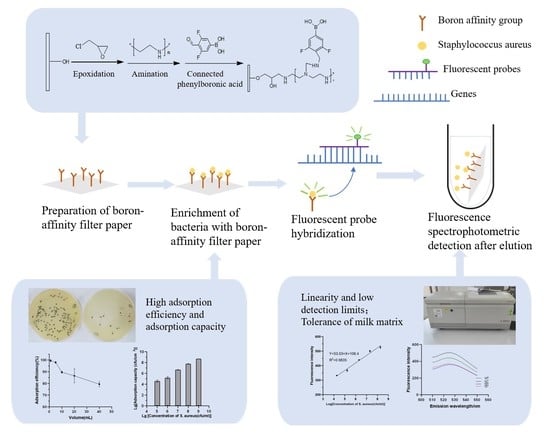
Rapid and Sensitive Fluorescence Detection of Staphylococcus aureus Based on Polyethyleneimine-Enhanced Boronate Affinity Isolation
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Apparatus
2.3. Bacterial Strains and Culture Conditions
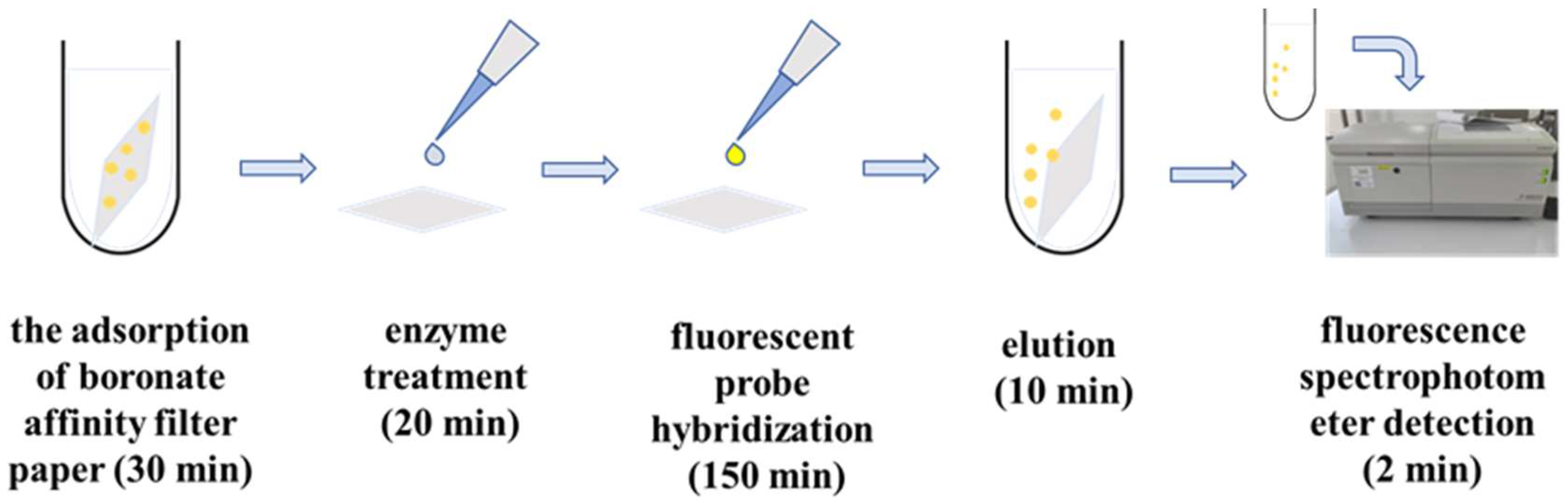
2.4. Preparation of Boronate Affinity Paper@epoxy@PEI-DFFPBA
2.5. Isolation of Paper@epoxy@PEI-DFFPBA to S. aureus
2.6. Fluorescent Detection of S. aureus
2.7. Validation in Food Matrix
2.8. Statistical Analysis
3. Results
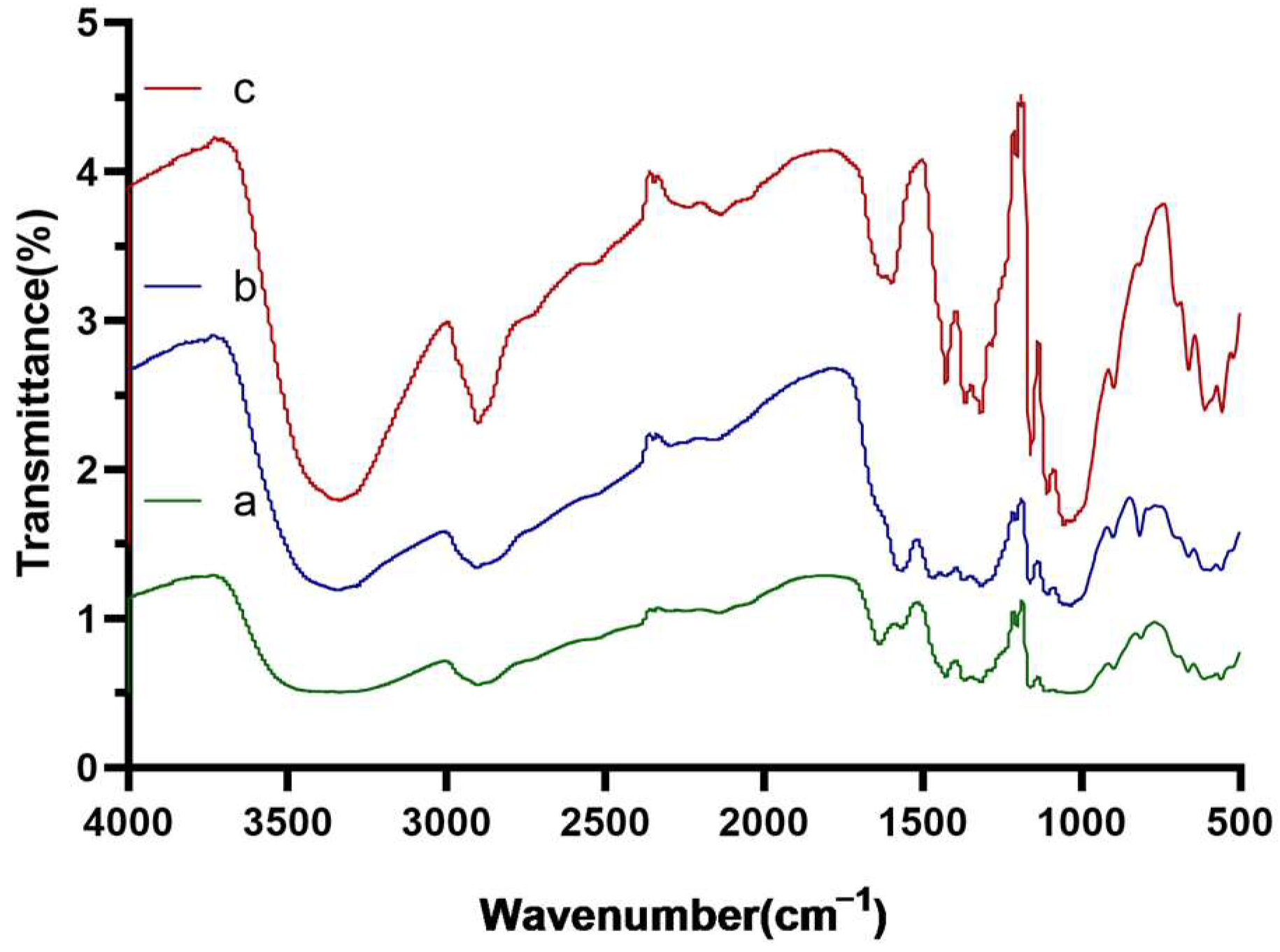
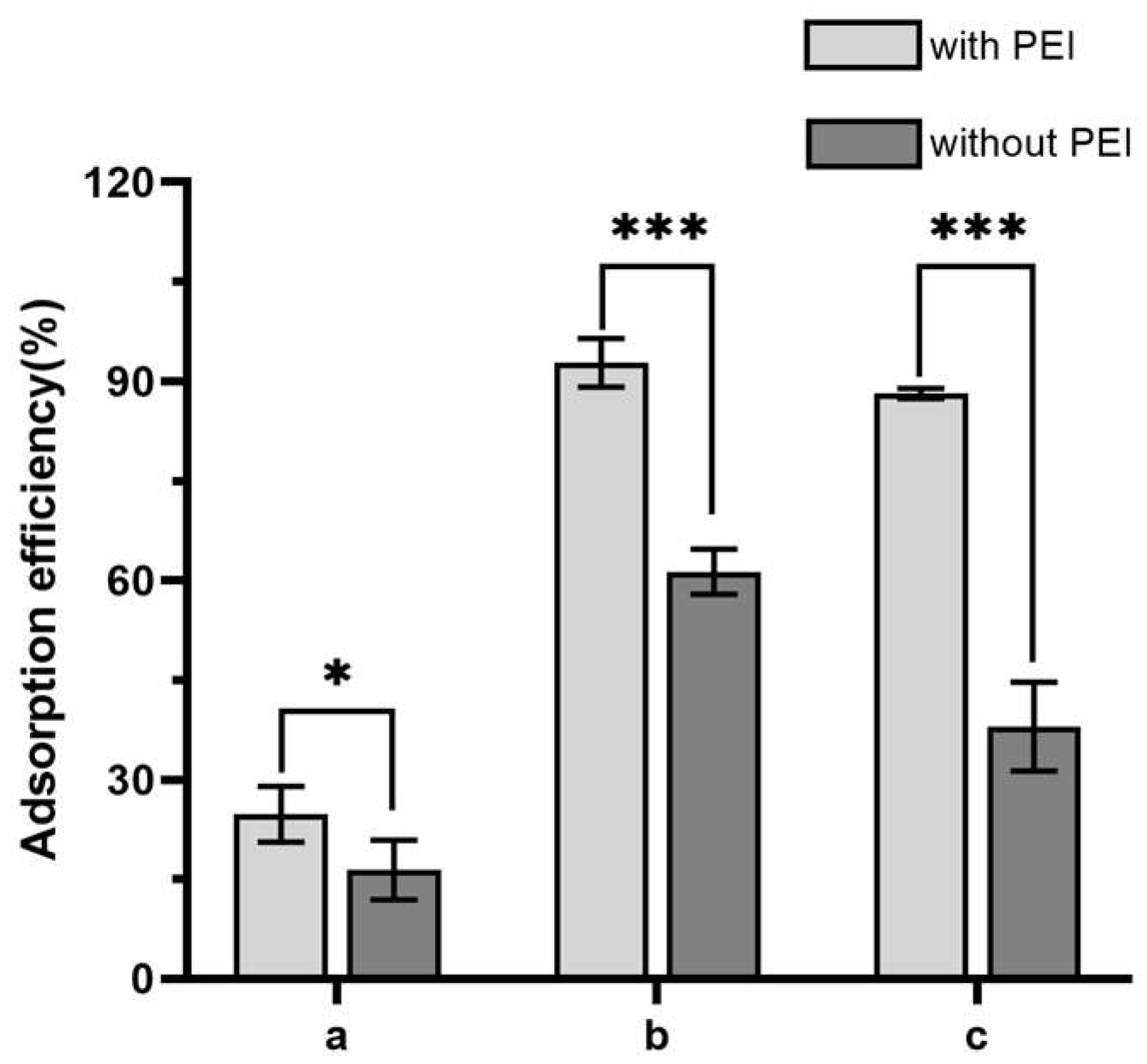
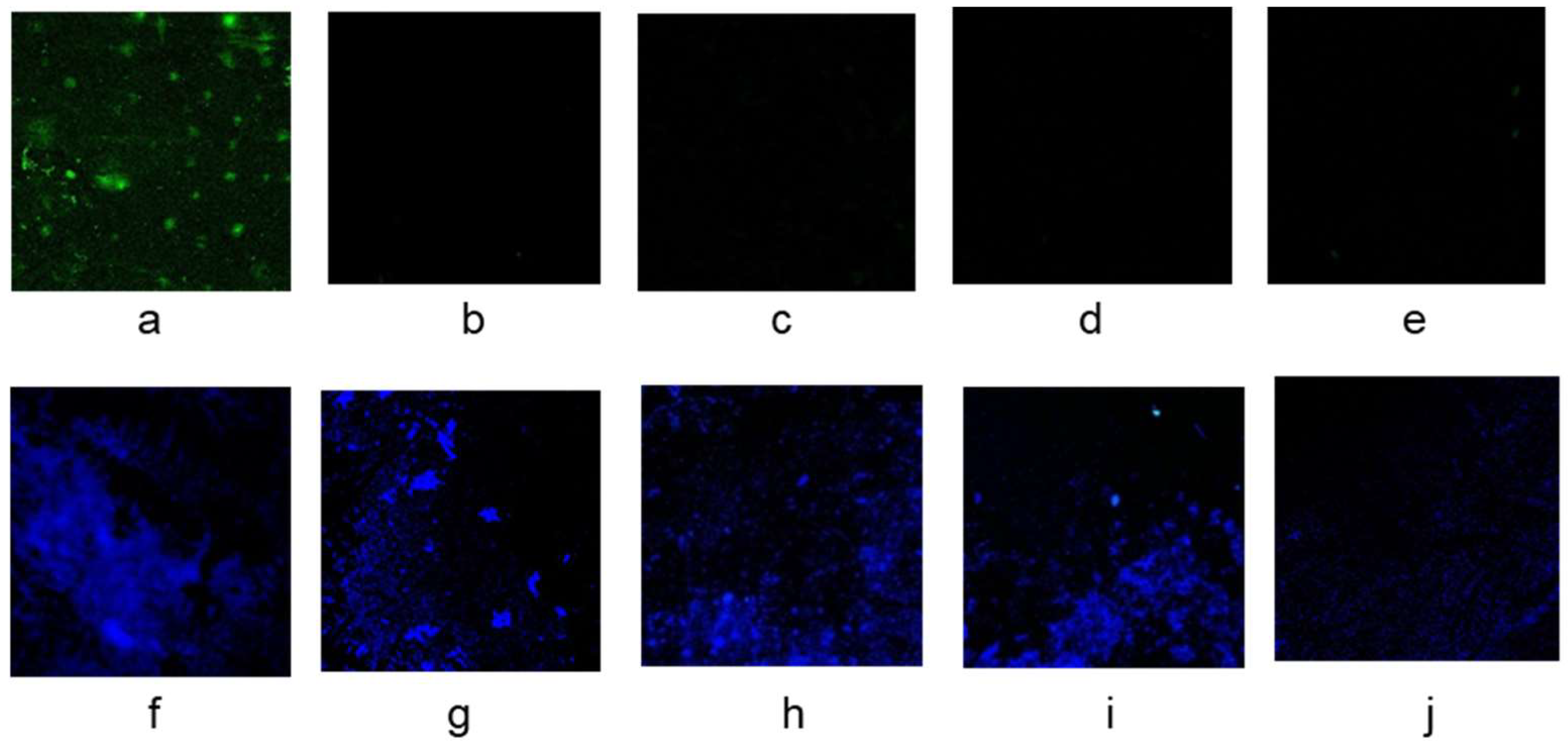
3.1. The Effect of Polyethyleneimine Modification to Improve the Boronate Affinity Ability of the Filter Paper
3.2. Optimization of Analytical Performance
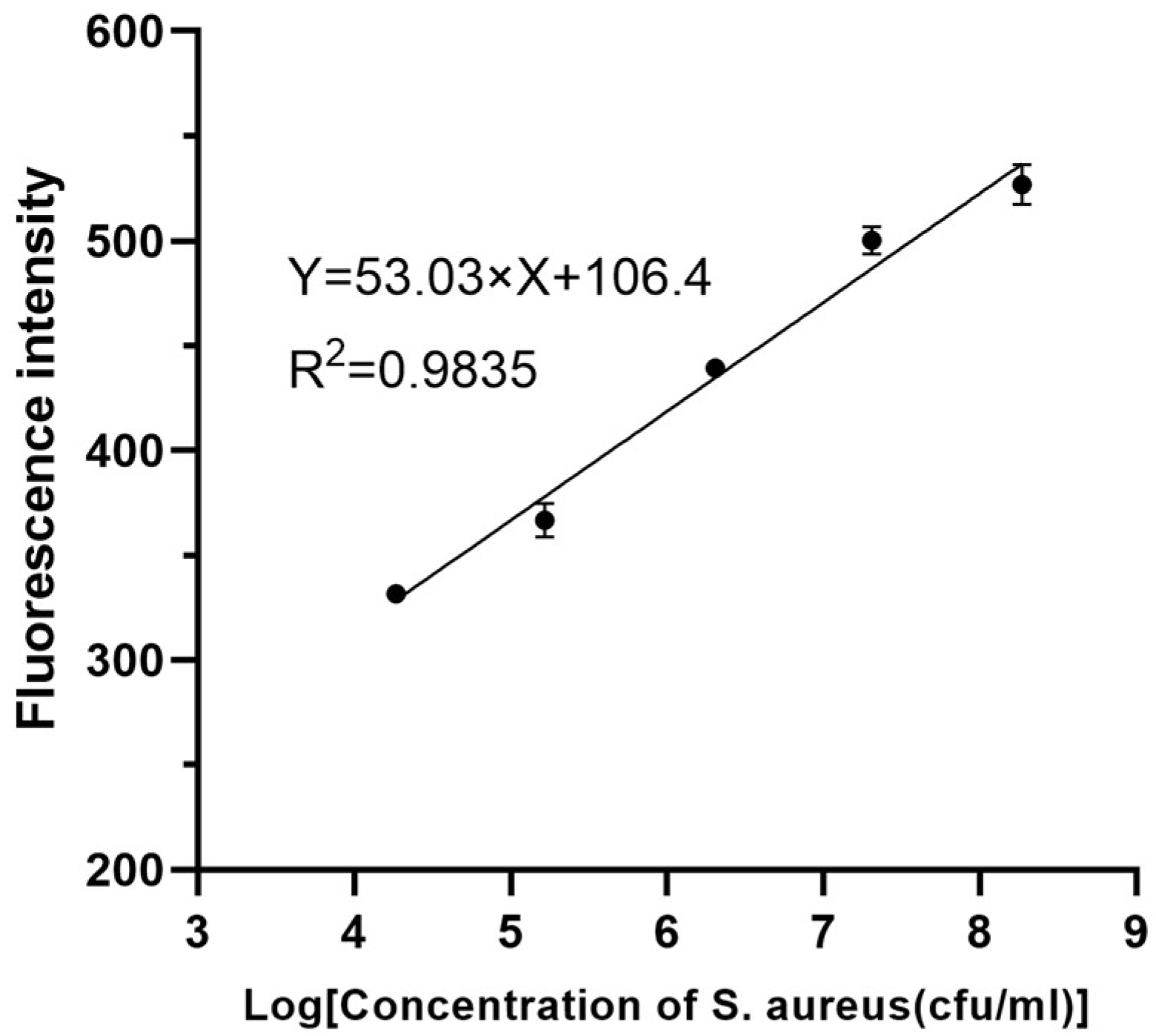
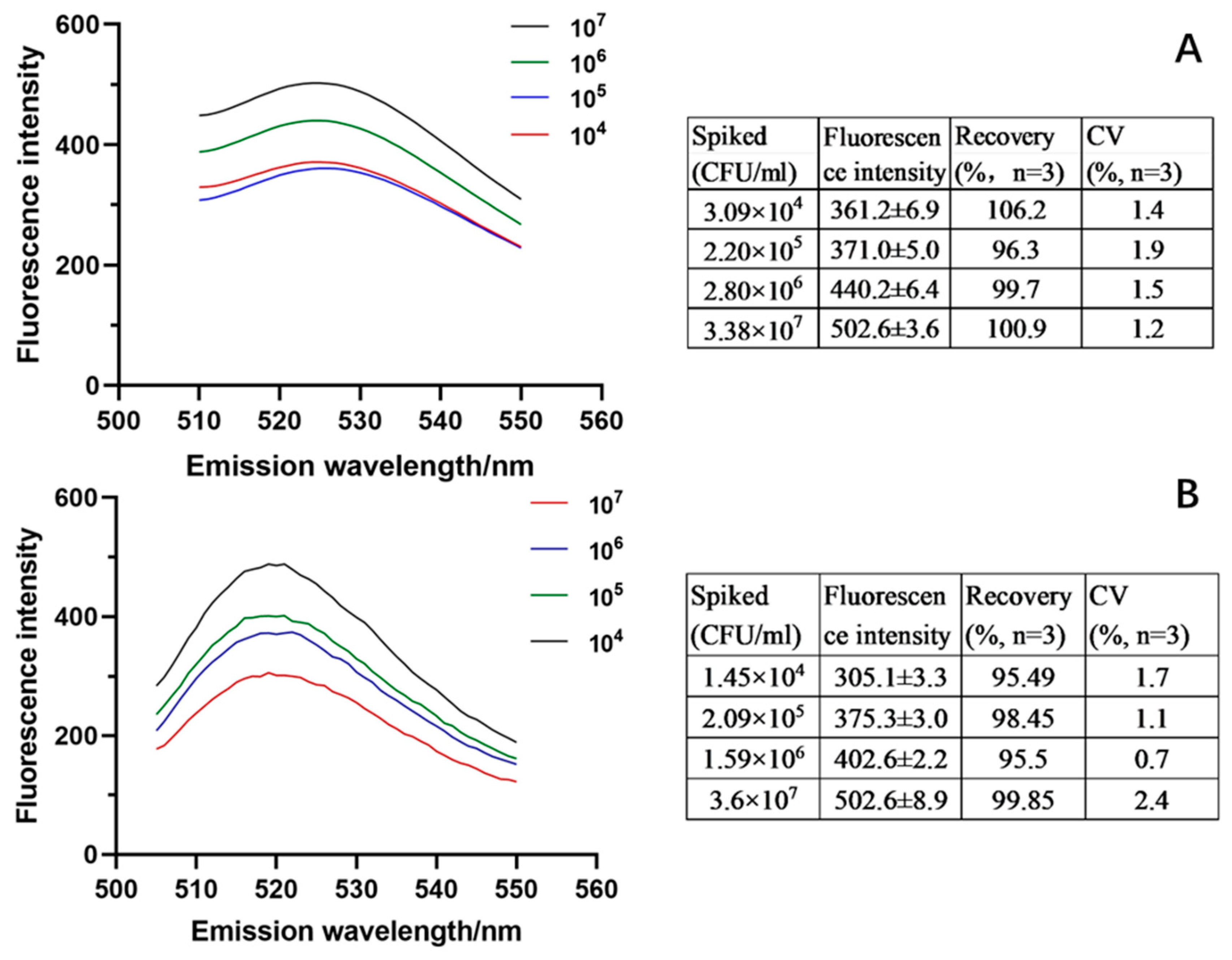
3.3. Evaluation of the Analytical Efficiency
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lipcsei, L.E.; Brown, L.G.; Coleman, E.W.; Kramer, A.; Masters, M.; Wittry, B.C.; Reed, K.; Radke, V.J. Foodborne illness outbreaks at retail establishments—National Environmental Assessment Reporting System, 16 state and local health departments, 2014–2016. MMWR Surveill. Summ. 2019, 68, 1–20. [Google Scholar] [CrossRef]
- Kalyantanda, G.; Shumyak, L.; Archibald, L.K. Cronobacter species contamination of powdered infant formula and the implications for neonatal health. Front. Pediatr. 2015, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.S.; Nielsen, L.T.; Leth, S. Staphylococcus aureus toxic shock syndrome originating from a split skin transplant. Ugeskr. Laeger 2019, 181, V08180580. [Google Scholar] [PubMed]
- Kim, H.J.; Kwon, C.; Lee, B.S.; Noh, H. One-step sensing of foodborne pathogenic bacteria using a 3D paper-based device. Analyst 2019, 144, 2248–2255. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Liu, C.; Wu, S.; Li, H.; Guo, H.; Yang, B.; Qiu, S.; Li, J.; Liu, L.; Zeng, H.; et al. Development of a lateral flow colloidal gold immunoassay strip for the simultaneous detection of Shigella boydii and Escherichia coli O157: H7 in bread, milk and jelly samples. Food Control 2016, 59, 345–351. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Shi, Q.; Huang, S.; Yu, T.; Zhang, L.; Yang, H. Multiplex PCR method for simultaneous detection of five pathogenic bacteria closely related to foodborne diseases. 3 Biotech 2021, 11, 219. [Google Scholar] [CrossRef]
- Fan, S.; Ma, C.; Tian, X.; Ma, X.; Qin, M.; Wu, H.; Tian, X.; Lu, J.; Lyu, M.; Wang, S. Detection of Vibrio vulnificus in seafood with a DNAzyme-based biosensor. Front. Microbiol. 2021, 12, 655845. [Google Scholar] [CrossRef]
- Yin, M.; Jing, C.; Li, H.; Deng, Q.; Wang, S. Surface chemistry modified upconversion nanoparticles as fluorescent sensor array for discrimination of foodborne pathogenic bacteria. J. Nanobiotechnol. 2020, 18, 41. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.F. Selective biosensing of Staphylococcus aureus using chitosan quantum dots. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 188, 50–56. [Google Scholar] [CrossRef]
- Choi, C.A.; Mazrad, Z.A.I.; Lee, G.; In, I.; Lee, K.D.; Park, S.Y. Boronate-based fluorescent carbon dot for rapid and selectively bacterial sensing by luminescence off/on system. J. Pharm. Biomed. Anal. 2018, 159, 1–10. [Google Scholar] [CrossRef]
- Rocha, R.; Sousa, J.M.; Cerqueira, L.; Vieira, M.J.; Almeida, C.; Azevedo, N.F. Development and application of peptide nucleic acid fluorescence in situ hybridization for the specific detection of Listeria monocytogenes. Food Microbiol. 2019, 80, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Oh, S.W. Pretreatment methods for nucleic acid-based rapid detection of pathogens in food: A review. Food Control 2021, 121, 107575. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, A.; Zhang, Y.; Bie, Z. Recent advances of boronate affinity materials in sample preparation. Anal. Chim. Acta 2019, 1076, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Pang, S.; Pearson, B.; Chujo, Y.; McLandsborough, L.; Fan, M.; He, L. Rapid concentration detection and differentiation of bacteria in skimmed milk using surface enhanced Raman scattering mapping on 4-mercaptophenylboronic acid functionalized silver dendrites. Anal. Bioanal. Chem. 2017, 409, 2229–2238. [Google Scholar] [CrossRef]
- Golabi, M.; Kuralay, F.; Jager, E.W.; Beni, V.; Turner, A.P. Electrochemical bacterial detection using poly (3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017, 93, 87–93. [Google Scholar] [CrossRef]
- Zheng, H.; Han, F.; Lin, H.; Cao, L.; Pavase, T.R.; Sui, J. Preparation of a novel polyethyleneimine functionalized sepharose-boronate affinity material and its application in selective enrichment of food borne pathogenic bacteria. Food Chem. 2019, 294, 468–476. [Google Scholar] [CrossRef]
- Zheng, H.; Hajizadeh, S.; Gong, H.; Lin, H.; Ye, L. Preparation of boronic acid-functionalized cryogels using modular and clickable building blocks for bacterial separation. J. Agric. Food Chem. 2020, 69, 135–145. [Google Scholar] [CrossRef]
- Zheng, H.; Lin, H.; Chen, X.; Sui, J.; Pavase, T.R.; Han, X.; Cao, L. Tailor-made magnetic nanocomposite with pH and thermo-dual responsive copolymer brush for bacterial separation. Food Chem. 2021, 358, 129907. [Google Scholar] [CrossRef]
- Amin, R.; Elfeky, S.A. Fluorescent sensor for bacterial recognition. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 108, 338–341. [Google Scholar] [CrossRef]
- Kwon, H.Y.; Liu, X.; Choi, E.G.; Lee, J.Y.; Choi, S.Y.; Kim, J.Y.; Wang, L.; Park, S.J.; Kim, B.; Lee, Y.A.; et al. Development of a universal fluorescent probe for gram-positive bacteria. Angew. Chem. 2019, 131, 8514–8519. [Google Scholar]
- Mikagi, A.; Tsurufusa, R.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Fast and sensitive bacteria detection by boronic acid modified fluorescent dendrimer. Sensors 2021, 21, 3115. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bie, Z.; Lü, C.; Liu, Z. Magnetic nanoparticles with dendrimer-assisted boronate avidity for the selective enrichment of trace glycoproteins. Chem. Sci. 2013, 4, 4298–4303. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, J.; Chen, G.; Feng, X.; Deng, M.; Mu, D.; Xu, Q.; Xu, H. An integrated system using phenylboronic acid functionalized magnetic beads and colorimetric detection for Staphylococcus aureus. Food Control 2022, 133, 108633. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, L.; Li, C.; Ye, B.; Xiong, J.; Shi, X. Immobilization of polyethyleneimine-templated silver nanoparticles onto filter paper for catalytic applications. Colloids Surf. A Physicochem. Eng. Asp. 2019, 571, 44–49. [Google Scholar] [CrossRef]
- He, Y.; Wu, Y.; Fu, J.Z.; Wu, W.B. Fabrication of paper-based microfluidic analysis devices: A review. Rsc Adv. 2015, 5, 78109–78127. [Google Scholar] [CrossRef]
- Peng, Y.B.; Mao, E.Q.; Fan, E.S.; Shen, H.P. Quantum dots labeled probes fluorescence in situ hybridization for rapid detection of Staphylococcus aureus. J. Diagn. Concepts Pact. 2010, 4, 327–330. [Google Scholar]
- Wu, Q.; Li, Y.; Wang, M.; Pan, X.P.; Tang, Y.F. Fluorescence in situ hybridization rapidly detects three different pathogenic bacteria in urinary tract infection samples. J. Microbiol. Methods 2010, 83, 175–178. [Google Scholar] [CrossRef]
- Xiao, C.; Li, H.; Zhao, Y.; Zhang, X.; Wang, X. Green synthesis of iron nanoparticle by tea extract (polyphenols) and its selective removal of cationic dyes. J. Environ. Manag. 2020, 275, 111262. [Google Scholar] [CrossRef]
- Han, X.; Lin, H.; Cao, L.; Chen, X.; Wang, L.; Zheng, H.; Zhang, Z.; Pavase, T.R.; Wang, S.; Sun, X.; et al. Hapten-branched polyethylenimine as a new antigen affinity ligand to purify antibodies with high efficiency and specificity. ACS Appl. Mater. Interfaces 2020, 12, 58191–58200. [Google Scholar] [CrossRef]
- Li, Q.; Lü, C.; Liu, Z. Preparation and characterization of fluorophenylboronic acid-functionalized monolithic columns for high affinity capture of cis-diol containing compounds. J. Chromatogr. A 2013, 1305, 123–130. [Google Scholar] [CrossRef]
- Li, D.; Chen, Y.; Liu, Z. Boronate affinity materials for separation and molecular recognition: Structure, properties and applications. Chem. Soc. Rev. 2015, 44, 8097–8123. [Google Scholar] [CrossRef]
- Yue, X.; Li, Z.; Zhang, T.; Yang, D.; Qiu, F. Design and fabrication of superwetting fiber-based membranes for oil/water separation applications. Chem. Eng. J. 2019, 364, 292–309. [Google Scholar] [CrossRef]
- Hajizadeh, S.; Ye, L. Hierarchical macroporous material with dual responsive copolymer brushes and phenylboronic acid ligands for bioseparation of proteins and living cells. Sep. Purif. Technol. 2019, 224, 95–105. [Google Scholar] [CrossRef]
- Lu, C.; Li, H.; Wang, H.; Liu, Z. Probing the interactions between boronic acids and cis-diol-containing biomolecules by affinity capillary electrophoresis. Anal. Chem. 2013, 85, 2361–2369. [Google Scholar] [CrossRef]
- Zheng, H.; Gong, H.; Cao, L.; Lin, H.; Ye, L. Photoconjugation of temperature-and pH-responsive polymer with silica nanoparticles for separation and enrichment of bacteria. Colloids Surf. B Biointerfaces 2021, 197, 111433. [Google Scholar] [CrossRef]
- Wang, J.; Gao, J.; Liu, D.; Han, D.; Wang, Z. Phenylboronic acid functionalized gold nanoparticles for highly sensitive detection of Staphylococcus aureus. Nanoscale 2012, 4, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Mustapha, A. Multiplex real-time PCR assays for detection of eight Shiga toxin-producing Escherichia coli in food samples by melting curve analysis. Int. J. Food Microbiol. 2015, 215, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Kitaguchi, A.; Nasu, M. Selective enumeration of viable Enterobacteriaceae and Pseudomonas spp. in milk within 7 h by multicolor fluorescence in situ hybridization following microcolony formation. J. Biosci. Bioeng. 2012, 113, 746–750. [Google Scholar] [CrossRef]
- Quirós, J.; Amaral, A.J.; Pasparakis, G.; Williams, G.R.; Rosal, R. Electrospun boronic acid-containing polymer membranes as fluorescent sensors for bacteria detection. React. Funct. Polym. 2017, 121, 23–31. [Google Scholar] [CrossRef]
- Li, Q.; Lü, C.; Li, H.; Liu, Y.; Wang, H.; Wang, X.; Liu, Z. Preparation of organic-silica hybrid boronate affinity monolithic column for the specific capture and separation of cis-diol containing compounds. J. Chromatogr. A 2012, 1256, 114–120. [Google Scholar] [CrossRef]
- Zeng, Y.; Liang, D.; Zheng, P.; Zhang, Y.; Wang, Z.; Mari, G.M.; Jiang, H. A simple and rapid immunochromatography test based on readily available filter paper modified with chitosan to screen for 13 sulfonamides in milk. J. Dairy Sci. 2021, 104, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Hankus, M.E.; Tian, L.; Pellegrino, P.M.; Singamaneni, S. Highly sensitive surface enhanced Raman scattering substrates based on filter paper loaded with plasmonic nanostructures. Anal. Chem. 2011, 83, 8953–8958. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Zhang, Y.; Liu, X.; Zhang, X.; Zhang, H. A filter paper coated with phenylboronic acid-modified mesoporous silica for enrichment of intracellular nucleosides prior to their quantitation by HPLC. Microchim. Acta 2017, 184, 4007–4013. [Google Scholar] [CrossRef]
- Qin, Q.; Li, H.; Shi, X.; Xu, G. Facile synthesis of Fe3O4@ polyethyleneimine modified with 4-formylphenylboronic acid for the highly selective extraction of major catecholamines from human urine. J. Sep. Sci. 2015, 38, 2857–2864. [Google Scholar] [CrossRef]
- Bao, W.; Hai, W.; Bao, L.; Yang, F.; Liu, Y.; Goda, T.; Liu, J. Poly (3, 4-ethylenedioxythiophene) bearing fluoro-containing phenylboronic acid for specific recognition of glucose. Mater. Chem. Front. 2021, 5, 7675–7683. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Li, X.; Bie, Z.; Pan, X.; Zhang, Q.; Liu, Z. A high boronate avidity monolithic capillary for the selective enrichment of trace glycoproteins. J. Chromatogr. A 2015, 1384, 88–96. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Ohba, H.; Nasu, M. Simple detection of small amounts of Pseudomonas cells in milk by using a microfluidic device. Lett. Appl. Microbiol. 2006, 43, 631–636. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.T.; Xia, X.; Liu, J.; Lu, Y.; Khan, M.R.; Deng, S.; Busquets, R.; He, G.; He, Q.; et al. Direct detection of foodborne pathogens via a proximal DNA probe-based CRISPR-Cas12 assay. J. Agric. Food Chem. 2021, 69, 12828–12836. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Augkarawaritsawong, S.; Santiwatanakul, S.; Chansiri, K. Rapid colorimetric assay for detection of Listeria monocytogenes in food samples using LAMP formation of DNA concatemers and gold nanoparticle-DNA probe complex. Front. Chem. 2018, 6, 90. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Fernandes, R.M.; Keevil, C.W.; Vieira, M.J. Fluorescence In Situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl. Environ. Microbiol. 2010, 76, 4476–4485. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, W.; Chung, J.; Yin, J.; Yoon, J. Recent progress in fluorescent probes for bacteria. Chem. Soc. Rev. 2021, 50, 7725–7744. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Aoi, R.; Osanai, Y.; Kawai, Y.; Yamazaki, K. Rapid quantitative detection of Salmonella enterica using fluorescence in situ hybridization with filter-cultivation (FISHFC) method. Food Sci. Technol. Res. 2013, 19, 59–67. [Google Scholar] [CrossRef]
- Jiang, L.; Messing, M.E.; Ye, L. Temperature and pH dual-responsive core-brush nanocomposite for enrichment of glycoproteins. ACS Appl. Mater. Interfaces 2017, 9, 8985–8995. [Google Scholar] [CrossRef] [PubMed]




| Materials and Methods | Targets | LODs | Time | Matrix Tolerance | References |
|---|---|---|---|---|---|
| Multiplex real-time PCR | E. coli | 1.4 × 102–4.3 × 102 CFU/mL | 6–8 h of enrichment time | Vulnerable to protein and fat and pre-enrichment required | [37] |
| Fluorescence in situ hybridization | Enterobacteriaceae/Pseudomonas spp. | 2 × 103 CFU/mL | 5–7 h | Enzymatic treatment of milk samples was required to remove digested milk lipids and proteins | [38] |
| Electrospun boronic acid-containing polymer membranes as fluorescent sensors for bacteria detection | E. coli, S. aureus and Pseudomonas aeruginosa | Not reported | 8–24 h | Not reported | [39] |
| Fluorescently labeled PAMAM with PBA; fluorescence spectrophotometry | S. aureus E. coli | 104 CFU/mL | 30 min | Not reported | [21] |
| Nanosensor consisting of a diol-modified fluorescent probe and phenylboronic acid-functionalized fluorescent carbon dots | S. aureus E. coli | 10 CFU/mL | Incubate for 1 h | No quantitative analysis of complex samples | [10] |
| Boronate affinity filter-paper-based fluorescence detection | S. aureus | 2.24 × 102 CFU/mL | 3.5 h | For 104 CFU/mL~107 CFU/mL S. aureus, excellent accuracy and precision were observed | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zheng, H.; Sui, J.; Lin, H.; Cao, L. Rapid and Sensitive Fluorescence Detection of Staphylococcus aureus Based on Polyethyleneimine-Enhanced Boronate Affinity Isolation. Foods 2023, 12, 1366. https://doi.org/10.3390/foods12071366
Xu Y, Zheng H, Sui J, Lin H, Cao L. Rapid and Sensitive Fluorescence Detection of Staphylococcus aureus Based on Polyethyleneimine-Enhanced Boronate Affinity Isolation. Foods. 2023; 12(7):1366. https://doi.org/10.3390/foods12071366
Chicago/Turabian StyleXu, Yujia, Hongwei Zheng, Jianxin Sui, Hong Lin, and Limin Cao. 2023. "Rapid and Sensitive Fluorescence Detection of Staphylococcus aureus Based on Polyethyleneimine-Enhanced Boronate Affinity Isolation" Foods 12, no. 7: 1366. https://doi.org/10.3390/foods12071366
APA StyleXu, Y., Zheng, H., Sui, J., Lin, H., & Cao, L. (2023). Rapid and Sensitive Fluorescence Detection of Staphylococcus aureus Based on Polyethyleneimine-Enhanced Boronate Affinity Isolation. Foods, 12(7), 1366. https://doi.org/10.3390/foods12071366