Evaluation of Alternative Colony Hybridization Methods for Pathogenic Vibrios
Abstract
1. Introduction
2. Materials and Methods
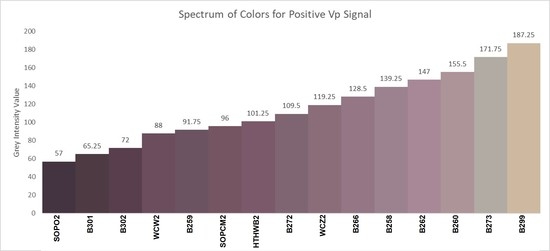
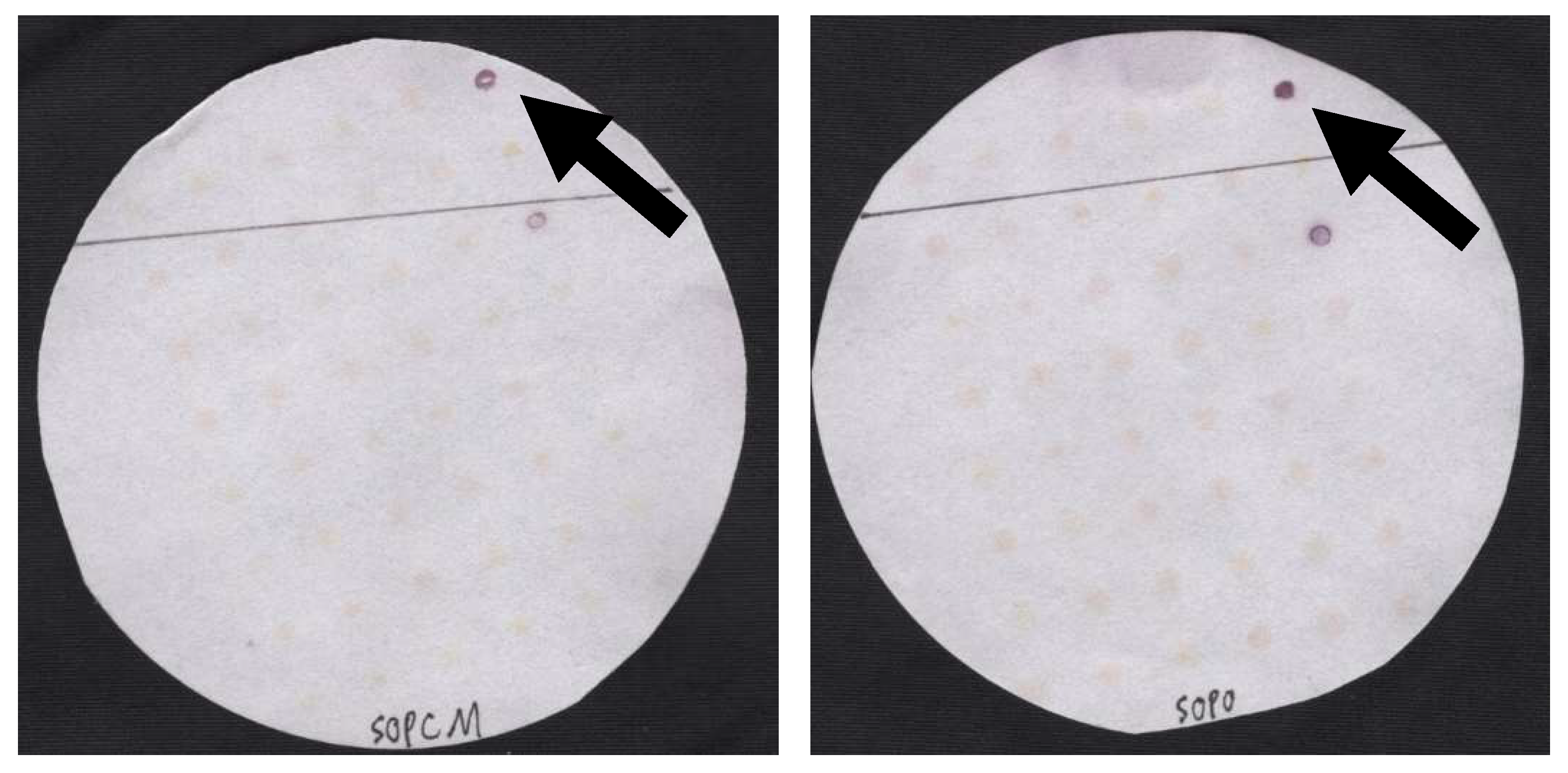
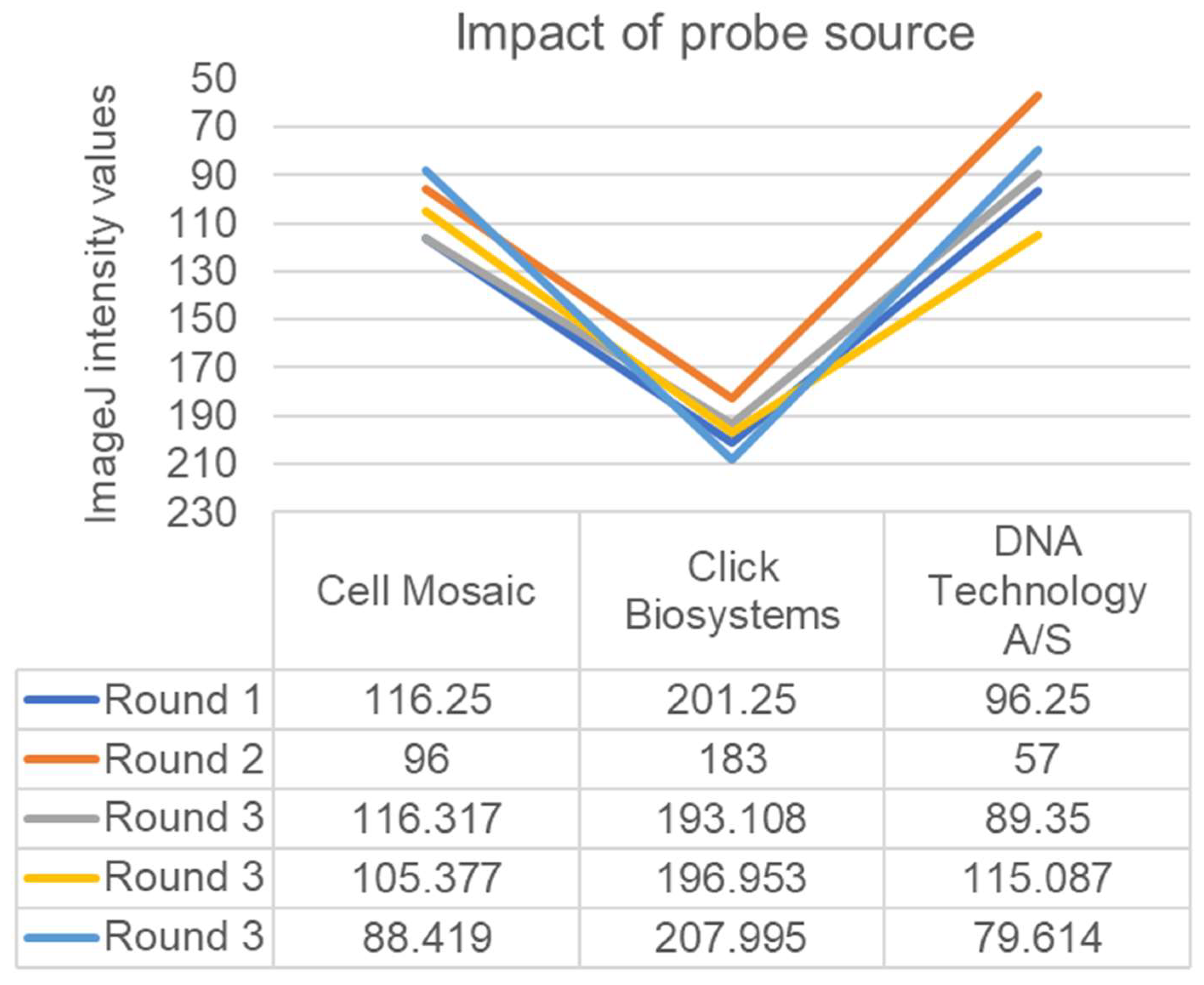
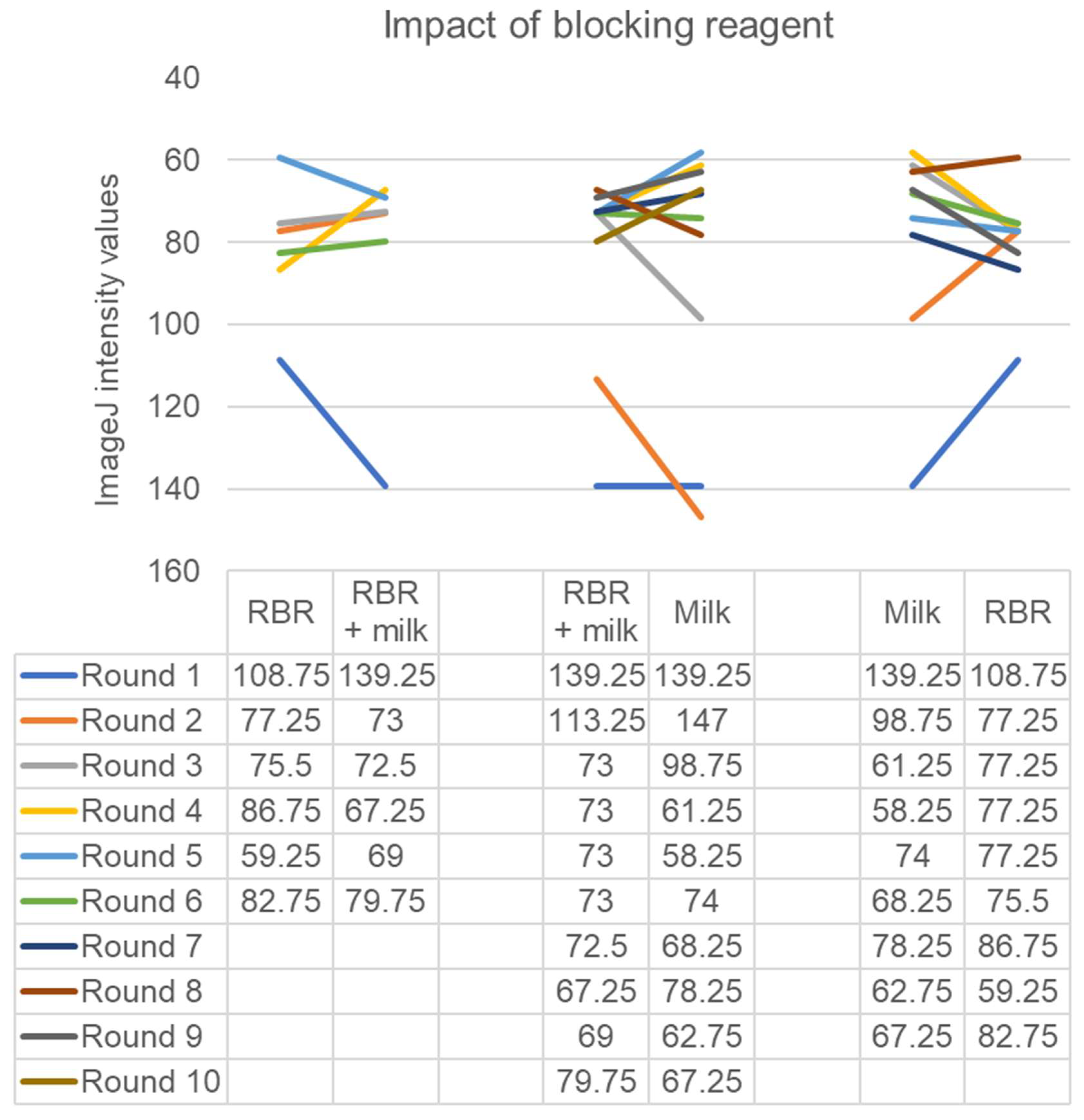
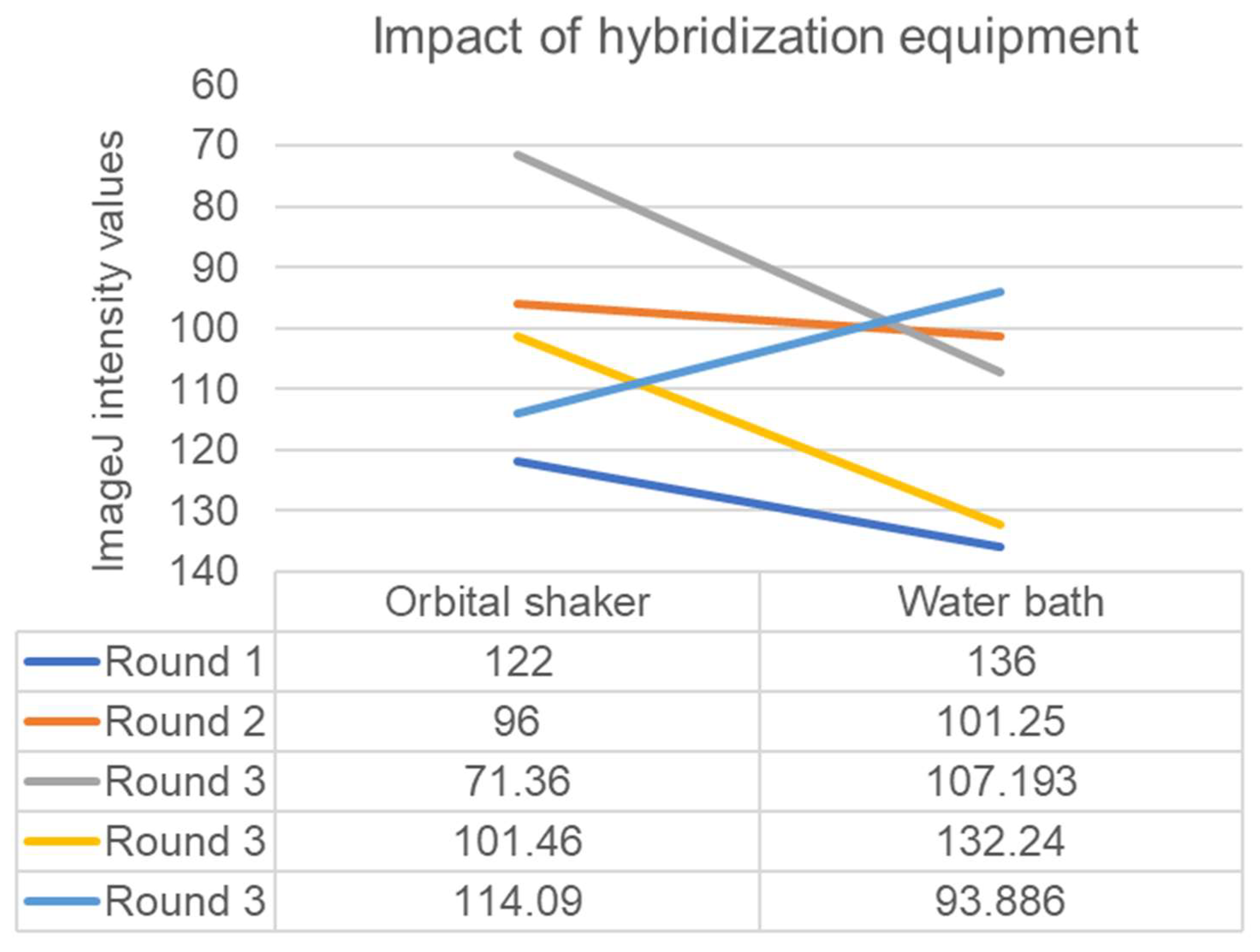
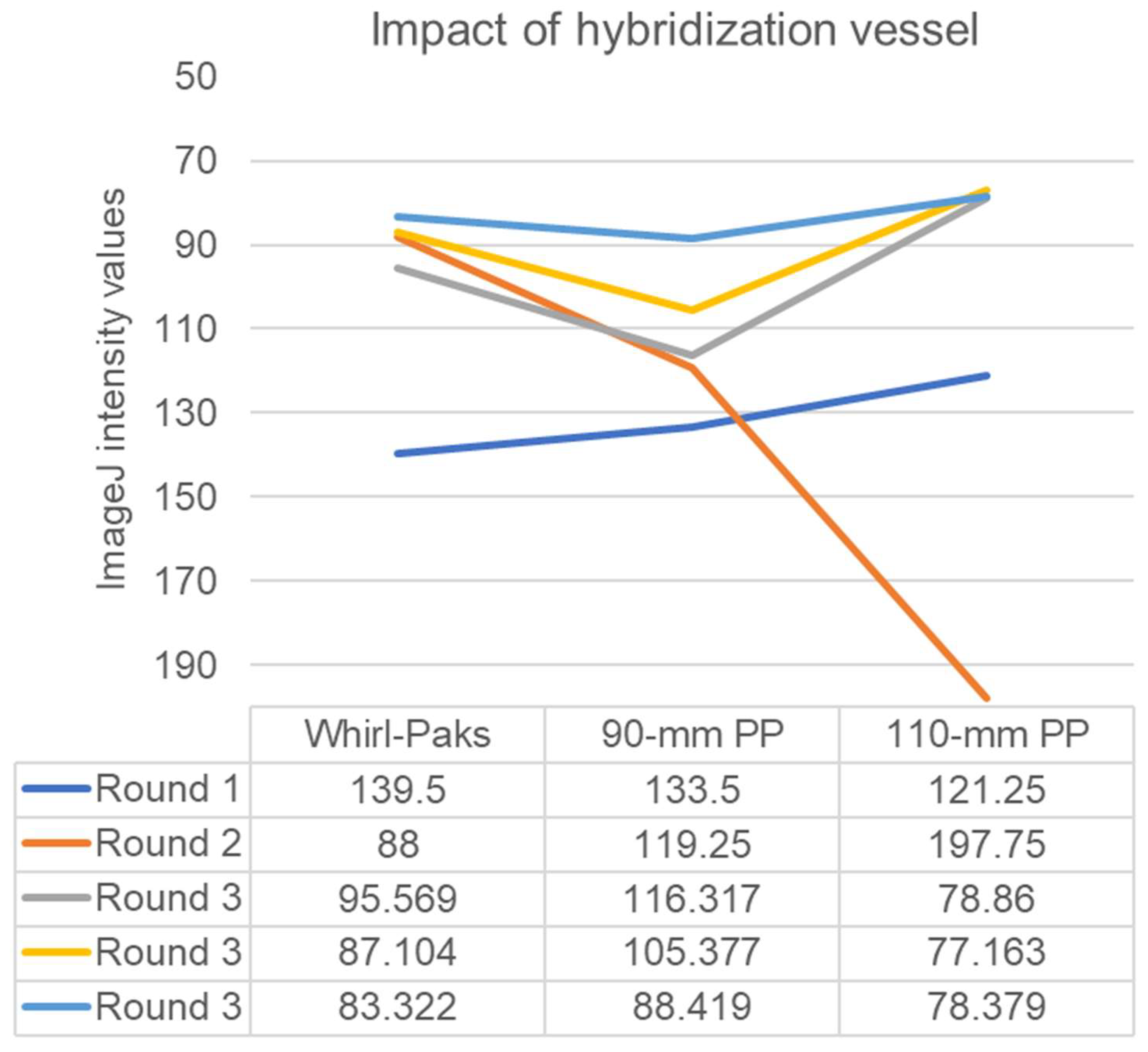
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCarthy, S.A.; DePaola, A.; Cook, D.W.; Kaysner, C.A.; Hill, W.E. Evaluation of alkaline phosphatase- and digoxigenin-labelled probes for detection of the thermolabile hemolysin (tlh) gene of Vibrio parahaemolyticus. Lett. Appl. Microbiol. 1999, 28, 66–70. [Google Scholar] [CrossRef]
- Grunstein, M.; Hogness, D.S. Colony hybridization: A method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 1975, 72, 3961–3965. [Google Scholar] [CrossRef]
- Blackstone, G.M.; Nordstrom, J.L.; Vickery, M.C.; Bowen, M.D.; Meyer, R.F.; DePaola, A. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 2003, 53, 149–155. [Google Scholar] [CrossRef]
- Honda, T.; Taga, S.; Takeda, T.; Hasibuan, M.; Takeda, Y.; Miwatani, T. Identification of lethal toxin with the thermostable direct hemolysin produced by Vibrio parahaemolyticus, and some physicochemical properties of the purified toxin. Infect. Immun. 1976, 13, 133–139. [Google Scholar] [CrossRef]
- Johnson, C.N.; Flowers, A.R.; Noriea, N.F.; Zimmerman, A.M., 3rd; Bowers, J.C.; DePaola, A.; Grimes, D.J. Relationships between environmental factors and pathogenic Vibrios in the Northern Gulf of Mexico. Appl. Environ. Microbiol. 2010, 76, 7076–7084. [Google Scholar] [CrossRef]
- Johnson, C.N.; Flowers, A.R.; Young, V.C.; Gonzalez-Escalona, N.; DePaola, A.; Noriea, N.F.; Grimes, D.J., 3rd. Genetic relatedness among tdh+ and trh+ Vibrio parahaemolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb. Ecol. 2009, 57, 437–443. [Google Scholar] [CrossRef]
- Kaysner, C.A.; DePaola, A. Vibrio. In Compendium of Methods for the Microbiological Examination of Foods; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001. [Google Scholar]
- Nishibuchi, M.; Kaper, J.B. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 1985, 162, 558–564. [Google Scholar] [CrossRef]
- Nordstrom, J.L.; Rangdale, R.; Vickery, M.C.; Phillips, A.M.; Murray, S.L.; Wagley, S.; DePaola, A. Evaluation of an alkaline phosphatase-labeled oligonucleotide probe for the detection and enumeration of the thermostable-related hemolysin (trh) gene of Vibrio parahaemolyticus. J. Food Prot. 2006, 69, 2770–2772. [Google Scholar] [CrossRef]
- Nordstrom, J.L.; Vickery, M.C.; Blackstone, G.M.; Murray, S.L.; DePaola, A. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl. Environ. Microbiol. 2007, 73, 5840–5847. [Google Scholar] [CrossRef]
- Panicker, G.; Bej, A.K. Real-time PCR detection of Vibrio vulnificus in oysters: Comparison of oligonucleotide primers and probes targeting vvhA. Appl. Environ. Microbiol. 2005, 71, 5702–5709. [Google Scholar] [CrossRef]
- DePaola, A.; Nordstrom, J.A.; Bowers, J.; Wells, J.G.; Cook, D.W. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 2003, 69, 1521–1526. [Google Scholar] [CrossRef]
- DePaola, A.; Ulaszek, J.; Kaysner, C.A.; Tenge, B.J.; Nordstrom, J.L.; Wells, J.; Puhr, N.; Gendel, S.M. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 2003, 69, 3999–4005. [Google Scholar] [CrossRef]
- Duhamel, R.C.; Johnson, D.D.A. Use of nonfat dry milk to block nonspecific nuclear and membrane staining by avidin conjugates. J. Histochem. Cytochem. 1985, 33, 711–714. [Google Scholar] [CrossRef]
- Boon, N.; De Windt, W.; Verstraete, W.; Top, E.M. Evaluation of nested PCR–DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 2002, 39, 101–112. [Google Scholar] [CrossRef]
- Kim, C.; Graves, L.M.; Swaminathan, B.; Mayer, L.; Weaver, R. Evaluation of hybridization characteristics of a cloned pRF106 probe for Listeria monocytogenes detection and development of a nonisotopic colony hybridization assay. Appl. Environ. Microbiol. 1991, 57, 289–294. [Google Scholar] [CrossRef]
- Church, G.M.; Gilbert, W. Genomic sequencing. Proc. Natl. Acad. Sci. USA 1984, 81, 1991–1995. [Google Scholar] [CrossRef]
- Johnson, C.N. Fitness factors in vibrios: A mini-review. Microb. Ecol. 2013, 65, 826–851. [Google Scholar] [CrossRef]
- Hlavsa, M.C.; Roberts, V.A.; Anderson, A.R.; Hill, V.R.; Kahler, A.M.; Orr, M.; Garrison, L.E.; Hicks, L.A.; Newton, A.; Hilborn, E.D. Surveillance for waterborne disease outbreaks and other health events associated with recreational water—United States, 2007–2008. MMWR Surveill. Summ. 2011, 60, 1–32. [Google Scholar]
- Namadi, P.; Deng, Z. Modeling and forecasting Vibrio parahaemolyticus concentrations in oysters. Water Res. 2021, 189, 116638. [Google Scholar] [CrossRef]
- Yoder, J.S.; Hlavsa, M.C.; Craun, G.F.; Hill, V.; Roberts, V.; Yu, P.A.; Hicks, L.A.; Alexander, N.T.; Calderon, R.L.; Roy, S.L. Surveillance for Waterborne Disease and Outbreaks Associated with Recreational Water Use and Other Aquatic Facility-Associated Health Events—United States, 2005–2006; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA; US Department of Health and Human Services: Washington, DC, USA, 2008; Volume 57, pp. 1–29.
- Johnson, C.N.; Bowers, J.C.; Griffitt, K.J.; Molina, V.; Clostio, R.W.; Pei, S.; Laws, E.; Paranjpye, R.N.; Strom, M.S.; Chen, A.; et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington, United States. Appl. Environ. Microbiol. 2012, 78, 7249–7257. [Google Scholar] [CrossRef]
- DePaola, A.; Hopkins, L.H.; Peeler, J.T.; Wentz, B.; McPhearson, R.M. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl. Environ. Microbiol. 1990, 56, 2299–2302. [Google Scholar] [CrossRef]
- DePaola, A.; Kaysner, C.A.; Bowers, J.; Cook, D.W. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 2000, 66, 4649–4654. [Google Scholar] [CrossRef]
- Kelly, M.T.; Stroh, E.M. Temporal relationship of Vibrio parahaemolyticus in patients and the environment. J. Clin. Microbiol. 1988, 26, 1754–1756. [Google Scholar] [CrossRef]
- Nordstrom, J.L.; Kaysner, C.A.; Blackstone, G.M.; Vickery, M.C.; Bowers, J.C.; DePaola, A. Effect of intertidal exposure on Vibrio parahaemolyticus levels in Pacific Northwest oysters. J. Food Prot. 2004, 67, 2178–2182. [Google Scholar] [CrossRef]
- Wang, J.; Deng, Z. Detection and forecasting of oyster norovirus outbreaks: Recent advances and future perspectives. Mar. Environ. Res. 2012, 80, 62–69. [Google Scholar] [CrossRef]
- Watkins, W.D.; Cabelli, V.J. Effect of fecal pollution on Vibrio parahaemolyticus densities in an estuarine environment. Appl. Environ. Microbiol. 1985, 49, 1307–1313. [Google Scholar] [CrossRef]
- Griffitt, K.J.; Noriea, N.F.; Johnson, C.N., 3rd; Grimes, D.J. Enumeration of Vibrio parahaemolyticus in the viable but nonculturable state using direct plate counts and recognition of individual gene fluorescence in situ hybridization. J. Microbiol. Methods 2011, 85, 114–118. [Google Scholar] [CrossRef]
- Oliver, J.D.; Bockian, R. In vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl. Environ. Microbiol. 1995, 61, 2620–2623. [Google Scholar] [CrossRef]
- Wong, H.C.; Wang, P. Induction of viable but nonculturable state in Vibrio parahaemolyticus and its susceptibility to environmental stresses. J. Appl. Microbiol. 2004, 96, 359–366. [Google Scholar] [CrossRef]
- Cook, D.W.; DePaola, A.; McCarthy, S.A. Direct Plating Procedure for the Enumeration of Total and Pathogenic Vibrio parahaemolyticus in Oyster Meats (Vp-ISSC-3); US Food and Drug Administration (FDA) Gulf Coast Seafood Laboratory: Dauphin Island, AL, USA, 2000.
- McCarthy, S.A.; DePaola, A.; Kaysner, C.A.; Hill, W.E.; Cook, D.W. Evaluation of nonisotopic DNA hybridization methods for detection of the tdh gene of Vibrio parahaemolyticus. J. Food Prot. 2000, 63, 1660–1664. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Risk Assessment on the Public. Health Impact of Vibrio parahaemolyticus in Raw Molluscan Shellfish; US Food and Drug Administration: Washington, DC, USA, 2001.
- US Food and Drug Administration. Quantitative Risk Assessment on the Public. Health Impact of Pathogenic Vibrio parahaemolyticus in Raw Oysters; US Food and Drug Administration: Washington, DC, USA, 2005.
- Kaysner, C.A.; DePaola, A.; Jones, J.L. Bacteriological Analytical Manual (BAM). BAM Chapter 9: Vibrio. 2004. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-9-vibrio (accessed on 29 March 2023).
- US Food and Drug Administration; Center for Food Safety and Applied Nutrition. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio spp. Bacteriol Anal Manual, Ch 9 URL. 2001. Available online: http://www.cfsan.fda.gov/~ebam/bam-9.html (accessed on 29 March 2023).
- Anupama, K.P.; Deeksha, K.; Deeksha, A.; Karunasagar, I.; Karunasagar, I.; Maiti, B. Comparative performance of TCBS and TSA for the enumeration of trh+ Vibrio parahaemolyticus by direct colony hybridization. J. Microbiol. Methods 2019, 157, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Betty, C.; Ann-Sofi, R.-H.; Stina-Mina, E.B.; Aidate, M.; Bodil, H. Characteristics of potentially pathogenic vibrios from subtropical Mozambique compared with isolates from tropical India and boreal Sweden. FEMS Microbiol. Ecol. 2013, 83, 255–264. [Google Scholar]
- Deepanjali, A.; Kumar, H.S.; Karunasagar, I.; Karunasagar, I. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 2005, 71, 3575–3580. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, P.; Acharya, S.; Bhanumathi, A.; Karunasagar, I.; Karunasagar, I. Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 2008, 25, 824–830. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. Bacteriological Analytical Manual (BAM). 2023. Available online: https://www.fda.gov/food/laboratory-methods-food/bacteriological-analytical-manual-bam (accessed on 29 March 2023).
- Wagley, S.; Koofhethile, K.; Rangdale, R. Prevalence and potential pathogenicity of Vibrio parahaemolyticus in Chinese mitten crabs (Eriocheir sinensis) harvested from the River Thames estuary, England. J. Food Prot. 2009, 72, 60–66. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations and World Health Organization. Risk Assessment Tools for Vibrio parahaemolyticus and Vibrio vulnificus Associated with Seafood. In Microbiological Risk Assessment Series No. 20; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2020; ISBN 978-92-5-132020-4. [Google Scholar]
- Food and Agriculture Organization of the United Nations and World Health Organization. Risk assessment of Vibrio vulnificus in raw oysters: Interprative summary and technical report. In Microbiological Risk Assessment Series; FAO: Rome, Italy, 2005; pp. 1–134. [Google Scholar]
- Reimer, C.; Kufs, J.E.; Rautschek, J.; Regestein, L.; Valiante, V.; Hillmann, F. Engineering the amoeba Dictyostelium discoideum for biosynthesis of a cannabinoid precursor and other polyketides. Nat. Biotechnol. 2022, 40, 751–758. [Google Scholar] [CrossRef]
- Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975, 98, 503–517. [Google Scholar] [CrossRef]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytom. J. Int. Soc. Anal. Cytol. 1993, 14, 136–143. [Google Scholar] [CrossRef]
- Paranjpye, R.N.; Johnson, A.B.; Baxter, A.E.; Strom, M.S. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl. Environ. Microbiol. 2007, 73, 5041–5044. [Google Scholar] [CrossRef]
- Pettis, G.S.; Mukerji, A.S. Structure, function, and regulation of the essential virulence factor capsular polysaccharide of Vibrio vulnificus. Int. J. Mol. Sci. 2020, 21, 3259. [Google Scholar] [CrossRef]
- Johnson, C.N.; Benjamin, W.H., Jr.; Moser, S.A., Jr.; Hollingshead, S.K.; Zheng, X.; Crain, M.J.; Nahm, M.H.; Waites, K.B. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 2003, 41, 2458–2464. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.N.; Briles, D.E.; Benjamin, W.H.; Hollingshead, S.K., Jr.; Waites, K.B. Relative fitness of fluoroquinolone-resistant Streptococcus pneumoniae. Emerg. Infect. Dis. 2005, 11, 814–820. [Google Scholar] [CrossRef] [PubMed]
| Isolate Name | tlh/tdh/trh/vvh | 16S rRNA Gene Sequence |
|---|---|---|
| AMS001 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGACCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS002 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTAGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS003 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATGCTGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS004 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCANGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATGCTGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS005 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGNAGTGTAGTTAATAGCTGCATTATTTGACGTTATCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS006 | None | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGCATATGCGTTAATAGCGCATGTGTTTGACGTTATGCTGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS007 | None | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTGGTGTAGTTAATAGCTGCATTACTTGACGTTATGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS008 | None | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGCATATGCGTTAATAGCGCATGTGTTTGACGTTAGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS009 | trh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAACGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS010 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTAGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS011 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGACCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGNNGTGTAGTTAATAGCTGCATTATTTGACGTTATCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| Haley | None | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAACGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTTTCAGTCGTGAGGAAGGGGGTNTCGTTAATAGCNGTATTCTTTGACGTTATCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS013 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS014 | None | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGTAGTGTAGTTAATAGCTGCATTATTTGACGTTATGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
| AMS015 | tlh+ | ACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGTCGTGAGGAAGGNNGTGTAGTTAATAGCTGCATTATTTGACGTTATGCGACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwartz, A.M.; Marcotte, H.A.; Johnson, C.N. Evaluation of Alternative Colony Hybridization Methods for Pathogenic Vibrios. Foods 2023, 12, 1472. https://doi.org/10.3390/foods12071472
Schwartz AM, Marcotte HA, Johnson CN. Evaluation of Alternative Colony Hybridization Methods for Pathogenic Vibrios. Foods. 2023; 12(7):1472. https://doi.org/10.3390/foods12071472
Chicago/Turabian StyleSchwartz, Andrew M., Haley A. Marcotte, and Crystal N. Johnson. 2023. "Evaluation of Alternative Colony Hybridization Methods for Pathogenic Vibrios" Foods 12, no. 7: 1472. https://doi.org/10.3390/foods12071472
APA StyleSchwartz, A. M., Marcotte, H. A., & Johnson, C. N. (2023). Evaluation of Alternative Colony Hybridization Methods for Pathogenic Vibrios. Foods, 12(7), 1472. https://doi.org/10.3390/foods12071472