A Machine Learning Method to Identify Umami Peptide Sequences by Using Multiplicative LSTM Embedded Features
Abstract
1. Introduction
2. Materials and Methods
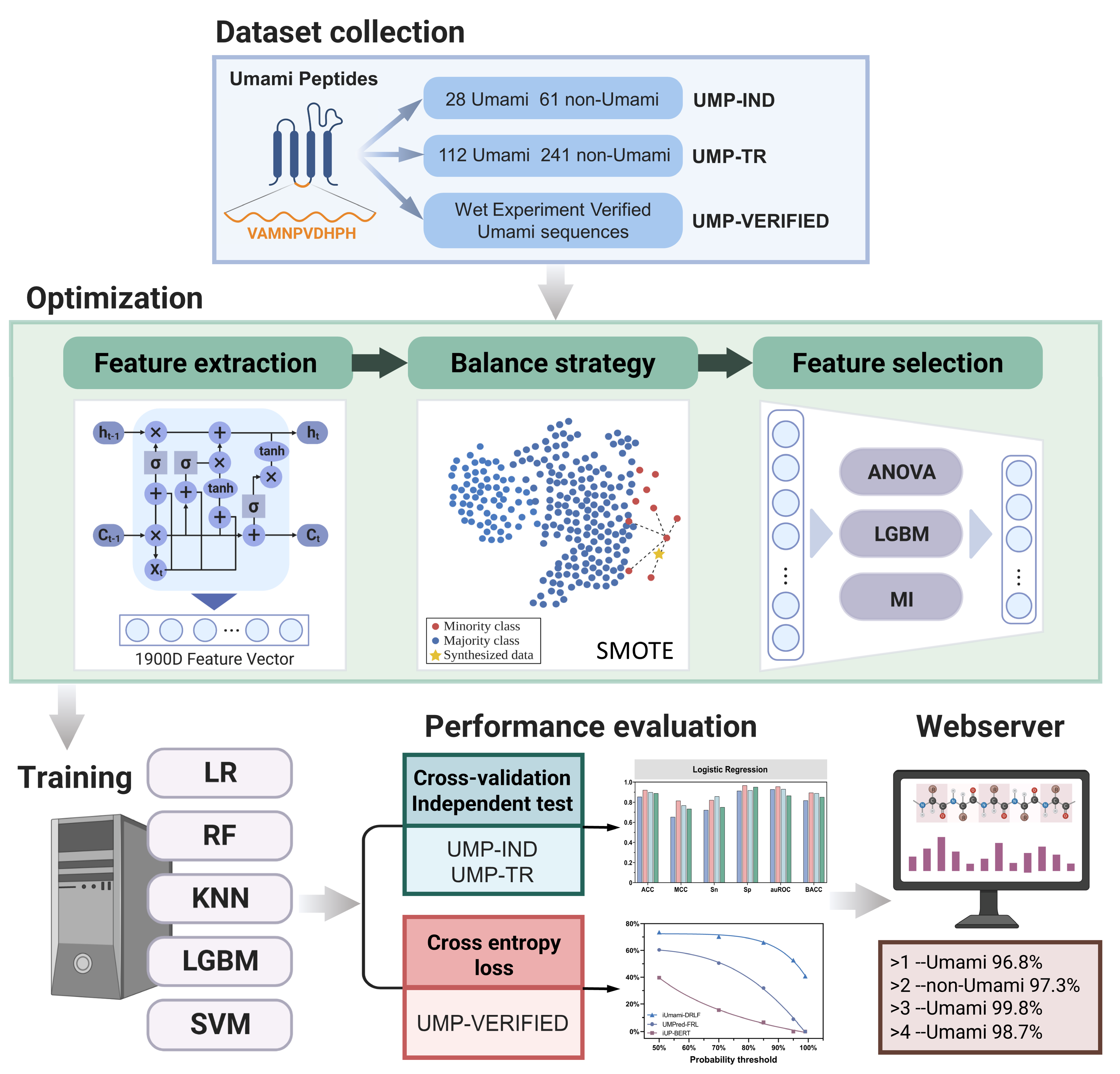
2.1. Benchmark Dataset
2.2. Feature Extraction
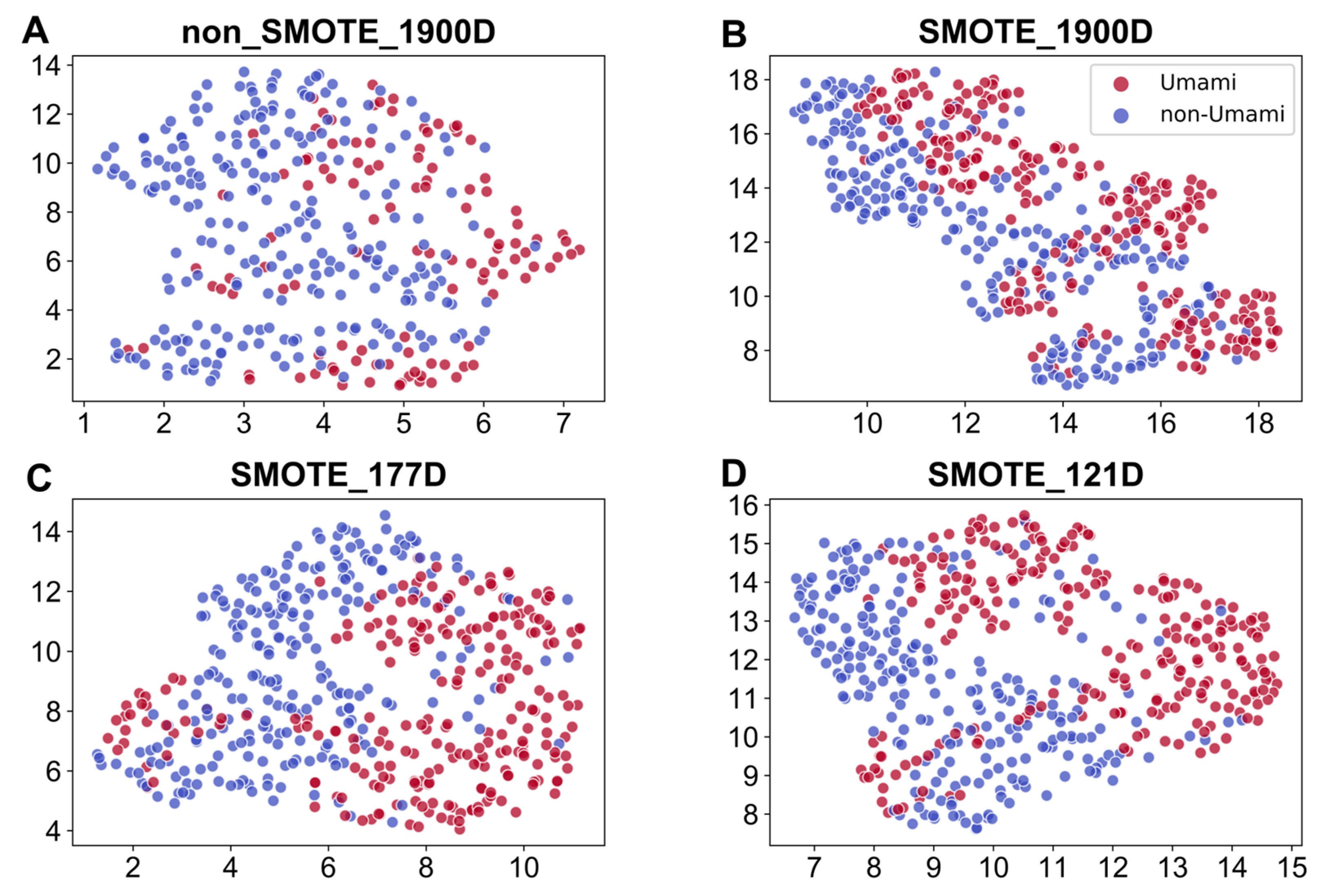
2.3. Balancing Strategy
2.4. Feature Selection Strategy
2.4.1. Analysis of Variance (ANOVA)
2.4.2. Lighting Gradient Boosting Machine (LGBM)
2.4.3. Mutual Information (MI)
2.5. Machine Learning Methods
2.6. Evaluation Metrics and Methods
2.7. Cross-Entropy Loss
3. Results and Discussion
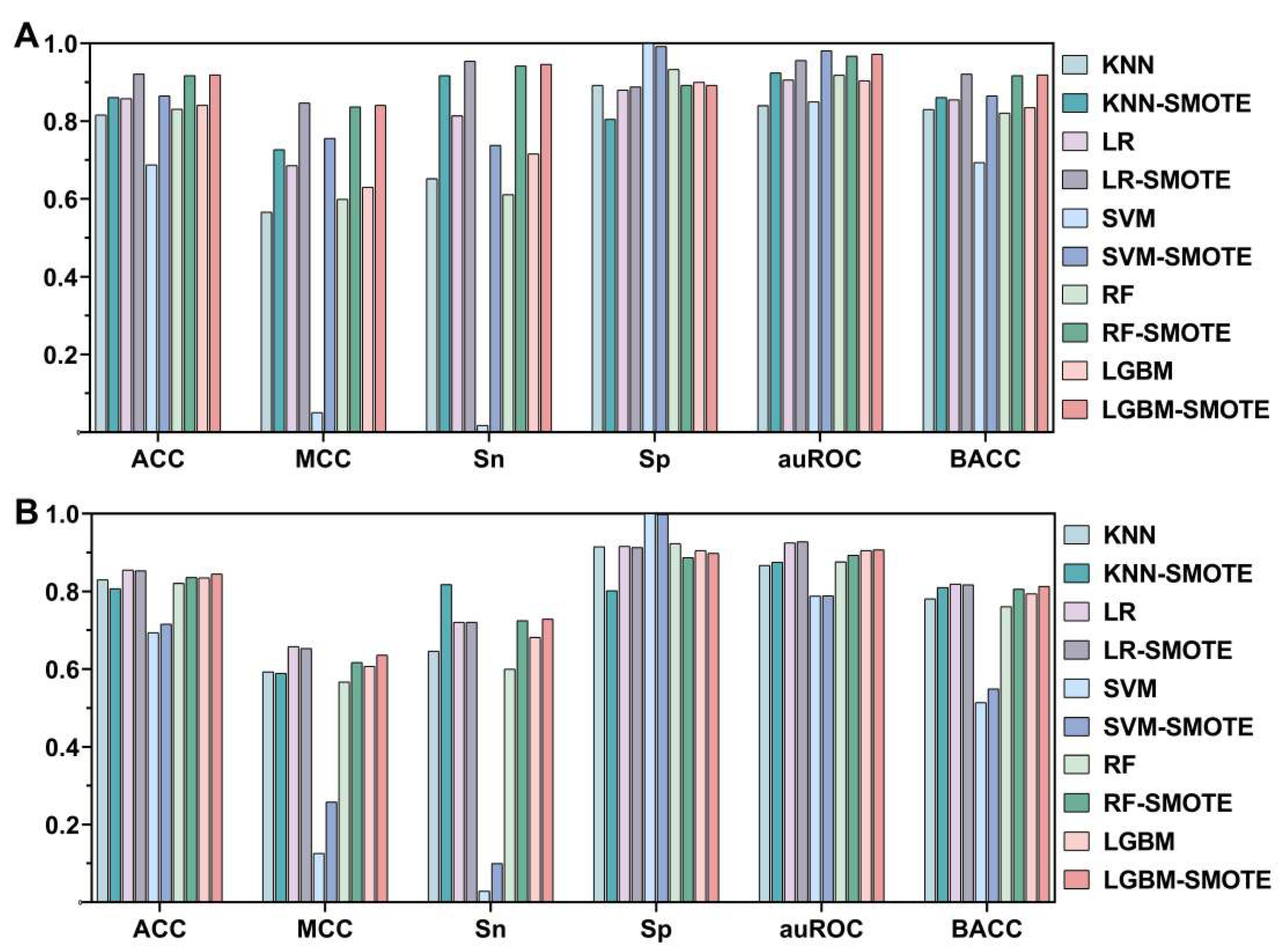
3.1. Effect of SMOTE
3.2. Effects of Different ML Models
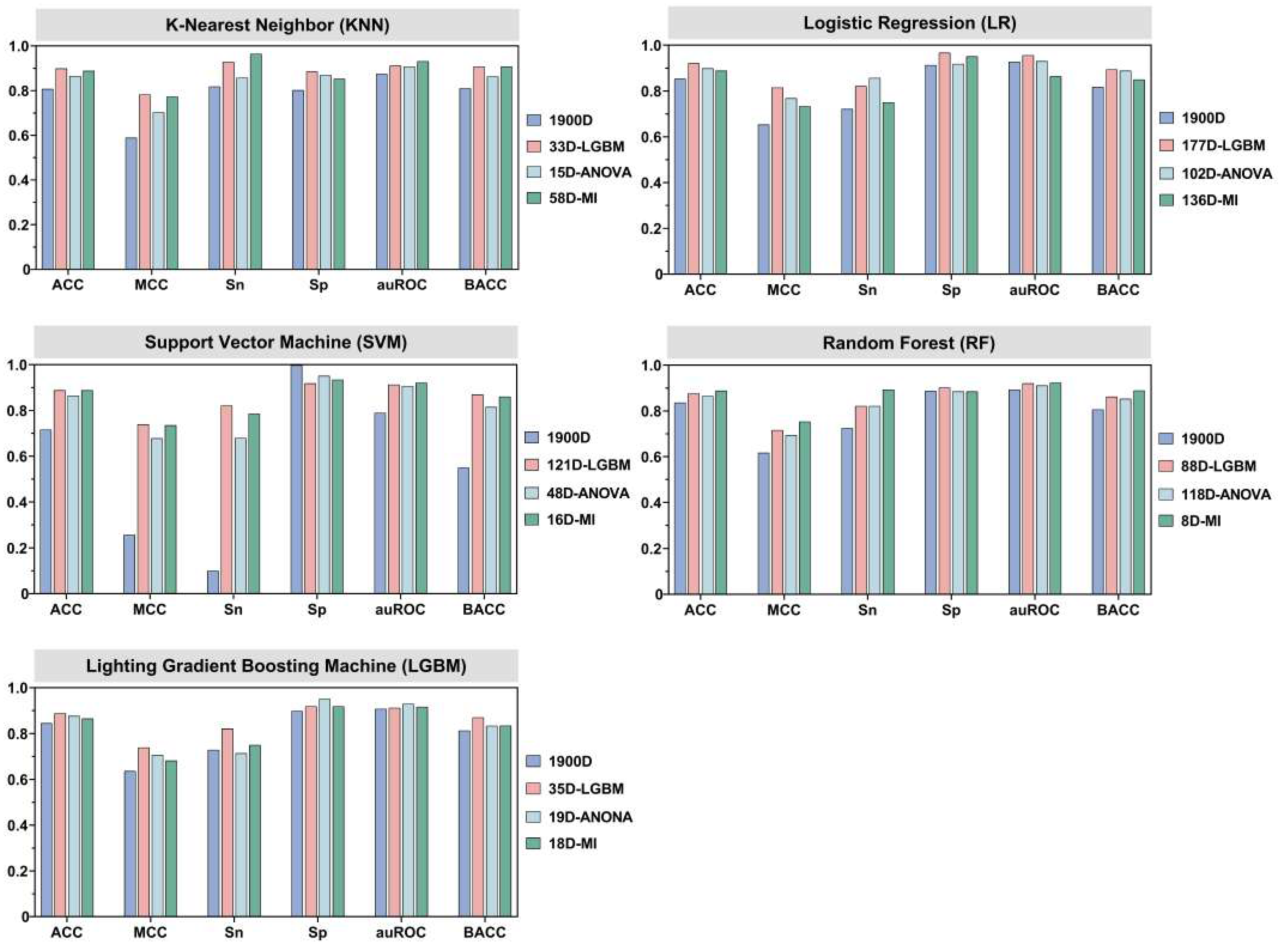
3.3. Effects of Different Feature Selection Methods
3.4. Comparison with Existing Methods
3.5. Feature Visualization
3.6. Web Server Development
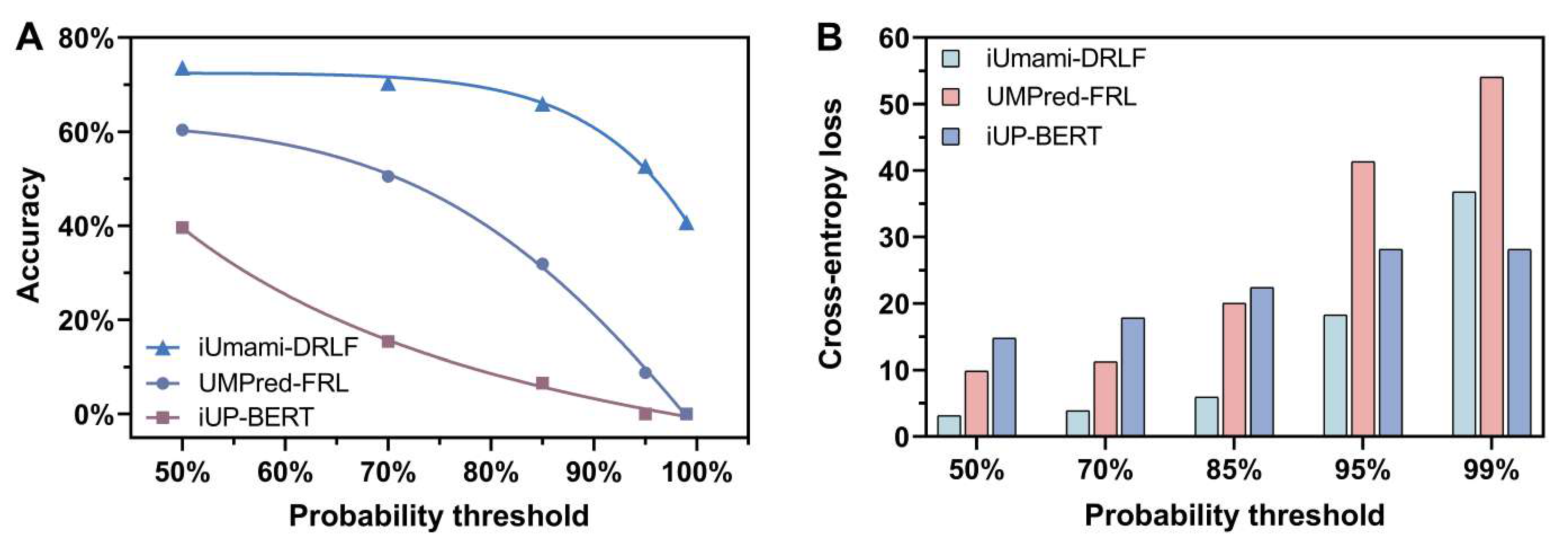
3.7. Methods’ Robustness
4. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Torii, K.; Uneyama, H.; Nakamura, E. Physiological roles of dietary glutamate signaling via gut–brain axis due to efficient digestion and absorption. J. Gastroenterol. 2013, 48, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Venkitasamy, C.; Pan, Z.; Liu, W.; Zhao, L. Novel Umami Ingredients: Umami Peptides and Their Taste. J. Food Sci. 2017, 82, 16–23. [Google Scholar] [CrossRef]
- Dang, Y.; Hao, L.; Cao, J.; Sun, Y.; Zeng, X.; Wu, Z.; Pan, D. Molecular docking and simulation of the synergistic effect between umami peptides, monosodium glutamate and taste receptor T1R1/T1R3. Food Chem. 2019, 271, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wang, J.; Kwok, D.; Cui, F.; Zhang, Z.; Zhao, D.; Li, M.J.; Zou, Q. webTWAS: A resource for disease candidate susceptibility genes identified by transcriptome-wide association study. Nucleic Acids Res. 2022, 50, D1123–D1130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Lin, X.; Jiang, Y.; Jiang, L.; Lv, Z. Identify Bitter Peptides by Using Deep Representation Learning Features. Int. J. Mol. Sci. 2022, 23, 7877. [Google Scholar] [CrossRef]
- Yan, N.; Lv, Z.; Hong, W.; Xu, X. Editorial: Feature Representation and Learning Methods With Applications in Protein Secondary Structure. Front. Bioeng. Biotechnol. 2021, 9, 748722. [Google Scholar] [CrossRef]
- Zhao, Q.; Ma, J.; Wang, Y.; Xie, F.; Lv, Z.; Xu, Y.; Shi, H.; Han, K. Mul-SNO: A Novel Prediction Tool for S-Nitrosylation Sites Based on Deep Learning Methods. IEEE J. Biomed. Health Informatics 2021, 26, 2379–2387. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Yana, J.; Nantasenamat, C.; Hasan, M.M.; Shoombuatong, W. iUmami-SCM: A Novel Sequence-Based Predictor for Prediction and Analysis of Umami Peptides Using a Scoring Card Method with Propensity Scores of Dipeptides. J. Chem. Inf. Model. 2020, 60, 6666–6678. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Yana, J.; Schaduangrat, N.; Nantasenamat, C.; Hasan, M.M.; Shoombuatong, W. iBitter-SCM: Identification and characterization of bitter peptides using a scoring card method with propensity scores of dipeptides. Genomics 2020, 112, 2813–2822. [Google Scholar]
- Charoenkwan, P.; Nantasenamat, C.; Hasan, M.; Moni, M.A.; Manavalan, B.; Shoombuatong, W. UMPred-FRL: A New Approach for Accurate Prediction of Umami Peptides Using Feature Representation Learning. Int. J. Mol. Sci. 2021, 22, 13124. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Jiang, J.; Wang, X.; Zhang, Y.; Zheng, B.; Liu, S.; Zhang, Y.; Liu, C.; Wan, Y.; Xiang, D.; et al. IUP-BERT: Identification of Umami Peptides Based on BERT Features. Foods 2022, 11, 3742. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; Courville, A.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [PubMed]
- Feifei, C. DeepMC-iNABP: Deep learning for multiclass identification and classification of nucleic acid-binding proteins. Comput. Struct. Biotechnol. J. 2022, 20, 2020–2028. [Google Scholar]
- Caro, M.C.; Huang, H.-Y.; Cerezo, M.; Sharma, K.; Sornborger, A.; Cincio, L.; Coles, P.J. Generalization in quantum machine learning from few training data. Nat. Commun. 2022, 13, 4919. [Google Scholar] [CrossRef]
- Cunningham, J.M.; Koytiger, G.; Sorger, P.K.; AlQuraishi, M. Biophysical prediction of protein–peptide interactions and signaling networks using machine learning. Nat. Methods 2020, 17, 175–183. [Google Scholar] [CrossRef]
- Lei, Y.; Li, S.; Liu, Z.; Wan, F.; Tian, T.; Li, S.; Zhao, D.; Zeng, J. A deep-learning framework for multi-level peptide–protein interaction prediction. Nat. Commun. 2021, 12, 5465. [Google Scholar] [CrossRef]
- Abbasi, A.; Miahi, E.; Mirroshandel, S.A. Effect of deep transfer and multi-task learning on sperm abnormality detection. Comput. Biol. Med. 2021, 128, 104121. [Google Scholar] [CrossRef]
- Arora, V.; Ng, E.Y.-K.; Leekha, R.S.; Darshan, M.; Singh, A. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput. Biol. Med. 2021, 135, 104575. [Google Scholar] [CrossRef]
- Cao, C.; He, J.; Mak, L.; Perera, D.; Kwok, D.; Wang, J.; Li, M.; Mourier, T.; Gavriliuc, S.; Greenberg, M.; et al. Reconstruction of Microbial Haplotypes by Integration of Statistical and Physical Linkage in Scaffolding. Mol. Biol. Evol. 2021, 38, 2660–2672. [Google Scholar] [CrossRef]
- Ao, C.; Jiao, S.; Wang, Y.; Yu, L.; Zou, Q. Biological Sequence Classification: A Review on Data and General Methods. Research 2022, 2022, 0011. [Google Scholar] [CrossRef]
- Harini, K.; Kihara, D.; Gromiha, M.M. PDA-Pred: Predicting the binding affinity of protein-DNA complexes using machine learning techniques and structural features. Methods 2023, 213, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Alnabati, E.; Aderinwale, T.W.; Maddhuri Venkata Subramaniya, S.R.; Terashi, G.; Kihara, D. Detecting protein and DNA/RNA structures in cryo-EM maps of intermediate resolution using deep learning. Nat. Commun. 2021, 12, 2302. [Google Scholar]
- Meier, F.; Köhler, N.D.; Brunner, A.-D.; Wanka, J.-M.H.; Voytik, E.; Strauss, M.T.; Theis, F.J.; Mann, M. Deep learning the collisional cross sections of the peptide universe from a million experimental values. Nat. Commun. 2021, 12, 1185. [Google Scholar] [CrossRef]
- Wilhelm, M.; Zolg, D.P.; Graber, M.; Gessulat, S.; Schmidt, T.; Schnatbaum, K.; Schwencke-Westphal, C.; Seifert, P.; de Andrade Krätzig, N.; Zerweck, J.; et al. Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat. Commun. 2021, 12, 3346. [Google Scholar]
- He, J.; Lin, P.; Chen, J.; Cao, H.; Huang, S.-Y. Model building of protein complexes from intermediate-resolution cryo-EM maps with deep learning-guided automatic assembly. Nat. Commun. 2022, 13, 4066. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Cheveralls, K.C.; Leonetti, M.D.; Royer, L.A. Self-supervised deep learning encodes high-resolution features of protein subcellular localization. Nat. Methods 2022, 19, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, K.; Bozdag, P.G.; Talo, M.; Yildirim, O.; Karabatak, M.; Acharya, U. Deep learning model for automated kidney stone detection using coronal CT images. Comput. Biol. Med. 2021, 135, 104569. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wu, J.; Lin, Z. Cancer diagnosis using generative adversarial networks based on deep learning from imbalanced data. Comput. Biol. Med. 2021, 135, 104540. [Google Scholar] [CrossRef]
- Jain, A.; Terashi, G.; Kagaya, Y.; Subramaniya, S.R.M.V.; Christoffer, C.; Kihara, D. Analyzing effect of quadruple multiple sequence alignments on deep learning based protein inter-residue distance prediction. Sci. Rep. 2021, 11, 7574. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Fu, H.; Ruan, X.; Tang, X. DeepFusion-RBP: Using Deep Learning to Fuse Multiple Features to Identify RNA-binding Protein Sequences. Curr. Bioinform. 2021, 16, 1089–1100. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Nantasenamat, C.; Hasan, M.; Manavalan, B.; Shoombuatong, W. BERT4Bitter: A bidirectional encoder representations from transformers (BERT)-based model for improving the prediction of bitter peptides. Bioinformatics 2021, 37, 2556–2562. [Google Scholar] [CrossRef]
- Bao, W.; Cui, Q.; Chen, B.; Yang, B. Phage_UniR_LGBM: Phage Virion Proteins Classification with UniRep Features and LightGBM Model. Comput. Math. Methods Med. 2022, 2022, 9470683. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Zou, Q.; Lin, C. prPred-DRLF: Plant R protein predictor using deep representation learning features. Proteomics 2022, 22, e2100161. [Google Scholar] [CrossRef]
- Villegas-Morcillo, A.; Gomez, A.M.; Sanchez, V. An analysis of protein language model embeddings for fold prediction. Briefings Bioinform. 2022, 23, bbac142. [Google Scholar] [CrossRef]
- Wei, Y.; Zou, Q.; Tang, F.; Yu, L. WMSA: A novel method for multiple sequence alignment of DNA sequences. Bioinformatics 2022, 38, 5019–5025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, F.; Lin, C.; Zhao, L.; Wang, C.; Zou, Q. Critical downstream analysis steps for single-cell RNA sequencing data. Briefings Bioinform. 2021, 22, bbab105. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, F.; Su, W.; Dou, L.; Xu, A.; Cao, C.; Zou, Q. webSCST: An interactive web application for single-cell RNA-sequencing data and spatial transcriptomic data integration. Bioinformatics 2022, 38, 3488–3489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, F.; Cao, C.; Wang, Q.; Zou, Q. Single-cell RNA analysis reveals the potential risk of organ-specific cell types vulnerable to SARS-CoV-2 infections. Comput. Biol. Med. 2022, 140, 105092. [Google Scholar] [CrossRef] [PubMed]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.; Schmidhuber, J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005, 18, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for Learning from Imbalanced Data: Progress and Challenges, Marking the 15-year Anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Kumar, M.; Rath, N.K.; Swain, A.; Rath, S.K. Feature Selection and Classification of Microarray Data using MapReduce based ANOVA and K-Nearest Neighbor. Procedia Comput. Sci. 2015, 54, 301–310. [Google Scholar] [CrossRef]
- Shaw, R.G.; Mitchell-Olds, T. Anova for Unbalanced Data: An Overview. Ecology 1993, 74, 1638–1645. [Google Scholar] [CrossRef]
- Lv, Z.; Cui, F.; Zou, Q.; Zhang, L.; Xu, L. Anticancer peptides prediction with deep representation learning features. Briefings Bioinform. 2021, 22, bbab008. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Jiang, Y.; Jin, J.; Li, Z.; Zhao, J.; Manavalan, B.; Su, R.; Gao, X.; Wei, L. Accelerating bioactive peptide discovery via mutual information-based meta-learning. Briefings Bioinform. 2022, 23, bbab499. [Google Scholar] [CrossRef]
- Zhao, D.; Teng, Z.; Li, Y.; Chen, D. iAIPs: Identifying Anti-Inflammatory Peptides Using Random Forest. Front. Genet. 2021, 12, 773202. [Google Scholar] [CrossRef]
- Battiti, R. Using mutual information for selecting features in supervised neural net learning. IEEE Trans. Neural Networks 1994, 5, 537–550. [Google Scholar] [CrossRef]
- Tripathi, V.; Tripathi, P. Detecting antimicrobial peptides by exploring the mutual information of their sequences. J. Biomol. Struct. Dyn. 2020, 38, 5037–5043. [Google Scholar] [CrossRef]
- Ao, C.; Zou, Q.; Yu, L. NmRF: Identification of multispecies RNA 2’-O-methylation modification sites from RNA sequences. Briefings Bioinform. 2022, 23, bbab480. [Google Scholar] [CrossRef]
- Chen, L.; Yu, L.; Gao, L. Potent antibiotic design via guided search from antibacterial activity evaluations. Bioinformatics 2023, 39, btad059. [Google Scholar] [CrossRef] [PubMed]
- Spindelböck, T.; Ranftl, S.; von der Linden, W. Cross-Entropy Learning for Aortic Pathology Classification of Artificial Multi-Sensor Impedance Cardiography Signals. Entropy 2021, 23, 1661. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Yao, L.; Zhao, X. Adaptive Margin Aware Complement-Cross Entropy Loss for Improving Class Imbalance in Multi-View Sleep Staging Based on EEG Signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 2927–2938. [Google Scholar] [CrossRef] [PubMed]
- Egusquiza, I.; Picon, A.; Irusta, U.; Bereciartua-Perez, A.; Eggers, T.; Klukas, C.; Aramendi, E.; Navarra-Mestre, R. Analysis of Few-Shot Techniques for Fungal Plant Disease Classification and Evaluation of Clustering Capabilities Over Real Datasets. Front. Plant Sci. 2022, 13, 813237. [Google Scholar] [CrossRef]
- Yu, L.; Xia, M.; An, Q. A network embedding framework based on integrating multiplex network for drug combination prediction. Briefings Bioinform. 2022, 23, bbab364. [Google Scholar] [CrossRef]
- Zhang, T.; Hua, Y.; Zhou, C.; Xiong, Y.; Pan, D.; Liu, Z.; Dang, Y. Umami peptides screened based on peptidomics and virtual screening from Ruditapes philippinarum and Mactra veneriformis clams. Food Chem. 2022, 394, 133504. [Google Scholar] [CrossRef]
- Liang, L.; Zhou, C.; Zhang, J.; Huang, Y.; Zhao, J.; Sun, B.; Zhang, Y. Characteristics of umami peptides identified from porcine bone soup and molecular docking to the taste receptor T1R1/T1R3. Food Chem. 2022, 387, 132870. [Google Scholar] [CrossRef]
- Bu, Y.; Liu, Y.; Luan, H.; Zhu, W.; Li, X.; Li, J. Characterization and structure–activity relationship of novel umami peptides isolated from Thai fish sauce. Food Funct. 2021, 12, 5027–5037. [Google Scholar] [CrossRef]
- Chen, W.; Li, W.; Wu, D.; Zhang, Z.; Chen, H.; Zhang, J.; Wang, C.; Wu, T.; Yang, Y. Characterization of novel umami-active peptides from Stropharia rugoso-annulata mushroom and in silico study on action mechanism. J. Food Compos. Anal. 2022, 110, 104530. [Google Scholar] [CrossRef]
- Zhu, X.; Sun-Waterhouse, D.; Chen, J.; Cui, C.; Wang, W. Comparative study on the novel umami-active peptides of the whole soybeans and the defatted soybeans fermented soy sauce. J. Sci. Food Agric. 2021, 101, 158–166. [Google Scholar] [CrossRef]
- Wang, W.; Huang, Y.; Zhao, W.; Dong, H.; Yang, J.; Bai, W. Identification and comparison of umami-peptides in commercially available dry-cured Spanish mackerels (Scomberomorus niphonius). Food Chem. 2022, 380, 132175. [Google Scholar] [CrossRef]
- Song, S.; Zhuang, J.; Ma, C.; Feng, T.; Yao, L.; Ho, C.-T.; Sun, M. Identification of novel umami peptides from Boletus edulis and its mechanism via sensory analysis and molecular simulation approaches. Food Chem. 2023, 398, 133835. [Google Scholar] [CrossRef]
- Yu, Z.; Kang, L.; Zhao, W.; Wu, S.; Ding, L.; Zheng, F.; Liu, J.; Li, J. Identification of novel umami peptides from myosin via homology modeling and molecular docking. Food Chem. 2021, 344, 128728. [Google Scholar] [CrossRef]
- Wang, Y.; Luan, J.; Tang, X.; Zhu, W.; Xu, Y.; Bu, Y.; Li, J.; Cui, F.; Li, X. Identification of umami peptides based on virtual screening and molecular docking from Atlantic cod (Gadus morhua). Food Funct. 2023, 14, 1510–1519. [Google Scholar] [CrossRef]
- Zhu, W.; Luan, H.; Bu, Y.; Li, X.; Li, J.; Zhang, Y. Identification, taste characterization and molecular docking study of novel umami peptides from the Chinese anchovy sauce. J. Sci. Food Agric. 2021, 101, 3140–3155. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Hu, X.; Xue, H.; Li, R.; Liu, H.; Han, T.; Ruan, D.; Tu, Y.; Zhao, Y. Isolation and screening of umami peptides from preserved egg yolk by nano-HPLC-MS/MS and molecular docking. Food Chem. 2022, 377, 131996. [Google Scholar] [CrossRef] [PubMed]
- Shiyan, R.; Liping, S.; Xiaodong, S.; Jinlun, H.; Yongliang, Z. Novel umami peptides from tilapia lower jaw and molecular docking to the taste receptor T1R1/T1R3. Food Chem. 2021, 362, 130249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; He, W.; Wang, F.; Bu, Y.; Li, X.; Li, J. Prediction, molecular docking and identification of novel umami hexapeptides derived from Atlantic cod (Gadus morhua ). Int. J. Food Sci. Technol. 2021, 56, 402–412. [Google Scholar] [CrossRef]
- Liu, Q.; Gao, X.; Pan, D.; Liu, Z.; Xiao, C.; Du, L.; Cai, Z.; Lu, W.; Dang, Y.; Zou, Y. Rapid screening based on machine learning and molecular docking of umami peptides from porcine bone. J. Sci. Food Agric. 2022. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Wang, W.; Zhou, X.; Chen, G.; Liu, Y. Seven novel umami peptides from Takifugu rubripes and their taste characteristics. Food Chem. 2020, 330, 127204. [Google Scholar] [CrossRef]
- Rajaraman, S.; Zamzmi, G.; Antani, S.K. Novel loss functions for ensemble-based medical image classification. PLoS ONE 2021, 16, e0261307. [Google Scholar] [CrossRef] [PubMed]
| Model | 10-Fold Cross-Validation | Independent Test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | MCC | Sn | Sp | auROC | BACC | ACC | MCC | Sn | Sp | auROC | BACC | |
| LR c | 0.921 a | 0.847 | 0.954 | 0.888 | 0.956 | 0.921 | 0.853 | 0.653 | 0.721 | 0.913 | 0.928 | 0.817 |
| KNN c | 0.861 b | 0.727 | 0.917 | 0.805 | 0.924 | 0.861 | 0.807 | 0.589 | 0.818 | 0.802 | 0.875 | 0.810 |
| SVM c | 0.865 | 0.756 | 0.738 | 0.992 | 0.981 | 0.865 | 0.716 | 0.258 | 0.100 | 0.998 | 0.789 | 0.549 |
| RFc | 0.917 | 0.837 | 0.942 | 0.892 | 0.967 | 0.917 | 0.836 | 0.617 | 0.725 | 0.887 | 0.893 | 0.806 |
| LGBM c | 0.919 | 0.841 | 0.946 | 0.892 | 0.972 | 0.919 | 0.845 | 0.636 | 0.729 | 0.898 | 0.907 | 0.813 |
| Model | Feature Selection Method | Dim | 10-Fold Cross-Validation | Independent Test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | MCC | Sn | Sp | auROC | BACC | ACC | MCC | Sn | Sp | auROC | BACC | |||
| LR c | LGBM d | 177 | 0.925 b | 0.853 | 0.959 | 0.892 | 0.957 | 0.925 | 0.921 a | 0.815 | 0.821 | 0.967 | 0.956 | 0.894 |
| ANOVA d | 102 | 0.882 | 0.764 | 0.896 | 0.867 | 0.938 | 0.882 | 0.899 | 0.768 | 0.857 | 0.918 | 0.930 | 0.888 | |
| MI d | 136 | 0.888 | 0.777 | 0.913 | 0.863 | 0.942 | 0.888 | 0.888 | 0.733 | 0.750 | 0.951 | 0.864 | 0.850 | |
| KNN c | LGBM d | 33 | 0.892 | 0.788 | 0.938 | 0.846 | 0.955 | 0.892 | 0.899 | 0.782 | 0.929 | 0.885 | 0.911 | 0.907 |
| ANOVA d | 15 | 0.873 | 0.748 | 0.896 | 0.851 | 0.934 | 0.873 | 0.865 | 0.703 | 0.857 | 0.869 | 0.907 | 0.863 | |
| MI d | 58 | 0.888 | 0.783 | 0.954 | 0.822 | 0.927 | 0.888 | 0.888 | 0.773 | 0.964 | 0.852 | 0.931 | 0.908 | |
| SVM c | LGBM d | 121 | 0.944 | 0.889 | 0.971 | 0.917 | 0.980 | 0.944 | 0.888 | 0.739 | 0.821 | 0.918 | 0.913 | 0.870 |
| ANOVA d | 48 | 0.925 | 0.854 | 0.967 | 0.884 | 0.977 | 0.925 | 0.865 | 0.678 | 0.679 | 0.951 | 0.906 | 0.815 | |
| MId | 16 | 0.919 | 0.841 | 0.959 | 0.880 | 0.968 | 0.919 | 0.888 | 0.735 | 0.786 | 0.934 | 0.921 | 0.860 | |
| RF c | LGBM d | 88 | 0.915 | 0.830 | 0.934 | 0.896 | 0.975 | 0.915 | 0.876 | 0.716 | 0.821 | 0.902 | 0.920 | 0.862 |
| ANOVA d | 118 | 0.898 | 0.797 | 0.913 | 0.884 | 0.961 | 0.898 | 0.865 | 0.694 | 0.821 | 0.885 | 0.911 | 0.853 | |
| MI d | 8 | 0.902 | 0.806 | 0.921 | 0.884 | 0.952 | 0.902 | 0.888 | 0.753 | 0.893 | 0.885 | 0.923 | 0.889 | |
| LGBM c | LGBM d | 35 | 0.938 | 0.877 | 0.971 | 0.905 | 0.988 | 0.938 | 0.888 | 0.739 | 0.821 | 0.918 | 0.912 | 0.870 |
| ANOVA d | 19 | 0.902 | 0.807 | 0.942 | 0.863 | 0.945 | 0.902 | 0.876 | 0.706 | 0.714 | 0.951 | 0.929 | 0.833 | |
| MI d | 18 | 0.888 | 0.777 | 0.917 | 0.859 | 0.953 | 0.888 | 0.865 | 0.682 | 0.750 | 0.918 | 0.916 | 0.834 | |
| Classifier | 10-Fold Cross-Validation | Independent Test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | MCC | Sn | Sp | auROC | BACC | ACC | MCC | Sn | Sp | auROC | BACC | |
| iUmami-DRLF(LR) | 0.925 b | 0.853 | 0.959 | 0.892 | 0.957 | 0.925 | 0.921 a | 0.815 | 0.821 | 0.967 | 0.956 | 0.894 |
| iUmami-DRLF(SVM) | 0.944 | 0.889 | 0.971 | 0.917 | 0.980 | 0.944 | 0.888 | 0.739 | 0.821 | 0.918 | 0.913 | 0.870 |
| iUP-BERT | 0.940 | 0.881 | 0.963 | 0.917 | 0.971 | 0.940 | 0.899 | 0.774 | 0.893 | 0.902 | 0.933 | 0.897 |
| UMPred-FRL | 0.921 | 0.814 | 0.847 | 0.955 | 0.938 | 0.901 | 0.888 | 0.735 | 0.786 | 0.934 | 0.919 | 0.860 |
| iUmami-SCM | 0.935 | 0.864 | 0.947 | 0.930 | 0.945 | 0.939 | 0.865 | 0.679 | 0.714 | 0.934 | 0.898 | 0.824 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Li, J.; Li, J.; Pei, H.; Li, M.; Zou, Q.; Lv, Z. A Machine Learning Method to Identify Umami Peptide Sequences by Using Multiplicative LSTM Embedded Features. Foods 2023, 12, 1498. https://doi.org/10.3390/foods12071498
Jiang J, Li J, Li J, Pei H, Li M, Zou Q, Lv Z. A Machine Learning Method to Identify Umami Peptide Sequences by Using Multiplicative LSTM Embedded Features. Foods. 2023; 12(7):1498. https://doi.org/10.3390/foods12071498
Chicago/Turabian StyleJiang, Jici, Jiayu Li, Junxian Li, Hongdi Pei, Mingxin Li, Quan Zou, and Zhibin Lv. 2023. "A Machine Learning Method to Identify Umami Peptide Sequences by Using Multiplicative LSTM Embedded Features" Foods 12, no. 7: 1498. https://doi.org/10.3390/foods12071498
APA StyleJiang, J., Li, J., Li, J., Pei, H., Li, M., Zou, Q., & Lv, Z. (2023). A Machine Learning Method to Identify Umami Peptide Sequences by Using Multiplicative LSTM Embedded Features. Foods, 12(7), 1498. https://doi.org/10.3390/foods12071498






