Abstract
The eating quality evaluation of rice is raising further concerns among researchers and consumers. This research is aimed to apply lipidomics in determining the distinction between different grades of indica rice and establishing effective models for rice quality evaluation. Herein, a high-throughput ultrahigh-performance liquid chromatography coupled with quadrupole time-of-flight (UPLC-QTOF/MS) method for comprehensive lipidomics profiling of rice was developed. Then, a total of 42 significantly different lipids among 3 sensory levels were identified and quantified for indica rice. The orthogonal partial least-squares discriminant analysis (OPLS-DA) models with the two sets of differential lipids showed clear distinction among three grades of indica rice. A correlation coefficient of 0.917 was obtained between the practical and model-predicted tasting scores of indica rice. Random forest (RF) results further verified the OPLS-DA model, and the accuracy of this method for grade prediction was 90.20%. Thus, this established approach was an efficient method for the eating grade prediction of indica rice.
1. Introduction
Rice is one of the staple food across the world [1]. With the tremendous development of society, rice eating quality attracts increasing attention among researchers and consumers [2]. Thus, the assessment of the eating quality of rice [3] is vital in variety cultivation and merchandise selection. Generally, rice palatability quality is identified by the standard sensory evaluation method in China, which is labor intensive and time-consuming [4], and the result is usually subjective due to the prejudice and alteration of the sensory system depending on the different daily psychological and physical states of human beings [5]. Hence, a novel high-throughput, sensitive, and comprehensive analytical tool was required.
Lipids are one of the three main compositions in rice [6,7] and play important roles in cell membrane components, energy storage, and signal transduction. Studies have shown that lipids affect the eating quality of rice [8,9], but the details remain unclear. Previous lipid research on rice eating quality were mainly fatty acid composition studied by gas chromatography/mass spectrometry (GC-MS) [10]. However, only limited lipids were studied before, and more detailed information on the lipid content and composition of rice relating to eating quality is still required. Comprehensive identification of lipids within biological systems, known as lipidomics, with tens of thousands of lipids, provides conceptions to lots of physiological activities [11,12] and diseases [13]. As for the sufficient resolution, sensitivity, mass accuracy, fragment ion scanning capability, and lipid profile information, ultrahigh-performance liquid chromatography coupled with quadrupole time-of-flight (UPLC-QTOF/MS) [14] has been applied for comprehensive lipidomics study in foods. Data processing is vital in lipidomics analysis, and useful information can be extracted by appropriate methods [15]; then, an effective prediction model is established. Many data processing ways such as principal component analysis (PCA) [16], linear discriminant analysis (LDA) [17], and orthogonal partial least-squares discriminant analysis (OPLS-DA) [18] have been utilized and reported in food analysis [19,20]. However, limited study on rice eating quality prediction by lipids has been reported.
In the present research, a high-throughput, high-sensitivity, and high-coverage UPLC-QTOF/MS method for comprehensive lipidomics profile analysis of rice was used. First, a comprehensive lipidomics study of the content and composition of rice was carried out by UPLC-QTOF/MS. Then, differential lipids were determined from three grades of indica rice. Finally, a novel lipid-based model was developed for predicting rice eating quality. This result provides details for which lipids are related to the eating quality of rice and serve as data for rice breeders and researchers to cultivate new rice varieties with improved eating quality.
2. Materials and Methods
2.1. Chemical and Reagents
Acetonitrile (ACN), isopropanol (IPA), methanol, and methyl tert-butyl ether were bought from Fisher (Chicago, IL, USA). Formic acid and ammonium acetate of LC-MS-grade were from CNW (CNW Technologies, Dusseldorf, Germany). Ultra-pure water was obtained from Milli-Q instrument (Milipore, Burlington, MA, USA).
The internal standards d7-monoglyceride (MG, 18:1), d7-diacylglycerol (DG, 15:0/18:1), d7-triacylglycerol (TG,15:0/18:1/15:0), d7-glycerophosphatidic acid (PA, 15:0/18:1), d7-glycerophosphatidylcholine (PC, 15:0/18:1), d7-glycerophosphatidylethanolamine (PE, 15:0/18:1), d7-glycerophosphatidylglycerol (PG, 15:0/18:1), d7-glycerophosphatidylinositol (PI, 15:0/18:1), d7-glycerophosphatidylserine (PS, 15:0/18:1), d7-ceramide (Cer, C15), and d9-sphingomyelin (SM, 18:1/18:1) were purchased from Avanti (Birmingham, AL, USA). Mixed reagents of all internal standards with final concentration of 100 μg/mL were prepared in methanol and preserved at −20°C in the freezer until analysis.
2.2. Sample Preparation and Extraction Procedure
Based on the variety information issued by Ministry of Agriculture and Rural Affairs of China from 2019 to 2020, collected rice varieties by planting area account for more than 70% of the promotion area in 10 provinces. Indica paddy rice samples were collected during harvest season in 2020. Further details related to field trial characteristics such as variety and geographical location can be found in Table 1. Paddy rice samples were shelled by JDMZ-100 huller (DFJH, Beijing, China). After that, part of the rice was milled by CT-293-CyclotecTM mill (Foss, Suzhou, China). The milled rice and powder was kept at −80 °C until analysis.

Table 1.
Cultivation region, variety, grade, and average tasting scores with standard deviation (SD) of indica rice (IR) analyzed.
The extraction procedure refers to that in a previous study [21] with minor modifications. In short, 20 ± 0.01 mg rice powder was mixed with 200 μL methanol already containing lipid internal standards. The solution was mixed before and after addition of 540 μL MTBE for 30 s. Then, 360 μL ultrapure water was added and mixed for 30 s. After that, the tube was put at 4 °C for 10 min equilibration, then centrifuged at 4 °C with 15,000 rpm for 10 min. Extraction in two phases was transferred to new tubes separately. After evaporation, residue in the upper phase was redissolved in 1 mL solution (ACN/IPA/H2O = 65:30:5, v/v/v) for lipidomics study.
2.3. Sensory Test
Sensory test was conducted in compliance with the Chinese National Standard “Method for Sensory Evaluation of Paddy or Rice Cooking and Eating Quality” (GB/T 15682-2008) and “Rice” (GB/T 1354-2009). In short, rice was cooked in cooker with a 1.3 (w/w, water/milled rice) ratio after soaking for 30 min in water. Then, cooked rice was tested and scored by 20 skilled and qualified panelists. All evaluations were conducted in separate partitions with cooked rice served in random order. After that, average eating score of each sample was calculated and analyzed by SPSS 19.0 (International Business Machines Corporation, New York, NY, USA, USA). According to GB/T 1354–2009, rice with high (score ≥ 90), medium (score ≥ 80), and low (score ≥ 70) eating quality was evaluated and rated as grade high, medium, and low, respectively.
2.4. UPLC-QTOF/MS Analysis
For lipidomics, the UPLC I-Class Plus Instrument (Waters, Manchester, UK) was applied. A BEH C18 Column (130 A, 1.7 µm, 2.1 mm × 50 mm; Waters, Manchester, UK) was used. Separation was carried out with acetonitrile/ultra-pure water (phase A, 60/40, v/v) and isopropanol/acetonitrile (phase B, 90/10, v/v), both within 10 mM ammonium acetate and 0.1% formic acid for positive and negative ionization mode analysis. Following the LC analysis, lipids were instantly detected through a tandem QTOF Mass System (Waters, Manchester, UK). Both positive and negative ionization analysis were carried out with settings: capillary voltage, 2.0 kV for positive and 1.0 kV for negative ion mode; desolvation gas flow rate, 900 L/h; and detection range, 100–1500 m/z. Leu-enkephalin (0.2 µg/mL) with a fixed m/z of 556.2771 for positive mode and m/z for 554.2615 for negative mode was utilized as internal standard during the whole acquisition process.
To guarantee the instrument stability and data quality, quality control (QC) samples were prepared by equal pooling in all rice samples with internal standards added-in to monitor the quality and robustness of the data [22]. QC samples were engaged every 10 samples of rice through the entire analysis procedure.
2.5. Statistical Analysis
The raw data of UPLC-QTOF/MS was loaded on QI software (Waters, Manchester, UK) for analysis, which was carried out by noise setting, baseline correction, alignment, peak detection, and compound identification. Compound determination was performed by the in-house lipids and metabolites database. The accurate contents of lipids were analyzed through peak areas, fragment intensities, and stable-labeled internal standard compounds [23].
These processed data were then imported into EZinfo 3.0 software (Umetrics, Umeå, Sweden) for Student’s t-test, ANOVA analysis, and orthogonal partial least-squares discriminant analysis. OPLS-DA, with distinct predictive and orthogonal information indicating between and within group variance, has the ability to identify which variables include the class-separating information [24,25]. To reduce the data noise induced over-fitting, ANOVA was applied to identify the significantly different lipids among three groups with p value ≤ 0.05. For ANOVA analysis, after parameter setting, the data distribution and variance homogeneity check were automatically carried out through EZinfo 3.0 software, and then lipids with p value ≤ 0.05 were listed.
After being analyzed and filtered through ANOVA analysis with p value ≤ 0.05 and VIP ≥ 1, the filtered biomarkers were finally identified.
3. Results and Discussion
3.1. Rice Sensory Test Analysis
Among the 51 samples, the sensory testing scores of indica rice (IR) are listed in Table 1. In accordance with GB/T 15682-2008 and GB/T 1354-2009, out of 51 samples, 6, 38, and 7 were at grades high, medium, and low, respectively, and the top three varieties at the high level were Hengliangyou, Ezhong 5, and Guangliangx 2. Among all the samples, the medium eating levels gained more samples, and this result was in accordance with the current situation of paddy rice planting in China [26,27]. As for the same variety, such as Guangliangx 2 planted in different locations, rice gained multiple scores, even in different grades. Studies [28,29] have shown that environment and region affect rice quality, which may be the cause of the taste results in this study, but further studies still need to be developed to acquire a comprehensive explanation.
3.2. Identified Rice Lipids by UPLC-QTOF/MS
Rice samples (61 of IR) and 10 QC samples were detected through positive and negative modes. There was no significant variation of RT and m/z (CV < 19.8% on UPLC-QTOF/MS) for internal standards added in the lipid profiles of 10 QC samples [30]. Principal component analysis (PCA) indicated that QC samples were tightly clustered (Figure S1) under the whole MS collection procedure, indicating good stability and precision of the measurements through the total test duration [31].
For the positive mode (ESI+), DG, TG, PA, PG, and PI produced [M + NH4]+, and other lipids, such as PE, LPE, and PS, possessed primarily [M + H]+. PC, LPC, and SM generated [M + CH3COO]−, and other lipids produced [M − H]− in negative ion mode [32]. In addition, PC and LPC formed fragment ions such as 184.07, while other lipids produced [M + H − NL]+ or [M + NH4 − NL]+ ions mainly (NL, neutral loss). Different lipid classes usually possess fixed NL [33], such as PA (NL, 115.01), PE (NL, 141.02), PG (NL, 189.03), PI (NL, 277.04), etc. As for detected ion message (MS1 and MS2) and matching scores (≥98.5) with the lipid maps library, the unknown lipids were characterized. The recoveries of the 12 internal standards for lipids are listed in Table S1, which are between 78.39% and 106.3% at different added concentrations, exhibiting approving and satisfying results.
In the present study, the two representative matching images and compound structures from QI of DG (12:0/22:3) and Cer (d18:2/20:1) are shown in Figure 1A,B. PCA score plot of three grades of rice was presented in Figure 1C, a clear distinction among three grades could be seen basing on the lipids dataset. There were a total of 92 lipids identified in indica rice, including the 10 lipid classes DG, PA, PC, PE, PG, PI, PS, Cer, SM, and TG (Table S2). In Figure 1D, the % coverage of the pie plot is the percentage of each type of identified lipid number in the total identified lipids quantity of rice. Most identified lipids in rice were TGs and DGs, with proportions of 35% and 19% (Figure 1D), respectively, of total identified lipids.
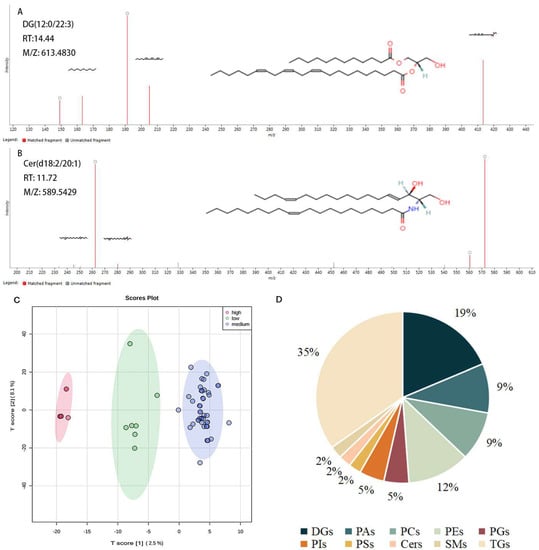
Figure 1.
Lipid determination of rice. (A,B) Representative lipids of DG (12:0/22:3) and Cer (d18:2/20:1) after mapping with QI. (C) PCA plot of three grades of rice, with eating scores high (score ≥ 90), medium (score ≥ 80), and low (score ≥ 70). (D) Distribution of differential lipids of rice.
3.3. Significantly Different Lipids among Three Eating Grades of Indica Rice
After filtering by t-test and ANOVA analysis, the details of potential lipid biomarkers with p-value ≤ 0.05 and VIP ≥ 1 are listed in Table 2, including category, retention time, and contents. In Table 2, forty-two lipids displayed significant difference of three sensory grades, containing eight DGs, four PAs, four PCs, five PEs, two PGs, two PIs, one PS, one Cer, one SM, and fourteen TGs. Lipids such as PC (18:1/18:2), PA (16:0), and PE (22:0) presented the highest contents in the high tasting value group compared to the medium and low ones and showed score dependence among three groups. The content of PC (18:1/18:2) ranged from 22.36 to 8.64 μg/kg, PA (16:0) from 171.41 to 136.59 μg/kg, and PE (22:0) from 100.15 to 44.41 μg/kg at high, medium and low sensory levels, respectively.

Table 2.
Annotations and contents (μg/kg) of significantly different lipids in three sensory grades (high, medium and low) of indica rice.
OPLS-DA was applied in view of lipids components to determine whether the three taste-level groups (high, medium, and low) of rice samples could be differentiated. Compared with PCA, OPLS-DA is a supervised method which obtains better classification results than PCA and results with reduced overfitting compared to PLS-DA. As shown in Figure 2A, an obvious distinction between the three groups could be seen in OPLS-DA. To further test the effectiveness of this OPLS-DA model, a permutation test (200 times) was carried out. Results show that this model was acceptable and valid with R2Y = 0.961 and Q2 = 0.928 (p < 0.005) [34]. In addition, OPLS-DA engaged VIP analysis to gain the most important lipids for the classification of the three groups. In the VIP analysis, the top 15 biomarkers with VIP values are presented in Figure 2B, and the first 3 VIP scores of lipids were 1.98, 1.69, and 1.67 for PE (22:0), PC (16:0/3:0), and PA (20:0), respectively. Glycerophospholipids, sphingolipids, and glycerolipids were highly responsible for the indica rice tasting scores. Since there were limited samples in each group, and sizes of the three groups were not inconsistent, the classification bias was inevitable. Thus, more analysis such as random forest analysis and correlation analysis should be performed to further test the OPLS-DA model. In addition, these differential lipids still need to be tested further with more samples in each group.
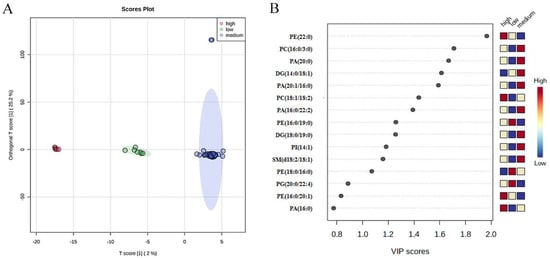
Figure 2.
Significantly different lipids for indica rice. (A) OPLS-DA plot of lipid biomarkers for three grades of rice. (B) VIP scores for significantly different lipid species of OPLS-DA for three groups. Red and blue colors indicate high and low contents, respectively.
Fatty acids are one of the main class of lipids in rice; however, there were no significant differences in the high, medium, and low levels of indica rice in the present study. This result was in accordance with previous research [10]. In general, lipids in rice contains starch lipids and non-starch lipids, playing significant roles in the cooking and eating quality of rice. Lipids are usually bound to shape complexes with amylose and amylopectin, and in turn, influence the viscosity and gel consistency of the rice texture and elasticity. TG is one of the main lipid classes in rice, which is generally located in the rice body of bran and germ fractions as storage lipids in seeds [35]. DG, as the degradation product and also the precursor of TG, was a signal factor in many physiological activities. TG and DG are the main components of non-starch lipids and also present in tiny amounts in the starch lipids in rice [35]. In the present study, glycerophospholipids and glycerolipids gained more weight than other lipids in distinguishing the three taste groups for indica rice (Table 2, Figure 2C), indicating their importance in deciding rice quality.
PLs are fundamental proportions of cell membranes, including mitochondrial and endoplasmic reticulum [36]. In rice, PLs are more abundant in starch lipids than the non-starch of rice bran and germ, with PC, PE and PI serving as the principal PLs [37]. Sphingolipids, playing a vital role in signal transduction processes, are important components of biological biomembranes [38]. Studies have shown that starch lipids in rice have more effect on rice tasting quality than non-starch lipids; however, whether more or less lipid starch increases the quality is still controversial [10]. In addition, to our knowledge, there have been few targeted studies on detailed structures and compositions of these starch-lipids within rice grains, so more information needs to be investigated further.
3.4. Validation of Grade Classification Model for Indica Rice
To assess the effectiveness of these lipid biomarkers for evaluating the grade level of indica rice further, random forest and correlation analysis were applied in this study. Random forest (RF) [39] is a classification method employing in lipidomics technology owing to the diverse rules of OPLS-DA. As presented in Figure 3A, the OOB error of the established model was only 0.255, and six samples were not classified correctly during RF analysis. RF data display the precision of this method for grade classification at 90.20%, indicating the accuracy of this model was greater than 90%.
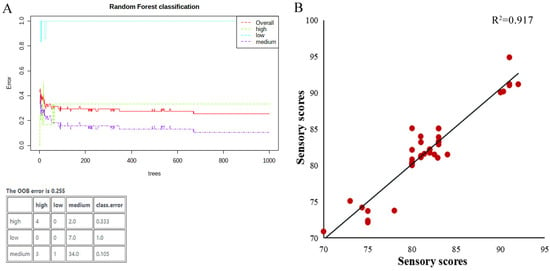
Figure 3.
Grade-predicted model validation of rice. (A) RF results with lipids biomarkers. (B) Correlation study of sensory scores between actual and predicted ones by potential lipid biomarkers.
During correlation analysis, sensory scores of rice were predicted by the lipid biomarkers model, and the scatter plot with practical and predicted sensory scores for indica rice is presented in Figure 3B. The correlation coefficient between the actual and predicted tasting scores was acquired for indica rice as 0.917. These results indicated this lipid-based model possesses high predictive ability and accuracy and could be used as a supplement for the current sensory evaluation of indica rice.
Screening the vital lipid points that related to rice tasting quality is necessary to ensure a desired method through biomarker-based models. Recently, researchers have focused on the impact of lipids on rice eating quality. Concepcion et al. [40] showed a clear distinction of lipid profiles between waxy and non-waxy rice. Researchers also studied the lipid components on rice cooking [41] and storage quality [9]. To our knowledge, there have been no studies concentrating on the lipids model by advanced UPLC-QTOF/MS for screening potential lipid biomarkers in identifying the taste quality of indica rice in China. In the present study, with the high-throughput information of lipid profiles and multivariate statistical analysis, obvious differentiation results (Figure 2 and Figure 3) showed lipids having exact relationships with indica rice eating quality and also presented which lipids (Table 2, Figure 2 and Figure 3) really affect it. However, more work is still required to assess the stability and effectiveness of this group of lipid biomarkers among other cultivars and different statuses of rice.
4. Conclusions
In this study, lipidomics was applied to identify the distinction of lipid composition at different grades (high, medium, and low) for indica rice. In total, 42 lipids displayed significant difference among 3 sensory grades, containing 8 DGs, 4 PAs, 4 PCs, 4 PEs, 2 PGs, 2 PIs, 1 PS, 1 Cer, 1 SM, and 14 TGs. A novel OPLS-DA model with this set of lipids for indica rice gradation was established. The RF result showed the accuracy of this model was greater than 90%. Thus, the developed lipid-based model could serve as a substitute tool for traditional sensory evaluation of indica rice in food and breeding departments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods12050944/s1, Figure S1: The PCA score plots of compound abundances based on QC samples; Table S1: Recovery of different lipids at low, medium and high concentrations; Table S2: All lipids including ten categories identified in indica rice.
Author Contributions
L.Z.: investigation, methodology, conceptualization, supervision, writing—original draft. X.D.: writing—review & editing. H.L.: methodology. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Special Fund of Chinese Central Government for Basic Scientific Research Operations in Commonweal Research Institutes (No. ZX2212).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
This study was supported by the Academy of National Food and Strategic Reserves Administration of the People’s Republic of China.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- MinJee, K.; Jongguk, L.; Sung Won, K.; Giyoung, K.; Moon, K. Geographical Origin Discrimination of White Rice Based on Image Pixel Size Using Hyperspectral Fluorescence Imaging Analysis. Appl. Sci. 2020, 235, 1794–1801. [Google Scholar]
- Lin, L.; Changyun, F.; Zhanqiang, H.; Xianqiao, H.; Zhiwei, Z.B. Grade classification model tandem BpNN method with multi-metal sensor for rice eating quality evaluation. Sens. Actuators B Chem. 2019, 2, 22–27. [Google Scholar]
- Ding, C.; Liu, Q.; Li, P.; Yongsheng, P.; Tingting, T.; Yan, W.; Guofeng, Y.; Xiaolong, S. Distribution and quantitative analysis of phenolic compounds in fractions of Japonica and Indica rice. Food Chem. 2019, 8, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Zhaomiao, L.; Xincheng, Z.; Zunxin, W.; Ganghua, L.; Shaohua, W.; Yanfeng, D. Metabolomic analysis of pathways related to rice grain chalkiness by a notched-belly mutant with high occurrence of white-belly grains. BMC Plant Biol. 2017, 39, 17–22. [Google Scholar]
- Zongkui, C.; Haiyu, C.; Yang, J. Metabolomic analysis reveals metabolites and pathways involved in grain quality traits of high-quality rice cultivars under a dry cultivation system. Food Chem. 2020, 2, 19–27. [Google Scholar]
- Shin, D.; Lee, S.; Kim, T.; Lee, J.; Park, J.; Lee, J.; Lee, J.Y.; Cho, L.; Choi, J.Y.; Lee, W.; et al. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 2020, 11, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Luyao, Z.; Xiaoliang, D.; Hongbin, L.; Dong, Z.; Qian, W.; Jianlei, L.; Hui, S. A panel of lipid markers for rice discrimination of Wuchang Daohuaxiang in China. Food Res. Int. 2022, 13, 111511–111520. [Google Scholar]
- Zhao, W.; Chung, J.; Kwon, S.; Lee, J.; Ma, K.; Park, Y. Association analysis of physicochemical traits on eating quality in rice (Oryza sativa L.). Euphytica 2013, 191, 9–21. [Google Scholar] [CrossRef]
- Yoon, M.; Rico, C.W.; Koh, H.; Kang, M. A study on the lipid components of rice in relation to palatability and storage. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 515–521. [Google Scholar] [CrossRef]
- Yoon, M.; Lee, S.; Kang, M. The lipid composition of rice cultivars with different eating qualities. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 291–295. [Google Scholar] [CrossRef]
- Huang, K.; Liu, H.; Chen, Z.; Xu, H. Role of selenium in cytoprotection against cholesterol oxide-induced vascular damage in rats. Atherosclerosis 2002, 162, 137–144. [Google Scholar] [CrossRef]
- Wang, J.; Uryga, A.K.; Reinhold, J.; Figg, N.; Baker, L.; Finigan, A.; Gray, K.; Kumar, S.; Clarke, M.; Bennett, M. Vascular Smooth Muscle Cell Senescence Promotes Atherosclerosis and Features of Plaque Vulnerability. Circulation 2015, 132, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Long, S.B.; Tao, X.; Campbell, E.B.; MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 2007, 450, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Gad, H.A.; Heiss, A.G.; Wessjohann, L.A. Metabolomics driven analysis of six Nigella species seeds via UPLC-qTOF-MS and GC–MS coupled to chemometrics. Food Chem. 2014, 151, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–738. [Google Scholar] [CrossRef]
- Moore, B.E.; Chen, G.; Nadakuditi, R.R. Panoramic Robust PCA for Foreground–Background Separation on Noisy, Free-Motion Camera Video. IEEE Trans. Comput. Imaging 2019, 8, 76–81. [Google Scholar] [CrossRef]
- Yang, J.; Yang, J.Y. Why can LDA be performed in PCA transformed space? Pattern Recognit. 2003, 36, 563–566. [Google Scholar] [CrossRef]
- Ildiz, G.O.; Karada, A.; Kaygsz, E.; Rui, F. PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra. Molecules 2021, 26, 133–139. [Google Scholar]
- Yang, Z.; Piironen, V.; Lampi, A. Epoxy and hydroxy fatty acids as non-volatile lipid oxidation products in oat. Food Chem. 2019, 295, 82–93. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Cao, P.; Liu, Y. Quantitative determination of epoxy stearic acids derived from oxidized frying oil based on solid-phase extraction and gas chromatography. LWT 2018, 92, 250–257. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, L.; Lu, X.; Zhao, C.; Zhu, Z.; Wang, F.; Zhang, J.; Chen, S.; Zhao, Y.; Xu, G. A simultaneous extraction method for metabolome and lipidome and its application in cry1Ac and sck-transgenic rice leaf treated with insecticide based on LC-MS analysis. Metabolomics 2014, 10, 1197–1209. [Google Scholar] [CrossRef]
- Broadhurst, D.I.; Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006, 2, 171–196. [Google Scholar] [CrossRef]
- Tu, J.; Yin, Y.; Xu, M.; Wang, R.; Zhu, Z. Absolute quantitative lipidomics reveals lipidome-wide alterations in aging brain. Metabolomics 2018, 14, 21–29. [Google Scholar] [CrossRef]
- Gennebäck, N.; Malm, L.; Hellman, U.; Waldenström, A.; Mörner, S. Using OPLS-DA to find new hypotheses in vast amounts of gene expression data-Studying the progression of cardiac hypertrophy in the heart of aorta ligated rat. Gene 2013, 522, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Luyao, Z.; Xiaoliang, D.; Can, H.; Hongbin, L.; Hui, S. Identification of key taste components in two aromatic rice varieties by widely targeted metabolomics in China. Cereal Chem. 2022, 109, 1–11. [Google Scholar]
- He, R.; Luo, H.; Li, Y.; Wang, X.; Zhang, L. Comparison and analysis of different rice planting methods in China. Trans. Chin. Soc. Agric. Eng. 2008, 24, 167–171. [Google Scholar]
- Han, Y.; Hou, M.; Wei, L.; Hao, L.; Tian, C.; Miao, S. Occurrence and management of rice insect pests in northern rice-planting regions: A review. Plant Prot. 2008, 245, 164–166. [Google Scholar]
- Anandan, A.; Sabesan, T.; Eswaran, R.; Rajiv, G.; Muthalagan, N.; Suresh, R. Appraisal of environmental interaction on quality traits of rice by additive main effects and multiplicative interaction analysis. Cereal Res. Commun. 2009, 37, 131–140. [Google Scholar] [CrossRef]
- Masashi, O.; Toshichika, I.; Yousay, H.; Masayuki, Y. A climatological analysis on the recent declining trend of rice quality in Japan. J. Agric. Meteorol. 2010, 65, 327–337. [Google Scholar]
- Bijlsma, S.; Bobeldijk, I.; Verheij, E.R.; Ramaker, R.; Kochhar, S.; Macdonald, I.A.; van Ommen, B.; Smilde, A.K. Large-Scale Human Metabolomics Studies: A Strategy for Data (Pre-) Processing and Validation. Anal. Chem. 2006, 78, 567–574. [Google Scholar] [CrossRef]
- Hu, A.; Wei, F.; Huang, F.; Xie, Y.; Wu, B.; Lv, X.; Chen, H. Comprehensive and High-Coverage Lipidomic Analysis of Oilseeds Based on Ultrahigh-Performance Liquid Chromatography Coupled with Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry. J. Agric. Food Chem. 2021, 69, 8964–8980. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, K.; Yang, J.; Cheng, H.; Gross, R.W. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J. Lipid Res. 2006, 47, 864–879. [Google Scholar] [CrossRef] [PubMed]
- Schwudke, D.; Oegema, J.; Burton, L.; Entchev, E.; Hannich, J.T.; Ejsing, C.S.; Kurzchalia, T.; Shevchenko, A. Lipid Profiling by Multiple Precursor and Neutral Loss Scanning Driven by the Data-Dependent Acquisition. Anal. Chem. 2006, 78, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Bazargani, H.S.; Bangdiwala, S.I.; Mohammad, K.; Maghsoudi, H.; Mohammadi, R. Compared application of the new OPLS-DA statistical model versus partial least squares regression to manage large numbers of variables in an injury case-control study. Sci. Res. Essays 2010, 6, 17–25. [Google Scholar]
- Yoshida, H.; Tanigawa, T.; Yoshida, N.; Kuriyama, I.; Tomiyama, Y.; Mizushina, Y. Lipid components, fatty acid distributions of triacylglycerols and phospholipids in rice brans. Food Chem. 2011, 129, 479–484. [Google Scholar] [CrossRef]
- Criado-Navarro, I.; Mena-Bravo, A.; Calderón-Santiago, M.; Priego-Capote, F. Determination of glycerophospholipids in vegetable edible oils: Proof of concept to discriminate olive oil categories. Food Chem. 2019, 299, 125–136. [Google Scholar] [CrossRef]
- Haisong, W.; Yiming, W.; Naifu, W.; Liping, Y.; Yibin, Y. Effect of water content of high-amylose corn starch and glutinous rice starch combined with lipids on formation of starch-lipid complexes during deep-fat frying. Food Chem. 2019, 12, 10–18. [Google Scholar]
- Huwiler, A.; Kolter, T.; Pfeilschifter, J.; Konrad, S. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim. Biophys. Acta 2000, 12, 63–99. [Google Scholar] [CrossRef]
- Bart, H.; Stefan, B.; Frans, V.R.; Pieter, D.B. A flexible integrative approach based on random forest improves prediction of transcription factor binding sites. Nucleic Acids Res. 2012, 40, 106–112. [Google Scholar]
- Concepcion, C.; Calingacion, M.; Garson, M.; Fitzgerald, M. Lipidomics reveals associations between rice quality traits. Metabolomics 2020, 16, 123–131. [Google Scholar] [CrossRef]
- Gu, D.; Liu, Z.; Liu, Y.; Wang, S.; Wang, Q.; Li, G.; Ding, Y. Effect of Lipid Content and Components on Cooking Quality and Their Responses to Nitrogen in Milled Japonica Rice. Acta Agron. Sin. 2011, 37, 2001–2010. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).