Hyperspectral Imaging Combined with Chemometrics Analysis for Monitoring the Textural Properties of Modified Casing Sausages with Differentiated Additions of Orange Extracts
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Image Acquisition and Processing
2.3. Textural Profile Analysis (TPA)
2.4. Selection of Important Wavelengths
2.5. Model Development and Evaluation
2.6. Statistical Analysis
3. Results and Discussion
3.1. Effects of SL, SO, RT, OE Addition, and Salt with LA on Textural Properties of Sausage Core
3.2. Spectra Overview
3.3. Calibration Model Using Full Wavelengths
3.4. Calibration Model Using Important Wavelengths
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamrruzzaman, M.; Kalita, D.; Ahmed, M.T.; ElMasry, G.; Makino, Y. Effect of variable selection algorithms on model performance for predicting moisture content in biological materials using spectral data. Anal. Chim. Acta 2022, 1202, 339390. [Google Scholar] [CrossRef]
- Siripatrawan, U. Hyperspectral imaging for rapid evaluation and visualization of quality deterioration index of vacuum packaged dry-cured sausages. Sens. Actuators B Chem. 2018, 254, 1025–1032. [Google Scholar] [CrossRef]
- Feng, C.H.; Makino, Y.; Yoshimura, M.; Thuyet, D.C.; García-Martín, J.F. Hyperspectral imaging in tandem with R statistics and image processing for detection and visualisation of pH in Japanese big sausages under different storage conditions. J. Food Sci. 2018, 83, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Parsimonious model development for real-time monitoring of moisture in red meat using hyperspectral imaging. Food Chem. 2016, 196, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.H.; Makino, Y.; Yoshimura, M.; Rodríguez-Pulido, F.J. Real-time prediction of pre-cooked Japanese sausages color with different storage days using hyperspectral imaging. J. Sci. Food Agric. 2018, 98, 2564–2572. [Google Scholar] [CrossRef]
- Cheng, J.H.; Sun, J.; Yao, K.S.; Xu, M.; Tian, Y.; Dai, C.X. A decision fusion method based on hyperspectral imaging and electronic nose techniques for moisture content prediction in frozen-thawed pork. LWT-Food Sci. Technol. 2022, 165, 113778. [Google Scholar] [CrossRef]
- Zhuang, Q.B.; Peng, Y.K.; Yang, D.Y.; Wang, Y.L.; Zhao, R.H.; Chao, K.L.; Guo, Q.H. Detection of frozen pork freshness by fluorescence hyperspectral image. J. Food Eng. 2022, 316, 110840. [Google Scholar] [CrossRef]
- Cheng, J.H.; Sun, J.; Yao, K.S.; Xu, M.; Zhou, X. Nondestructive detection and visualization of protein oxidation degree of frozen-thawed pork using fluorescence hyperspectral imaging. Meat Sci. 2022, 194, 108975. [Google Scholar] [CrossRef]
- Wan, G.-L.; Fan, S.-X.; Liu, G.S.; He, J.G.; Wang, W.; Li, Y.; Cheng, L.J.; Ma, C.; Guo, M. Fusion of spectra and texture data of hyperspectral imaging for prediction of myoglobin content in nitrite-cured mutton. Food Control 2023, 144, 109332. [Google Scholar] [CrossRef]
- Zhang, J.J.; Ma, Y.H.; Liu, G.S.; Fan, N.Y.; Li, Y.; Sun, Y.R. Rapid evaluation of texture parameters of Tan mutton using hyperspectral imaging with optimization algorithms. Food Control 2022, 135, 108815. [Google Scholar] [CrossRef]
- Zhang, J.J.; Liu, G.S.; Li, Y.; Guo, M.; Pu, F.N.; Wang, H. Rapid identification of lamb freshness grades using visible and near-infrared spectroscopy (Vis-NIR). J. Food Compos. Anal. 2022, 111, 104590. [Google Scholar] [CrossRef]
- León-Ecay, S.; López-Maestresalas, A.; Murillo-Arbizu, M.T.; Beriain, M.J.; Mendizabal, J.A.; Arazuri, S.; Carmen, J.; Bass, P.D.; Colle, M.J.; García, D.; et al. Classification of beef longissimus thoracis muscle tenderness using hyperspectral imaging and chemometrics. Foods 2022, 11, 3105. [Google Scholar] [CrossRef]
- Song, K.; Wang, S.-H.; Yang, D.; Shi, T.-Y. Combination of spectral and image information from hyperspectral imaging for the prediction and visualization of the total volatile basic nitrogen content in cooked beef. J. Food Meas. Charact. 2021, 15, 4006–4020. [Google Scholar] [CrossRef]
- Xie, A.G.; Sun, J.; Wang, T.M.; Liu, Y.H. Visualized detection of quality change of cooked beef with condiments by hyperspectral imaging technique. Food Sci. Biotechnol. 2022, 31, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Reed, D.D.; Young, J.M.; Eshkabilov, S.; Berg, E.P.; Sun, X. Beef quality grade classification based on intramuscular fat content using hyperspectral imaging technology. Appl. Sci. 2021, 11, 4588. [Google Scholar] [CrossRef]
- Qiu, R.C.; Zhao, Y.L.; Kong, D.D.; Wu, N.; He, Y. Development and comparison of classification models on VIS-NIR hyperspectral imaging spectra for qualitative detection of the Staphylococcus aureus in fresh chicken breast. Spectrochim. Acta A Mol. 2023, 285, 121838. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.D.; Li, P.; Ma, J.C.; Tian, X.G.; Zhong, N. Identification of four chicken breeds by hyperspectral imaging combined with chemometrics. Processes 2022, 10, 1484. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, W.; Zhuang, H.; Yoon, S.-C.; Jiang, H.Z. Fusion of spectra and texture data of hyperspectral imaging for the prediction of the water-holding capacity of fresh chicken breast filets. Appl. Sci. 2018, 8, 640. [Google Scholar] [CrossRef]
- Cernadas, E.; Fernández-Delgado, M.; Fulladosa, E.; Munoz, I. Automatic marbling prediction of sliced dry-cured ham using image segmentation, texture analysis and regression. Expert Syst. Appl. 2022, 206, 117765. [Google Scholar] [CrossRef]
- ElMasry, G.; Gou, P.; Al-Rejaie, S. Effectiveness of specularity removal from hyperspectral images on the quality of spectral signatures of food products. J. Food Eng. 2021, 289, 110148. [Google Scholar] [CrossRef]
- Feng, C.H.; Makino, Y.; García-Martín, J.F. Hyperspectral imaging coupled with multivariate analysis and image processing for detection and visualisation of colour in cooked sausages stuffed with different modified casings. Foods 2020, 9, 1089. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.H.; Makino, Y.; Yoshimura, M.; Rodríguez-Pulido, F.J. Estimation of adenosine triphosphate content in ready-to-eat sausages with different storage days, using hyperspectral imaging coupled with R statistics. Food Chem. 2018, 264, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.H.; Arai, H.; Rodríguez-Pulido, F.J. Evaluation of pH in sausages stuffed in a modified casing with orange extracts by hyperspectral imaging coupled with response surface methodology. Foods 2022, 11, 2797. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, Y. Rapid and nondestructive classification of Cantonese sausage degree using hyperspectral images. Appl. Sci. 2019, 9, 822. [Google Scholar] [CrossRef]
- Siripatrawan, U.; Makino, Y. Simultaneous assessment of various quality attributes and shelf life of packaged bratwurst using hyperspectral imaging. Meat Sci. 2018, 146, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.H.; Drummond, L.; Zhang, Z.H. Evaluation of innovative immersion vacuum cooling with different pressure reduction rates and agitation for cooked sausages stuffed in natural or artificial casing. LWT-Food Sci. Technol. 2014, 59, 77–85. [Google Scholar] [CrossRef]
- Feng, C.H.; Drummond, L.; Sun, D.W.; Zhang, Z.-H. Evaluation of natural hog casings modified by surfactant solutions combined with lactic acid by response surface methodology. LWT-Food Sci. Technol. 2014, 58, 427–438. [Google Scholar] [CrossRef]
- Sammani, M.S.; Cerdὰ, V. Sample pre-treatment and flavonoids analytical methodologies for the quality control of foods and pharmaceutical matrices. In The Book of Flavonoids, 1st ed.; Feng, C.-H., García Martín, J.F., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2021; Chapter 1; pp. 1–130. [Google Scholar]
- Feng, C.-H.; Otani, C.; Ogawa, Y. Innovatively identifying naringin and hesperidin by using terahertz spectroscopy and evaluating flavonoids extracts from waste orange peels by coupling with multivariate analysis. Food Control 2022, 137, 108897. [Google Scholar] [CrossRef]
- Bellavite, P.; Donzelli, A. Hesperidin and SARS-CoV-2: New light on the healthy function of citrus fruits. Antioxidants 2020, 9, 742. [Google Scholar] [CrossRef]
- Feng, C.H.; Otani, C.; García-Martín, J.F. Flavonoids as a starting point for therapeutics against COVID-19: Current state-of-the art research advances. In The Book of Flavonoids, 1st ed.; Feng, C.-H., García Martín, J.F., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2021; Chapter 12; pp. 340–350. [Google Scholar]
- Guemes-Vera, N.; Nicanor, A.B.; Gonzalez, L.; Totosaus-Sanchez, A. Effect of the addition of orange peel flour on the physicochemical characteristics of texture profile analysis and sensory in bakery products and sausages. In Citric Acid: Synthesis, Properties and Applications; Vargas, D.A., Medina, J.V., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2012; Chapter 9; pp. 1–10. [Google Scholar]
- Díaz-Vela, J.; Totosaus, A.; Pérez-Chabela, M.L. Integration of agroindustrial co-products as functional food ingredients: Cactus pear (opuntia ficus indica) flour and pineapple (ananas comosus) peel flour as fiber source in cooked sausages inoculated with lactic acid bacteria. J. Food Process. Preserv. 2015, 39, 2630–2638. [Google Scholar] [CrossRef]
- Shin, S.-H.; Choi, W.-S. Variation in significant difference of sausage textural parameters measured by texture profile analysis (TPA) under changing measurement conditions. Food Sci. Anim. Resour. 2021, 41, 739–747. [Google Scholar] [CrossRef]
- Herrero, A.M.; Hoz, L.D.L.; Ordónez, J.A.; Herranz, B.; Romero de Ávila, M.D.; Cambero, M.I. Tensile properties of cooked meat sausages and their correlation with texture profile analysis (TPA) parameters and physico-chemical characteristics. Meat Sci. 2008, 80, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, M.D.D.; Derks, J.A.M.; Ketel, E.C.; Wijk, R.A.D.; Stieger, M. Eating behaviour explains differences between individuals in dynamic texture perception of sausages. Food Qual. Prefer. 2015, 41, 189–200. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Hyperspectral imaging for real-time monitoring of water holding capacity in red meat. LWT-Food Sci. Technol. 2016, 66, 685–691. [Google Scholar] [CrossRef]
- Sanz, J.A.; Fernandes, A.M.; Barrenechea, E.; Silva, S.; Santos, V.; Gonçalves, N.; Paternain, D.; Jurio, A.; Melo-Pinto, P. Lamb muscle discrimination using hyperspectral imaging: Comparison of various machine learning algorithms. J. Food Eng. 2016, 174, 92–100. [Google Scholar] [CrossRef]
- ElMasry, G.; Kamruzzaman, M.; Sun, D.-W.; Allen, P. Principles and applications of hyperspectral imaging in quality evaluation of agro-food products: A review. Crit. Rev. Food Sci. Nutr. 2012, 52, 999–1023. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.B.; Yoon, S.-C.; Zhuang, H.; Wang, W.; Li, C.Y. Prediction of pH of fresh chicken breast fillets by VNIR hyperspectral imaging. J. Food Eng. 2017, 8, 57–65. [Google Scholar] [CrossRef]
- Helland, I.S.; Næs, T.; Isaksson, T. Related versions of the multiplicative scatter correction method for preprocessing spectroscopic data. Chemom. Intell. Lab. Syst. 1995, 29, 233–241. [Google Scholar] [CrossRef]
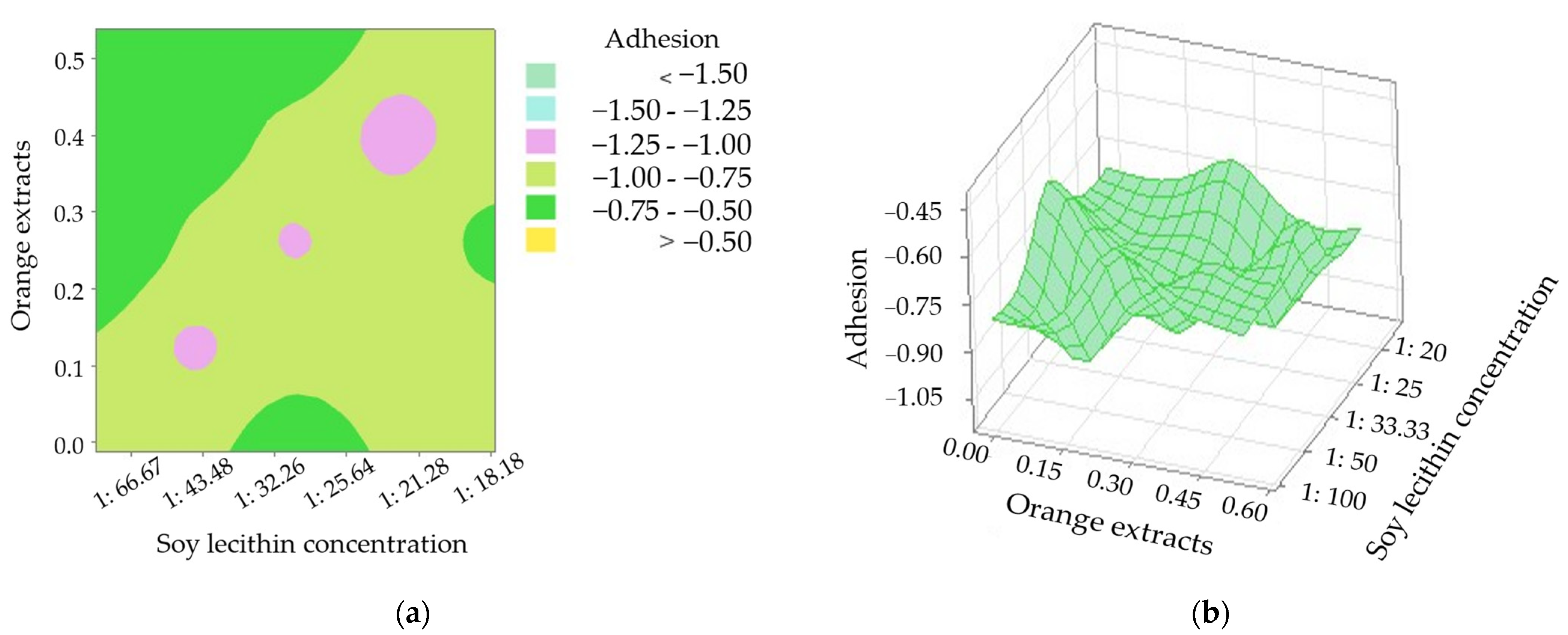
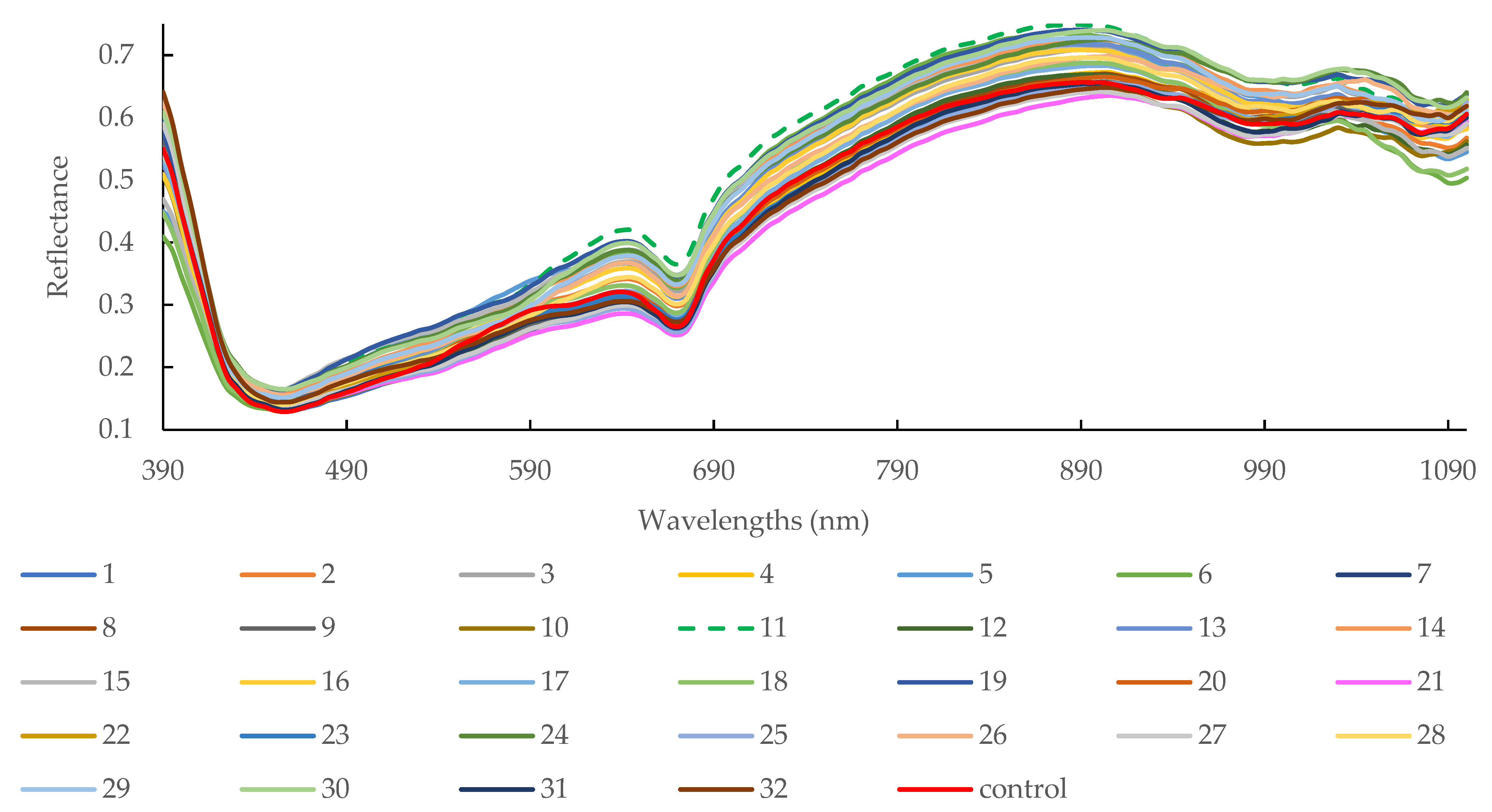
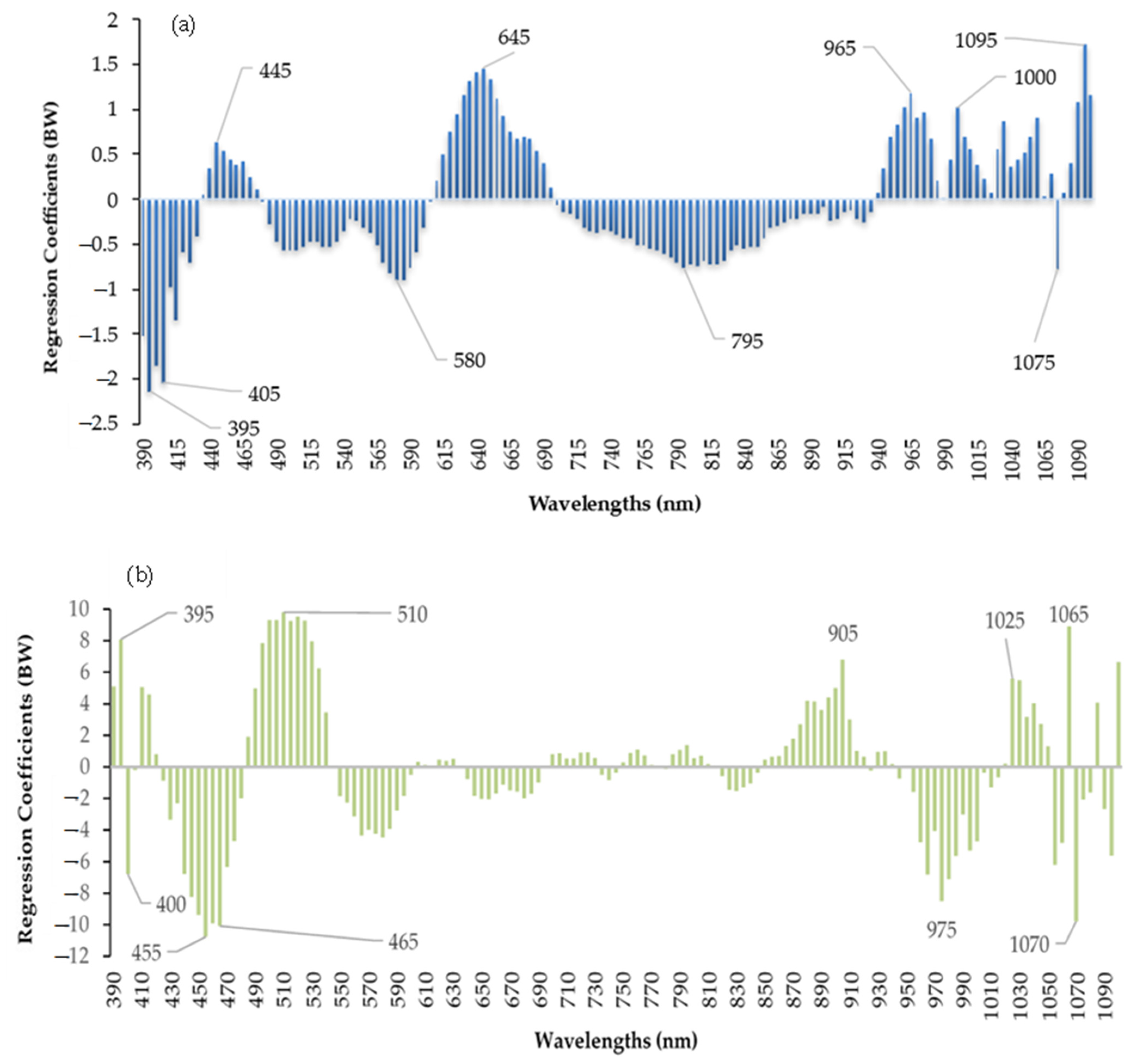
| Point | Symbol | Coded (Xi) Variable Level | ||||
|---|---|---|---|---|---|---|
| Star | Low | Center | High | Star | ||
| −2 | −1 | 0 | 1 | 2 | ||
| SL concentration 1 | X1 | 1:90 | 1:45 | 1:30 | 1:22.5 | 1:18 |
| SO concentration (%, w/w) | X2 | 0.625 | 1.25 | 1.875 | 2.5 | 3.125 |
| RT (min) | X3 | 45 | 60 | 75 | 90 | 105 |
| OE (%, w/w) | X4 | 0 | 0.12 | 0.26 | 0.4 | 0.54 |
| LA (mL/kg NaCl) | X5 | 16.5 | 18 | 19.5 | 21 | 22.5 |
| Samples | Surfactant Solution with OE | Salt with LA | ||||
|---|---|---|---|---|---|---|
| SL Concentration (X1, w/w) b | SO Concentration (X2, %, w/w) | RT (X3, min) | OE (X4, %, w/w) | LA (mL/kg NaCl, X5) | RT (X3, min) | |
| 1 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 2 | 1:30 | 1.875 | 75 | 0.26 | 22.50 | 75 |
| 3 | 1:22.5 | 1.250 | 90 | 0.40 | 18.00 | 90 |
| 4 | 1:30 | 1.875 | 75 | 0.26 | 16.50 | 75 |
| 5 | 1:45 | 2.500 | 90 | 0.12 | 21.00 | 90 |
| 6 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 7 | 1:45 | 2.500 | 60 | 0.40 | 21.00 | 60 |
| 8 | 1:90 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 9 | 1:30 | 1.875 | 105 | 0.26 | 19.50 | 105 |
| 10 | 1:30 | 1.875 | 45 | 0.26 | 19.50 | 45 |
| 11 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 12 | 1:45 | 1.250 | 90 | 0.40 | 21.00 | 90 |
| 13 | 1:22.5 | 2.500 | 60 | 0.12 | 21.00 | 60 |
| 14 | 1:22.5 | 2.500 | 60 | 0.40 | 18.00 | 60 |
| 15 | 1:22.5 | 1.250 | 60 | 0.40 | 21.00 | 60 |
| 16 | 1:45 | 1.250 | 90 | 0.12 | 18.00 | 90 |
| 17 | 1:30 | 3.125 | 75 | 0.26 | 19.50 | 75 |
| 18 | 1:22.5 | 1.250 | 90 | 0.12 | 21.00 | 90 |
| 19 | 1:45 | 1.250 | 60 | 0.12 | 21.00 | 60 |
| 20 | 1:22.5 | 2.500 | 90 | 0.40 | 21.00 | 90 |
| 21 | 1:45 | 1.250 | 60 | 0.40 | 18.00 | 60 |
| 22 | 1:22.5 | 1.250 | 60 | 0.12 | 18.00 | 60 |
| 23 | 1:30 | 1.875 | 75 | 0.53 | 19.50 | 75 |
| 24 | 1:18 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 25 | 1:45 | 2.500 | 90 | 0.40 | 18.00 | 90 |
| 26 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 27 | 1:22.5 | 2.500 | 90 | 0.12 | 18.00 | 90 |
| 28 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 29 a | 1:30 | 1.875 | 75 | 0.26 | 19.50 | 75 |
| 30 | 1:45 | 2.500 | 60 | 0.12 | 18.00 | 60 |
| 31 | 1:30 | 1.875 | 75 | 0.00 | 19.50 | 75 |
| 32 | 1:30 | 0.625 | 75 | 0.26 | 19.50 | 75 |
| Analysis of Variance | Df | Response Variables | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Hardness (Yh) | Springiness (Ys) | Gumminess (Yg) | Adhesion (Ya) | ||||||
| Source | Adj SS | F-Value | Adj SS | F-Value | Adj SS | F-Value | Adj SS | F-Value | |
| Model | 20 | 2195.12 | 0.87 | 0.52324 | 0.44 | 1.38276 | 1.29 | 2.19927 | 1.90 |
| Linear | 5 | 261.61 | 0.42 | 0.0711 | 0.24 | 0.17593 | 0.65 | 0.3688 | 1.28 |
| Soy lecithin (X1) | 1 | 10.34 | 0.08 | 0.00279 | 0.05 | 0.00831 | 0.15 | 0.10545 | 1.82 |
| Soy oil (X2) | 1 | 51.58 | 0.41 | 0.02982 | 0.50 | 0.00073 | 0.01 | 0.22761 | 3.94 |
| Residence time (X3) | 1 | 90.29 | 0.72 | 0.00156 | 0.03 | 0.01124 | 0.21 | 0.00472 | 0.08 |
| Addition of orange extracts (X4) | 1 | 54.83 | 0.44 | 0.03693 | 0.62 | 0.03286 | 0.61 | 0.0276 | 0.48 |
| Lactic acid (X5) | 1 | 54.57 | 0.43 | 0.00001 | 0.00 | 0.12279 | 2.28 | 0.00342 | 0.06 |
| Square | 5 | 759.30 | 1.21 | 0.33969 | 1.14 | 0.56663 | 2.11 | 0.74679 | 2.58 |
| 1 | 92.41 | 0.74 | 0.03333 | 0.56 | 0.16329 | 3.04 | 0.21651 | 3.74 | |
| 1 | 523.37 | 4.17 | 0.07666 | 1.28 | 0.1684 | 3.13 | 0.08971 | 1.55 | |
| 1 | 175.70 | 1.40 | 0.00137 | 0.02 | 0.01316 | 0.24 | 0.00012 | 0.00 | |
| 1 | 113.35 | 0.90 | 0.14656 | 2.45 | 0.12764 | 2.37 | 0.22977 | 3.97 | |
| 1 | 3.66 | 0.03 | 0.08727 | 1.46 | 0.08375 | 1.56 | 0.18835 | 3.26 | |
| 2-Way Interaction | 10 | 1174.21 | 0.94 | 0.11245 | 0.19 | 0.6402 | 1.19 | 1.08368 | 1.87 |
| X1 × X2 | 1 | 50.52 | 0.40 | 0.00671 | 0.11 | 0.18471 | 3.44 | 0.00277 | 0.05 |
| X1 × X3 | 1 | 81.41 | 0.65 | 0.00776 | 0.13 | 0.00134 | 0.02 | 0.24642 | 4.26 |
| X1 × X4 | 1 | 550.21 | 4.39 | 0.00053 | 0.01 | 0.12008 | 2.23 | 0.34024 | 5.88 * |
| X1 × X5 | 1 | 36.25 | 0.29 | 0.03108 | 0.52 | 0.00096 | 0.02 | 0.00082 | 0.01 |
| X2 × X3 | 1 | 56.47 | 0.45 | 0.04032 | 0.67 | 0.12035 | 2.24 | 0.06547 | 1.13 |
| X2 × X4 | 1 | 89.04 | 0.71 | 0.01603 | 0.27 | 0.00143 | 0.03 | 0.00024 | 0.00 |
| X2 × X5 | 1 | 116.42 | 0.93 | 0.00007 | 0.00 | 0.01488 | 0.28 | 0.23318 | 4.03 |
| X3 × X4 | 1 | 0.41 | 0.00 | 0.00459 | 0.08 | 0.09796 | 1.82 | 0.088 | 1.52 |
| X3 × X5 | 1 | 0.00 | 0.00 | 0.00083 | 0.01 | 0.00091 | 0.02 | 0.03097 | 0.54 |
| X4 × X5 | 1 | 193.48 | 1.54 | 0.00453 | 0.08 | 0.09758 | 1.82 | 0.07558 | 1.31 |
| Error | 11 | 1380.02 | 0.65748 | 0.59121 | 0.63611 | ||||
| Lack of Fit | 6 | 1058.46 | 2.74 | 0.31435 | 0.76 | 0.22919 | 0.53 | 0.34281 | 0.97 |
| Pure Error | 5 | 321.56 | 0.34313 | 0.36202 | 0.2933 | ||||
| R2 (%) | 61.40 | 44.32 | 70.05 | 77.57 | |||||
| Sample | Hardness (N) | Springiness (mm) | Gumminess (N) | Adhesion (N s) |
|---|---|---|---|---|
| 1 | 65.25 ± 18.75 ab | 0.51 ± 0.02 a | 0.97 ± 0.05 a | −1.14 ± 0.01 abcdefg |
| 2 | 66.47 ± 7.42 ab | 0.53 ± 0.05 a | 0.54 ± 0.00 a | −1.19 ± 0.17 abcdefg |
| 3 | 47.31 ± 10.05 ab | 0.96 ± 0.76 a | 0.59 ± 0.59 a | −0.87 ± 0.32 abcdefg |
| 4 | 58.06 ± 1.67 ab | 0.48 ± 0.07 a | 0.98 ± 0.04 a | −1.36 ± 0.06 bcdefg |
| 5 | 44.00 ± 12.58 b | 0.48 ± 0.03 a | 0.54 ± 0.05 a | −1.51 ± 0.03 fg |
| 6 | 43.59 ± 11.75 b | 1.19 ± 0.44 a | 0.20 ± 0.04 a | −0.79 ± 0.08 abcdefg |
| 7 | 66.84 ± 4.37 ab | 0.51 ± 0.07 a | 0.68 ± 0.37 a | −0.93 ± 0.16 abcdefg |
| 8 | 79.66 ± 1.13 ab | 1.06 ± 0.62 a | 0.19 ± 0.04 a | −0.51 ± 0.07 abcdefg |
| 9 | 78.96 ± 5.79 ab | 0.51 ± 0.06 a | 0.92 ± 0.06 a | −1.18 ± 0.11 abcd |
| 10 | 62.32 ± 8.90 ab | 0.98 ± 0.20 a | 0.34 ± 0.17 a | −0.75 ± 0.47 abcdefg |
| 11 | 45.67 ± 5.69 b | 0.70 ± 0.38 a | 0.57 ± 0.26 a | −0.94 ± 0.04 abcdefg |
| 12 | 68.94 ± 14.13 ab | 0.76 ± 0.27 a | 0.17 ± 0.04 a | −0.44 ± 0.02 a |
| 13 | 60.46 ± 4.33 ab | 0.44 ± 0.03 a | 0.31 ± 0.13 a | −1.36 ± 0.57 defg |
| 14 | 54.05 ± 6.19 ab | 0.52 ± 0.02 a | 0.55 ± 0.04 a | −1.50 ± 0.26 g |
| 15 | 62.99 ± 2.94 ab | 0.50 ± 0.02 a | 0.68 ± 0.37 a | −0.97 ± 0.11 abcdefg |
| 16 | 57.13 ± 11.36 ab | 0.49 ± 0.06 a | 0.76 ± 0.03 a | −1.04 ± 0.14 abcdefg |
| 17 | 71.07 ± 12.90 ab | 0.51 ± 0.22 a | 0.17 ± 0.04 a | −0.80 ± 0.31 abcdefg |
| 18 | 74.55 ± 5.38 ab | 0.66 ± 0.25 a | 0.47 ± 0.37 a | −0.74 ± 0.10 abcdefg |
| 19 | 61.27 ± 13.01 ab | 0.44 ± 0.02 a | 0.15 ± 0.08 a | −0.65 ± 0.20 abcd |
| 20 | 70.65 ± 14.63 ab | 0.50 ± 0.00 a | 0.25 ± 0.02 a | −0.99 ± 0.17 abcdefg |
| 21 | 64.11 ± 10.55 ab | 0.48 ± 0.03 a | 0.23 ± 0.02 a | −0.65 ± 0.19 abcdefg |
| 22 | 67.95 ± 7.68 ab | 0.47 ± 0.02 a | 0.54 ± 0.33 a | −0.90 ± 0.20 abcdefg |
| 23 | 77.12 ± 11.28 ab | 0.48 ± 0.02 a | 0.22 ± 0.04 a | −0.58 ± 0.04 abcd |
| 24 | 56.24 ± 21.73 ab | 0.66 ± 0.07 a | 0.30 ± 0.08 a | −0.71 ± 0.30 abcdefg |
| 25 | 66.46 ± 7.29 ab | 0.51 ± 0.02 a | 0.19 ± 0.02 a | −0.56 ± 0.06 ab |
| 26 | 52.61 ± 4.10 ab | 0.49 ± 0.10 a | 0.40 ± 0.04 a | −1.23 ± 0.15 abcdefg |
| 27 | 77.62 ± 1.73 ab | 0.57 ± 0.04 a | 0.38 ± 0.12 a | −0.55 ± 0.03 abcdefg |
| 28 | 58.14 ± 10.65 ab | 0.54 ± 0.10 a | 0.75 ± 0.30 a | −1.38 ± 0.01 abcd |
| 29 | 51.25 ± 1.80 ab | 0.67 ± 0.26 a | 0.67 ± 0.05 a | −0.80 ± 0.30 cdefg |
| 30 | 55.71 ± 8.72 ab | 0.42 ± 0.01 a | 0.84 ± 0.11 a | −0.92 ± 0.31 abcdefg |
| 31 | 60.31 ± 3.82 ab | 0.39 ± 0.03 a | 0.34 ± 0.39 a | −0.61 ± 0.13 abcd |
| 32 | 84.43 ± 1.01 a | 0.52 ± 0.06 a | 0.31 ± 0.12 a | −0.66 ± 0.25 abcdefg |
| Control | 64.47 ± 2.57 ab | 0.46 ± 0.02 a | 0.66 ± 0.60 a | −1.52 ± 0.38 efg |
| Parameters | Raw | Normalization | 1st Derivative | 2nd Derivative | SNV | MSC | ||
|---|---|---|---|---|---|---|---|---|
| Hardness (N) | Calibration group | Rc2 | 0.6840 | 0.4760 | 0.4183 | 0.5772 | 0.4557 | 0.4552 |
| RMSEC (%) | 7.0092 | 9.0264 | 9.5102 | 8.1079 | 9.2000 | 9.2039 | ||
| Validation group | Rv2 | Na | Na | Na | Na | Na | Na | |
| RMSEV (%) | 9.5719 | 6.8142 | 5.6889 | 6.4465 | 6.1000 | 6.0803 | ||
| Springiness | Calibration group | Rc2 | 0.2083 | 0.9820 | 0.1485 | 0.2955 | 0.2653 | 0.2609 |
| RMSEC (%) | 0.1782 | 0.2685 | 0.1848 | 0.1681 | 0.1716 | 0.1721 | ||
| Validation group | Rv2 | 0.1785 | 0.2859 | 0.1267 | Na | 0.2014 | 0.1917 | |
| RMSEV (%) | 0.1615 | 0.1506 | 0.1666 | 0.2060 | 0.1593 | 0.1602 | ||
| Gumminess (N) | Calibration group | Rc2 | 0.4128 | 0.4005 | 0.9994 | 0.3245 | 0.6210 | 0.5919 |
| RMSEC (%) | 0.1679 | 0.1697 | 0.0052 | 0.1801 | 0.1349 | 0.1400 | ||
| Validation group | Rv2 | 0.3908 | 0.3462 | 0.1397 | Na | 0.5042 | 0.5091 | |
| RMSEV (%) | 0.2359 | 0.2444 | 0.2803 | 0.3360 | 0.2128 | 0.2117 | ||
| Adhesion (N s) | Calibration group | Rv2 | 0.8591 | 0.8494 | 0.9999 | 0.4825 | 0.8744 | 0.8451 |
| RMSEC (%) | 0.1189 | 0.1229 | 0.0030 | 0.2279 | 0.1123 | 0.1247 | ||
| Validation group | Rv2 | 0.5556 | 0.5742 | 0.3370 | 0.1706 | 0.6837 | 0.5949 | |
| RMSEV (%) | 0.2249 | 0.1964 | 0.2451 | 0.2742 | 0.1693 | 0.1916 |
| Parameters | Raw | Normalization | 1st Derivative | 2nd Derivative | SNV | MSC | ||
|---|---|---|---|---|---|---|---|---|
| Gumminess (N) | Calibration group | Rc2 | 0.5301 | 0.4807 | 0.5063 | 0.5450 | 0.4984 | 0.4892 |
| RMSEC (%) | 0.1502 | 0.1579 | 0.1540 | 0.1478 | 0.1552 | 0.1557 | ||
| Validation group | RV2 | 0.3620 | 0.4228 | 0.3862 | 0.3731 | 0.4354 | 0.4417 | |
| RMSEV (%) | 0.2414 | 0.2296 | 0.2368 | 0.2393 | 0.2271 | 0.2258 | ||
| Adhesion (N s) | Calibration group | Rc2 | 0.7746 | 0.7530 | 0.6877 | 0.7042 | 0.7311 | 0.6665 |
| RMSEC (%) | 0.1504 | 0.1574 | 0.1770 | 0.1723 | 0.1643 | 0.1829 | ||
| Validation group | RV2 | 0.4427 | 0.4748 | 0.5440 | 0.3765 | 0.5248 | 0.4877 | |
| RMSEV (%) | 0.2247 | 0.2182 | 0.2033 | 0.2377 | 0.2075 | 0.2155 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, C.-H.; Arai, H.; Rodríguez-Pulido, F.J. Hyperspectral Imaging Combined with Chemometrics Analysis for Monitoring the Textural Properties of Modified Casing Sausages with Differentiated Additions of Orange Extracts. Foods 2023, 12, 1069. https://doi.org/10.3390/foods12051069
Feng C-H, Arai H, Rodríguez-Pulido FJ. Hyperspectral Imaging Combined with Chemometrics Analysis for Monitoring the Textural Properties of Modified Casing Sausages with Differentiated Additions of Orange Extracts. Foods. 2023; 12(5):1069. https://doi.org/10.3390/foods12051069
Chicago/Turabian StyleFeng, Chao-Hui, Hirofumi Arai, and Francisco J. Rodríguez-Pulido. 2023. "Hyperspectral Imaging Combined with Chemometrics Analysis for Monitoring the Textural Properties of Modified Casing Sausages with Differentiated Additions of Orange Extracts" Foods 12, no. 5: 1069. https://doi.org/10.3390/foods12051069
APA StyleFeng, C.-H., Arai, H., & Rodríguez-Pulido, F. J. (2023). Hyperspectral Imaging Combined with Chemometrics Analysis for Monitoring the Textural Properties of Modified Casing Sausages with Differentiated Additions of Orange Extracts. Foods, 12(5), 1069. https://doi.org/10.3390/foods12051069







