021, 21, 767. [Google Scholar] [CrossRef]
- Uchil, P.D.; Hinz, A.; Siegel, S.; Coenen-Stass, A.; Pertel, T.; Luban, J.; Mothes, W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 2013, 87, 257–272. [Google Scholar] [CrossRef]
- Alderdice, M.; Craig, S.G.; Humphries, M.P.; Gilmore, A.; Johnston, N.; Bingham, V.; Coyle, V.; Senevirathne, S.; Longley, D.B.; Loughrey, M.B.; et al. Evolutionary genetic algorithm identifies IL2RB as a potential predictive biomarker for immune-checkpoint therapy in colorectal cancer. NAR Genom. Bioinform. 2021, 3, lqab016. [Google Scholar] [CrossRef]
- Veiga, R.N.; de Oliveira, J.C.; Gradia, D.F. PBX1: A key character of the hallmarks of cancer. J. Mol. Med. 2021, 99, 1667–1680. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-C.; Liu, S.-Y.; Li, H.-R.; Meng, F.-B.; Qiu, J.; Qian, Y.-Z.; Xu, Y.-Y. Use of Transcriptomics to Reveal the Joint Immunotoxicity Mechanism Initiated by Difenoconazole and Chlorothalonil in the Human Jurkat T-Cell Line. Foods 2024, 13, 34. https://doi.org/10.3390/foods13010034
Li Y-C, Liu S-Y, Li H-R, Meng F-B, Qiu J, Qian Y-Z, Xu Y-Y. Use of Transcriptomics to Reveal the Joint Immunotoxicity Mechanism Initiated by Difenoconazole and Chlorothalonil in the Human Jurkat T-Cell Line. Foods. 2024; 13(1):34. https://doi.org/10.3390/foods13010034
Chicago/Turabian StyleLi, Yun-Cheng, Shu-Yan Liu, Hou-Ru Li, Fan-Bing Meng, Jing Qiu, Yong-Zhong Qian, and Yan-Yang Xu. 2024. "Use of Transcriptomics to Reveal the Joint Immunotoxicity Mechanism Initiated by Difenoconazole and Chlorothalonil in the Human Jurkat T-Cell Line" Foods 13, no. 1: 34. https://doi.org/10.3390/foods13010034
APA StyleLi, Y.-C., Liu, S.-Y., Li, H.-R., Meng, F.-B., Qiu, J., Qian, Y.-Z., & Xu, Y.-Y. (2024). Use of Transcriptomics to Reveal the Joint Immunotoxicity Mechanism Initiated by Difenoconazole and Chlorothalonil in the Human Jurkat T-Cell Line. Foods, 13(1), 34. https://doi.org/10.3390/foods13010034






