CSGNN: Contamination Warning and Control of Food Quality via Contrastive Self-Supervised Learning-Based Graph Neural Network
Abstract
1. Introduction
- 1.
- They rely on supervised learning [13]. Still, manual labeling of detection data labels will significantly increase the time cost and require operators to have an explicit knowledge of data category classification. Once a casual error in classifying data categories occurs, it will lead to a series of subsequent tasks with persistent interference from subjective factors, which is fatal in practical application scenarios. The supervised learning process on raw data is shown in (a) in Figure 1.
- 2.
- They use balanced training data or do not consider the category imbalance in the training data. Data category imbalance is a significant quantitative difference in the sample size of different labels in the data, which is common in practical scenarios. Category imbalance can limit the model’s performance to varying degrees [14,15], so it is critical to investigate how to adopt strategies to address the data category imbalance while ensuring relatively good performance [16].
- 3.
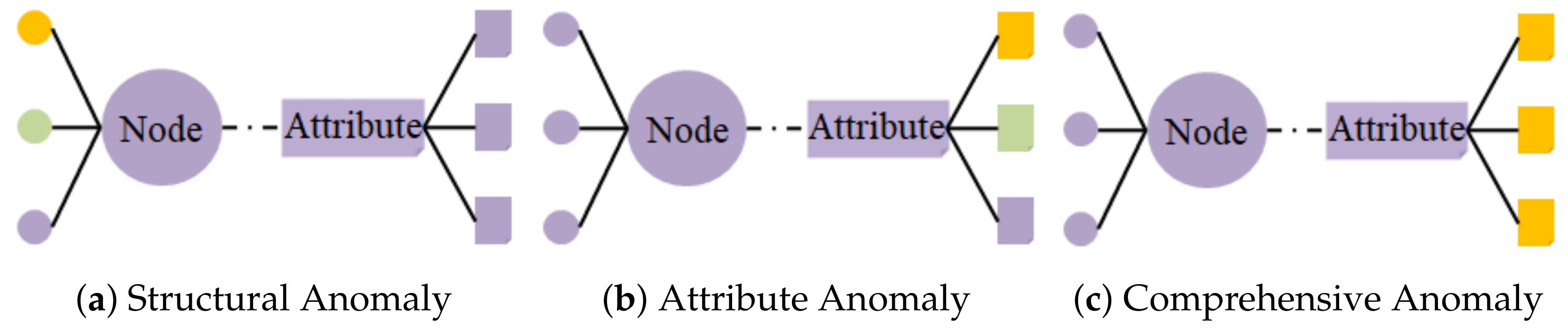
- They do not adequately capture topological information between detection samples. The data obtained in the detection process has the characteristics of complexity, nonlinearity and discreteness, which means that we need to pay attention to the detection data’s attribute information and topology information as much as possible to realize the contamination warning of food quality more accurately.
- 1.
- An end-to-end contamination warning and control framework for food quality is proposed by us, which can efficiently mine quality-contaminated unqualified samples and potentially contaminated qualified samples in food detection data by obtaining contamination values from food detection data.
- 2.
- A contrastive self-supervised learning scheme for contamination warning and control of food quality is proposed by us. Contrastive learning solves the dependence of the previous method on the balance of data categories. Meanwhile, self-supervised learning efficiently solves the problems of being easily interfered with by subjective factors and high time costs in the manual labeling operation of data categories in practical applications.
- 3.
- GNNs are used for information transfer. The CSGNN considers the data nodes’ attributes and structural information by constructing an attribute graph. To the best of our knowledge, it is also the first time graph algorithms have been applied to food safety risk assessment-related tasks.
- 4.
- The data in the actual scenarios verify that the contamination warning effect of the proposed algorithm is better than that of the current mainstream model. On a batch of dairy product detection data in a specific province in China, we compared the CSGNN framework with the mainstream model. Under self-supervision, the recall of unqualified samples of CSGNN reached 1.0000, which was more than 13% higher than the sub-optimal model. In addition, CSGNN completed the contamination classification of the food detection data based on the contamination value of each sample.
2. Related Work
2.1. Contamination Assessment Models for Food Quality
2.2. Contrastive Learning
3. Materials and Methods
3.1. Problem Definition and Data Source
3.1.1. Problem Definition
3.1.2. Data Source
3.2. Contamination Warning of Food Quality Based on Contrastive Self-Supervised Learning
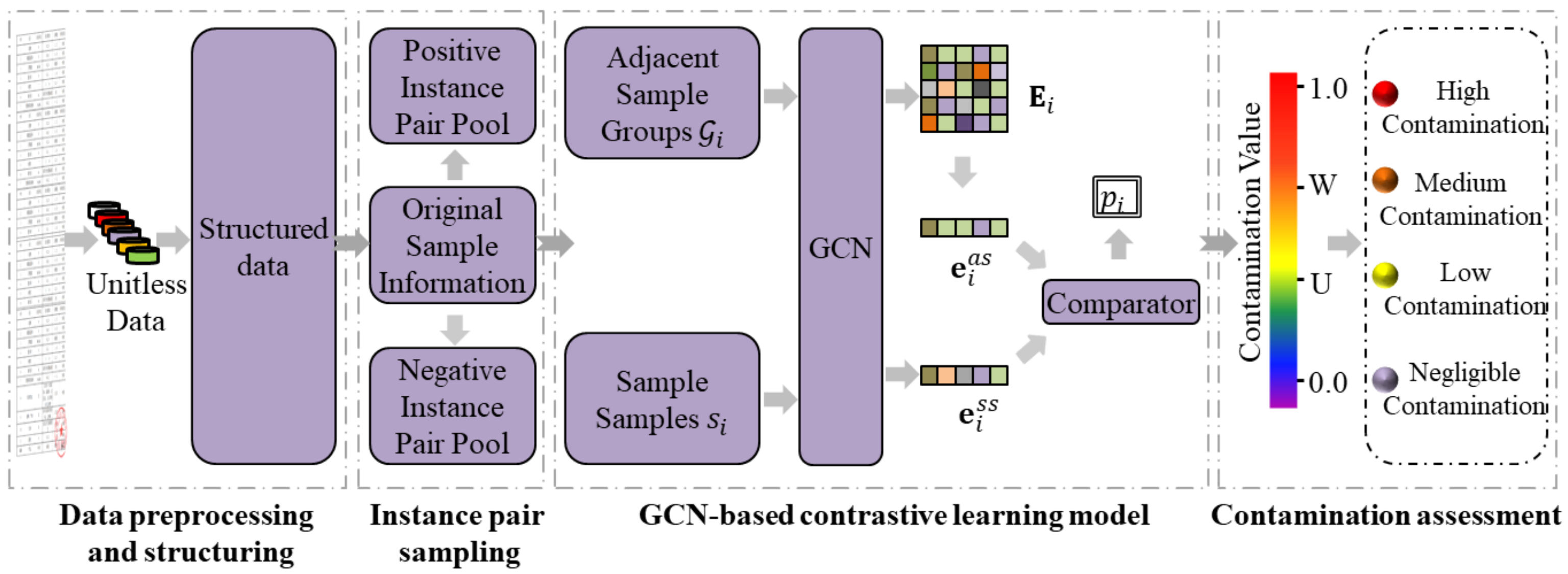
3.2.1. Data Preprocessing and Structuring
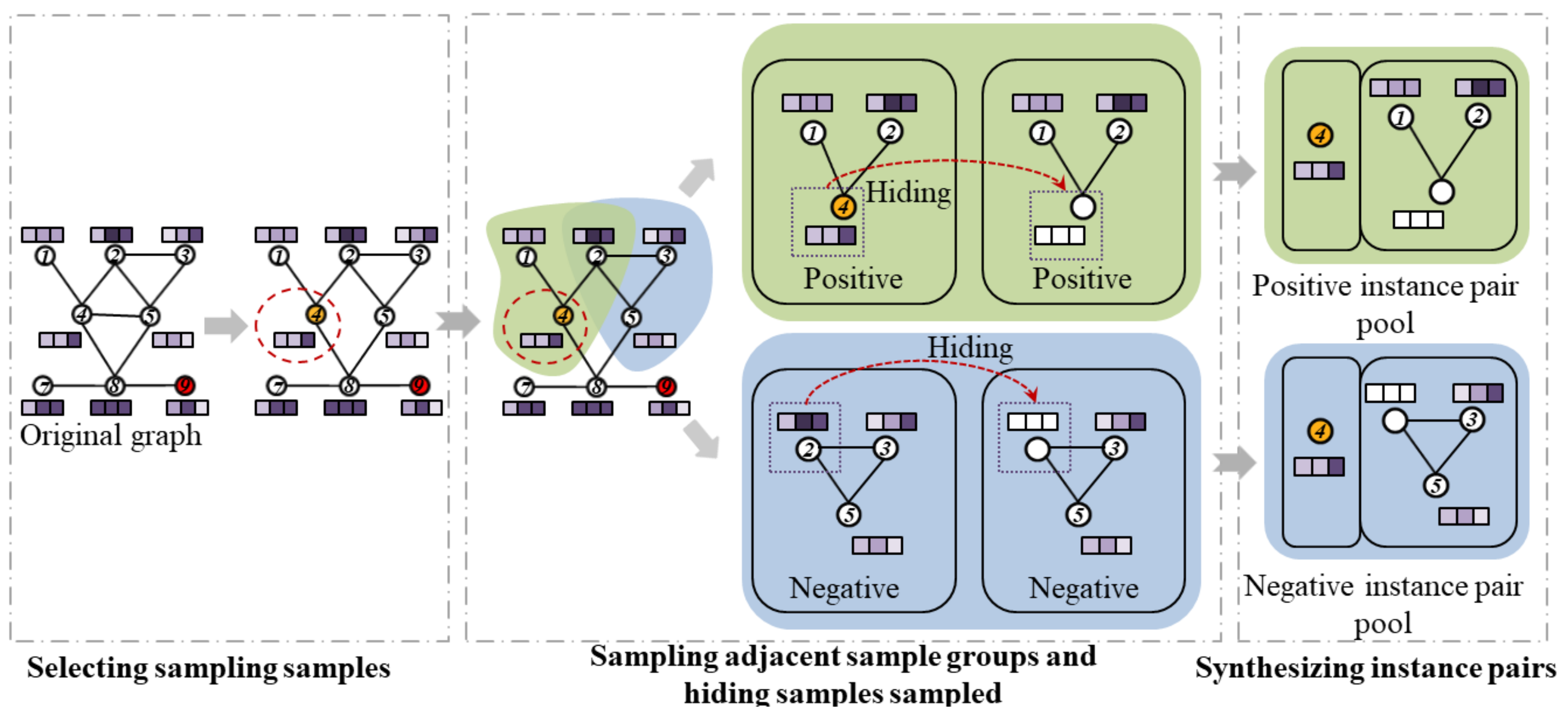
3.2.2. Contrastive Instance Pair Sampling
- 1.
- Selecting the sampled samples. All samples in the detection data are traversed randomly within each epoch, and the sampling samples are determined randomly.
- 2.
- Sampling adjacent sample groups. We set their initial samples as sampling samples and random sampling samples for the adjacent sample groups of positive and negative instance pairs, respectively. Inspired by the paper [43], we use RWR [44] as the sampling strategy for local sample groups to make the sampling strategy for adjacent sample groups more efficient.
- 3.
- Hiding sampled samples. To avoid the contrastive learning module to quickly identify the presence of sampled samples in the adjacent sample groups, we zeroed out the attribute features of the initial samples. That is, the attribute information of the sampled samples is hidden.
- 4.
- Synthesizing instance pairs. Combine sampled and adjacent samples into instance pairs and save them to the positive and negative instance pairs sample pool, respectively.
3.2.3. GCN-Based Contrastive Learning
3.2.4. Contamination Assessment
3.3. Evaluation Metrics
3.4. Baseline Model
3.4.1. NNLM
3.4.2. CNN
3.4.3. GCN
3.4.4. LOF
3.4.5. GAN
3.5. Parameter Settings
4. Experiments and Analysis of Results
4.1. Analysis of Results
4.1.1. For Q1 (What Are the Advantages of the CSGNN Framework over the Baseline Model? What Performance Will These Advantages Demonstrate in Real-World Application Scenarios?)
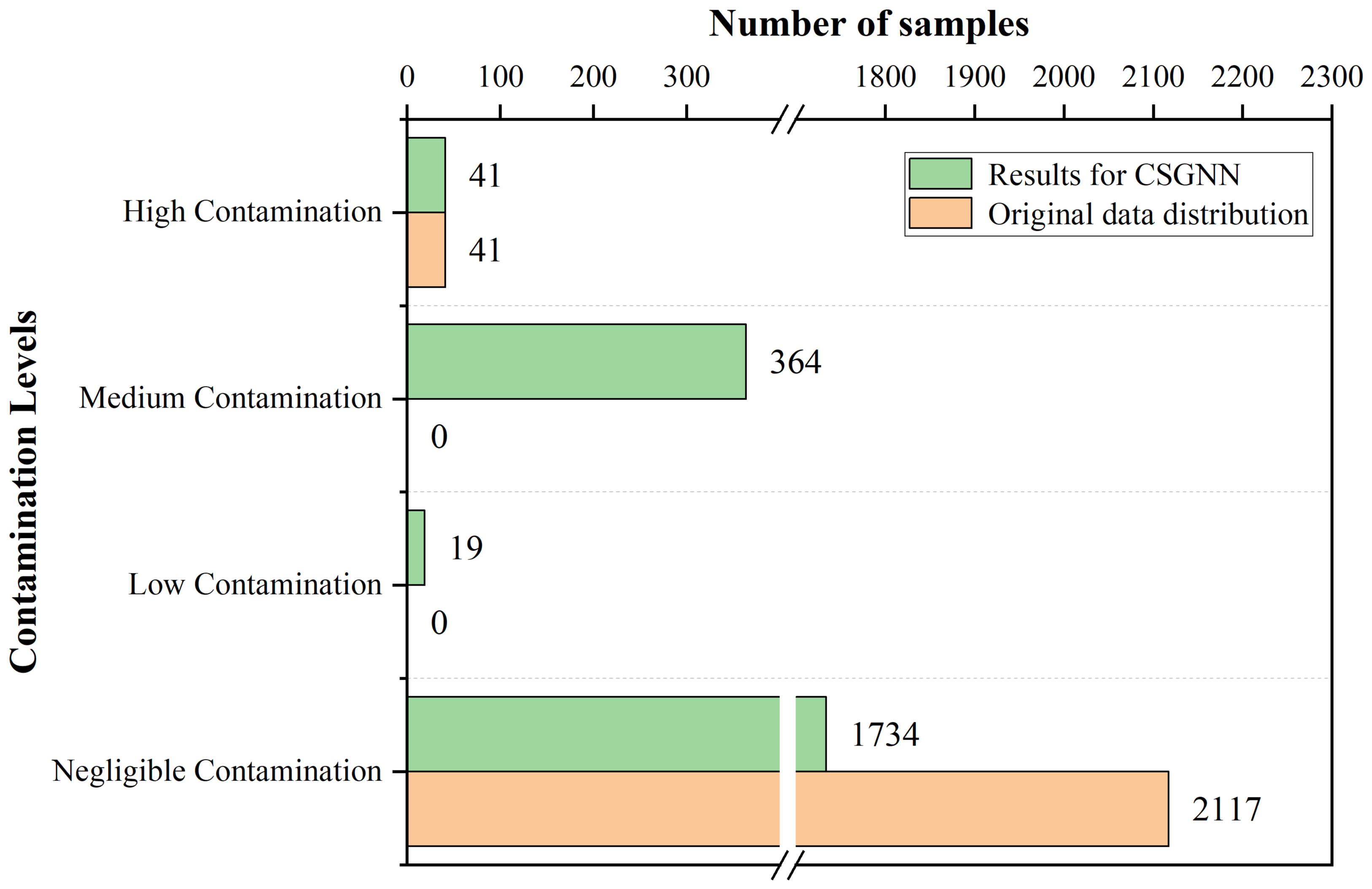
4.1.2. For Q2 (What Kind of Contamination Warning Does the CSGNN Framework Enable in Contamination Assessment Applications for Food Quality? How Does It Do It?)
- •
- Negligible contamination: negligible contamination level, samples are basically risk-free. 0 ⩽ contamination value <U for qualified samples.
- •
- Low contamination: Low contamination level, samples with a low probability of being at risk. U⩽ contamination value <W for qualified samples.
- •
- Medium contamination: Medium contamination level, samples with a high probability of being at risk. W⩽ contamination value ⩽ 1 for qualified samples.
- •
- High contamination: High contamination level, all unqualified samples belong to this category.
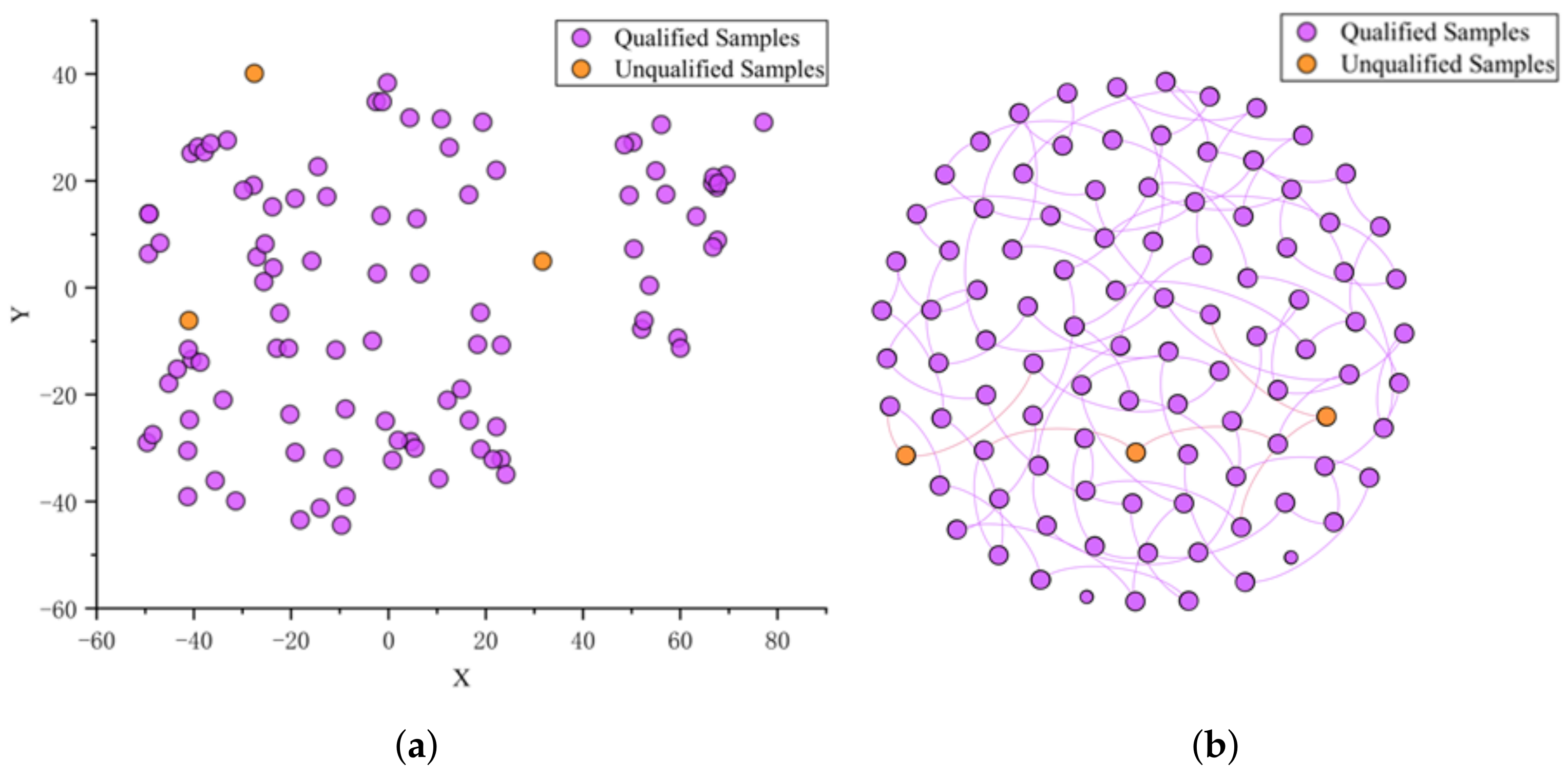
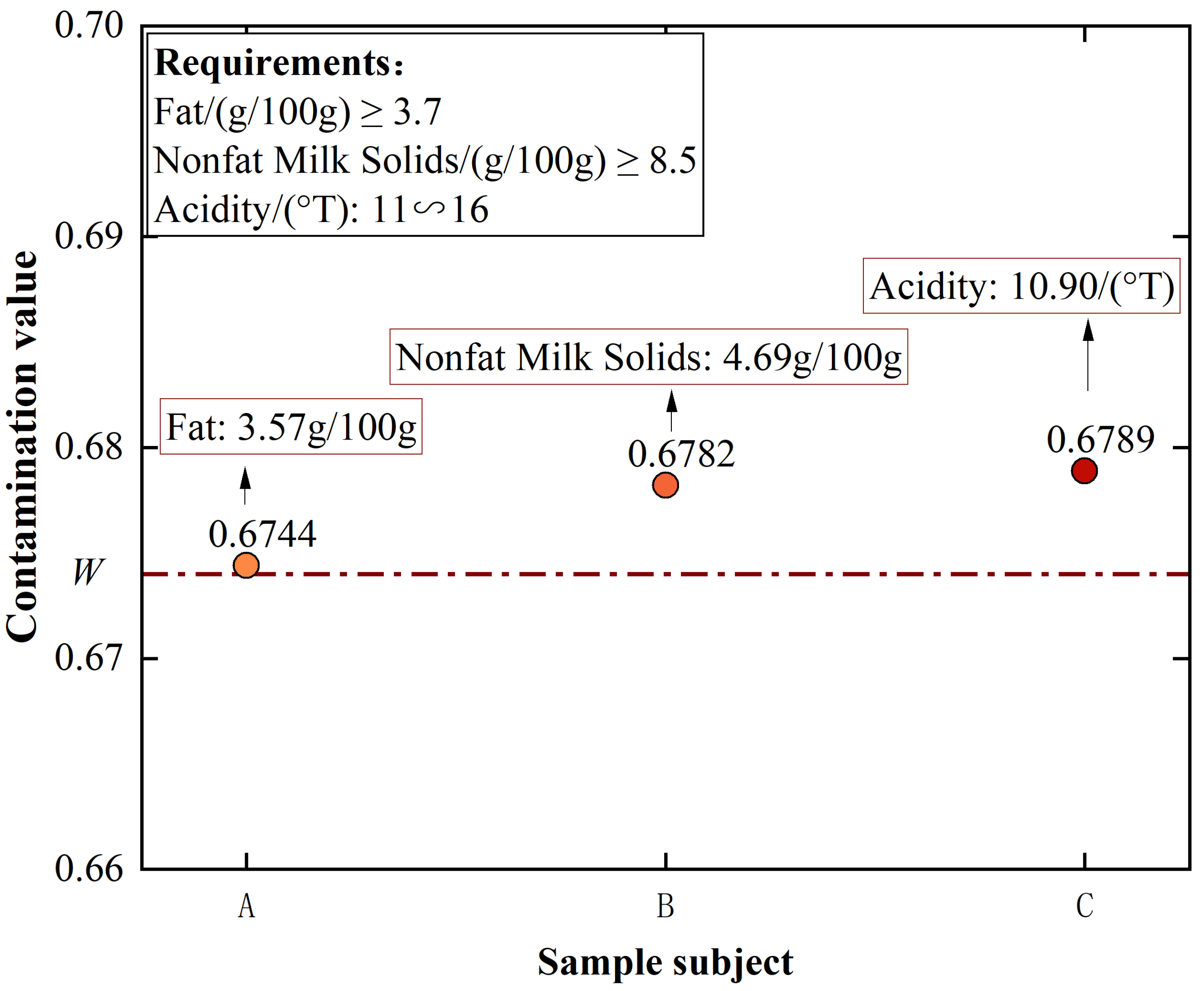
4.1.3. For Q3 (Is There a Reasonable and Feasible Explanation for the Contamination Classification of the CSGNN Framework in Contamination Assessment for Food Quality?)
4.2. Application and Optimization of the CSGNN
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization. Maximum Residue Limits (mrls) and Risk Management Recommendations (rmrs) for Residues of Veterinary Drugs in Foods-cx/mrl 2-2018. 2018. Available online: https://dokumen.tips/documents/maximumresiduelimitsmrlsandriskmanagement.html (accessed on 12 August 2022).
- Health Canada. List of Maximum Residue Limits (mrls) for Veterinary Drugs in Foods. 2022. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/maximum-residue-limits-mrls/list-maximum-residue-limits-mrls-veterinary-drugs-foods.html (accessed on 1 September 2022).
- Mensah, L.D.; Julien, D. Implementation of food safety management systems in the uk. Food Control 2011, 22, 1216–1225. [Google Scholar] [CrossRef]
- Neltner, T.G.; Kulkarni, N.R.; Alger, H.M.; Maffini, M.V.; Bongard, E.D.; Fortin, N.D.; Olson, E.D. Navigating the us food additive regulatory program. Compr. Rev. Food Sci. Food Saf. 2011, 10, 342–368. [Google Scholar] [CrossRef]
- Jen, J.J.-S.; Chen, J. Food Safety in China: Science, Technology, Management and Regulation; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Wu, Y.-N.; Liu, P.; Chen, J.-S. Food safety risk assessment in china: Past, present and future. Food Control 2018, 90, 212–221. [Google Scholar] [CrossRef]
- Han, Y.; Cui, S.; Geng, Z.; Chu, C.; Chen, K.; Wang, Y. Food quality and safety risk assessment using a novel hmm method based on gra. Food Control 2019, 105, 180–189. [Google Scholar] [CrossRef]
- Geng, Z.; Shang, D.; Han, Y.; Zhong, Y. Early warning modeling and analysis based on a deep radial basis function neural network integrating an analytic hierarchy process: A case study for food safety. Food Control 2019, 96, 329–342. [Google Scholar] [CrossRef]
- Geng, Z.; Liu, F.; Shang, D.; Han, Y.; Shang, Y.; Chu, C. Early warning and control of food safety risk using an improved ahc-rbf neural network integrating ahp-ew. J. Food Eng. 2021, 292, 110239. [Google Scholar] [CrossRef]
- Smid, J.H.; Verloo, D.; Barker, G.C.; Havelaar, A.H. Strengths and weaknesses of monte carlo simulation models and bayesian belief networks in microbial risk assessment. Int. J. Food Microbiol. 2010, 139, S57–S63. [Google Scholar] [CrossRef]
- Soon, J.M. Application of bayesian network modelling to predict food fraud products from china. Food Control 2020, 114, 107232. [Google Scholar] [CrossRef]
- Geng, Z.Q.; Zhao, S.S.; Tao, G.C.; Han, Y.M. Early warning modeling and analysis based on analytic hierarchy process integrated extreme learning machine (ahp-elm): Application to food safety. Food Control 2017, 78, 33–42. [Google Scholar] [CrossRef]
- Goldberg, D.M.; Khan, S.; Zaman, N.; Gruss, R.J.; Abrahams, A.S. Text mining approaches for postmarket food safety surveillance using online media. Risk Anal. 2020. [Google Scholar] [CrossRef]
- Weller, D.L.; Love, T.M.T.; Wiedmann, M. Comparison of resampling algorithms to address class imbalance when developing machine learning models to predict foodborne pathogen presence in agricultural water. front. Environ. Sci 2021, 9, 701288. [Google Scholar] [CrossRef]
- Seiffert, C.; Khoshgoftaar, T.M.; Hulse, J.V.; Napolitano, A. Rusboost: A hybrid approach to alleviating class imbalance. IEEE Trans. Syst. Man Cybern.-Part A Syst. Hum. 2009, 40, 185–197. [Google Scholar] [CrossRef]
- Sun, Y.; Kamel, M.S.; Wong, A.K.C.; Wang, Y. Cost-sensitive boosting for classification of imbalanced data. Pattern Recognit. 2007, 40, 3358–3378. [Google Scholar] [CrossRef]
- Pöppelbaum, J.; Chadha, G.S.; Schwung, A. Contrastive learning based self-supervised time-series analysis. Appl. Soft Comput. 2022, 117, 108397. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, S.; Jin, M.; Zhou, C.; Xia, F.; Yu, P.S. Graph self-supervised learning: A survey. arXiv 2021, arXiv:2103.00111. [Google Scholar] [CrossRef]
- Velickovic, P.; Fedus, W.; Hamilton, W.L.; Liò, P.; Bengio, Y.; Hjelm, R.D. Deep graph infomax. ICLR (Poster) 2019, 2, 4. [Google Scholar]
- Hassani, K.; Khasahmadi, A.H. Contrastive multi-view representation learning on graphs. In International Conference on Machine Learning; PMLR: New York, NY, USA, 2020; pp. 4116–4126. [Google Scholar]
- Liu, Y.; Li, Z.; Pan, S.; Gong, C.; Zhou, C.; Karypis, G. Anomaly detection on attributed networks via contrastive self-supervised learning. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 2378–2392. [Google Scholar] [CrossRef]
- Moayedikia, A. Multi-objective community detection algorithm with node importance analysis in attributed networks. Appl. Soft Comput. 2018, 67, 434–451. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Li, J.; Zhang, Q. Deep attributed network representation learning of complex coupling and interaction. Knowl.-Based Syst. 2021, 212, 106618. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhou, H.; Li, C.; Tang, J.; Zeng, Q. Deepemlan: Deep embedding learning for attributed networks. Inf. Sci. 2021, 543, 382–397. [Google Scholar] [CrossRef]
- Zuo, E.; Du, X.; Aysa, A.; Lv, X.; Muhammat, M.; Zhao, Y.; Ubul, K. Anomaly score-based risk early warning system for rapidly controlling food safety risk. Foods 2022, 11, 2076. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; He, X.; Zhang, H.; Chua, T.-S. Attributed social network embedding. IEEE Trans. Knowl. Data Eng. 2018, 30, 2257–2270. [Google Scholar] [CrossRef]
- Pan, S.; Hu, R.; Fung, S.-F.; Long, G.; Jiang, J.; Zhang, C. Learning graph embedding with adversarial training methods. IEEE Trans. Cybern. 2019, 50, 2475–2487. [Google Scholar] [CrossRef]
- Wang, X.; Bouzembrak, Y.; Lansink, A.G.J.M.O.; van der Fels-Klerx, H.J. Application of machine learning to the monitoring and prediction of food safety: A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 416–434. [Google Scholar] [CrossRef] [PubMed]
- Widodo, A.; Yang, B.-S. Support vector machine in machine condition monitoring and fault diagnosis. Mech. Syst. Signal Process. 2007, 21, 2560–2574. [Google Scholar] [CrossRef]
- Bornn, L.; Farrar, C.R.; Park, G.; Farinholt, K. Structural health monitoring with autoregressive support vector machines. J. Vib. Acoust. 2009, 131. [Google Scholar] [CrossRef]
- Yang, Y.; Wei, L.; Pei, J. Application of bayesian modelling to assess food quality & safety status and identify risky food in china market. Food Control 2019, 100, 111–116. [Google Scholar]
- Wei, L.; Pei, J.; Zhang, A.; Wu, X.; Xie, Y.; Yang, Y. Application of stochastic bayesian modeling to assess safety status of baby formulas and quantify factors leading to unsafe products in china market. Food Control 2020, 108, 106826. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Nogales, A.; Díaz-Morón, R.; García-Tejedor, Á.J. A comparison of neural and non-neural machine learning models for food safety risk prediction with european union rasff data. Food Control 2022, 134, 108697. [Google Scholar] [CrossRef]
- Parisot, S.; Ktena, S.I.; Ferrante, E.; Lee, M.; Guerrero, R.; Glocker, B.; Rueckert, D. Disease prediction using graph convolutional networks: Application to autism spectrum disorder and alzheimer’s disease. Med. Image Anal. 2018, 48, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, F.; Hou, Z.; Mian, L.; Wang, Z.; Zhang, J.; Tang, J. Self-supervised learning: Generative or contrastive. IEEE Trans. Knowl. Data Eng. 2021, 35, 857–876. [Google Scholar] [CrossRef]
- Hafidi, H.; Ghogho, M.; Ciblat, P.; Swami, A. Negative sampling strategies for contrastive self-supervised learning of graph representations. Signal Process. 2022, 190, 108310. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Y.; Zhang, D.; Chen, H.; Peng, H.; Pan, S. Towards unsupervised deep graph structure learning. arXiv 2022, arXiv:2201.06367. [Google Scholar]
- Van den Oord, A.; Li, Y.; Vinyals, O. Representation learning with contrastive predictive coding. arXiv 2018, arXiv:1807.03748. [Google Scholar]
- Chen, X.; He, K. Exploring simple siamese representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 15750–15758. [Google Scholar]
- Jintao, H. Food safety law of the people’s republic of china. Chin. L. Gov’t 2012, 45, 10. [Google Scholar]
- Qiu, J.; Chen, Q.; Dong, Y.; Zhang, J.; Yang, H.; Ding, M.; Wang, K.; Tang, J. Gcc: Graph contrastive coding for graph neural network pre-training. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Long Beach, CA, USA, 6–10 July 2020; pp. 1150–1160. [Google Scholar]
- Tong, H.; Faloutsos, C.; Pan, J.Y. Fast random walk with restart and its applications. In Proceedings of the Sixth International Conference on Data Mining (ICDM’06), Hong Kong, China, 18–22 December 2006; pp. 613–622. [Google Scholar]
- Saha, P.; Mukherjee, D.; Singh, P.K.; Ahmadian, A.; Ferrara, M.; Sarkar, R. Retracted article: Graphcovidnet: A graph neural network based model for detecting COVID-19 from ct scans and x-rays of chest. Sci. Rep. 2021, 11, 8304. [Google Scholar] [CrossRef]
- Jiang, P.; Huang, S.; Fu, Z.; Sun, Z.; Lakowski, T.M.; Hu, P. Deep graph embedding for prioritizing synergistic anticancer drug combinations. Comput. Struct. Biotechnol. J. 2020, 18, 427–438. [Google Scholar] [CrossRef]
- Zhao, L.; Song, Y.; Zhang, C.; Liu, Y.; Wang, P.; Lin, T.; Deng, M.; Li, H. T-gcn: A temporal graph convolutional network for traffic prediction. IEEE Trans. Intell. Transp. Syst. 2019, 21, 3848–3858. [Google Scholar] [CrossRef]
- Monti, F.; Bronstein, M.; Bresson, X. Geometric matrix completion with recurrent multi-graph neural networks. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Yao, L.; Mao, C.; Luo, Y. Graph convolutional networks for text classification. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 7370–7377. [Google Scholar]
- Shi, L.; Zhang, Y.; Cheng, J.; Lu, H. Two-stream adaptive graph convolutional networks for skeleton-based action recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 12026–12035. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Darliansyah, A.; Naeem, M.A.; Mirza, F.; Pears, R. Sentipede: A smart system for sentiment-based personality detection from short texts. J. Univ. Comput. Sci. 2019, 25, 1323–1352. [Google Scholar]
- Vo, S.A.; Scanlan, J.; Turner, P. An application of convolutional neural network to lobster grading in the southern rock lobster supply chain. Food Control 2020, 113, 107184. [Google Scholar] [CrossRef]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph convolutional networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 5966–5978. [Google Scholar] [CrossRef]
- Breunig, M.M.; Kriegel, H.; Ng, R.T.; Sander, J. Lof: Identifying density-based local outliers. In Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data, Dallas, TX, USA, 16–18 May 2000; pp. 93–104. [Google Scholar]
- Kim, J.-Y.; Bu, S.-J.; Cho, S.-B. Zero-day malware detection using transferred generative adversarial networks based on deep autoencoders. Inf. Sci. 2018, 460, 83–102. [Google Scholar] [CrossRef]
- Zuo, E.; Aysa, A.; Muhammat, M.; Zhao, Y.; Chen, B.; Ubul, K. A food safety prescreening method with domain-specific information using online reviews. J. Consum. Prot. Food Saf. 2022, 17, 163–175. [Google Scholar] [CrossRef]


| Notation | Description and Explanation |
|---|---|
| The minimum value of the original value of the v-th indicator for all samples. | |
| The maximum value of the original value of the v-th indicator for all samples. | |
| The average value of the original value of the v-th indicator for all samples. | |
| The original value of the v-th indicator of the n-th sample. | |
| N | The number of samples in the detection data. |
| V | The number of testing indicators in the detection data. |
| n | The number of nodes in . |
| d | The dimension of the attribute in . |
| Attribute networks constructed from the detection data. | |
| The set of nodes of . | |
| The set of edges of . | |
| The attribute matrix of . | |
| Instance pairs for each batch with a total batch size of M. | |
| Sampled samples in instance pair . | |
| The group of adjacent samples in instance pair . | |
| True label of the sampled sample . | |
| The representation matrix is learned by the ℓ-th implicit layer. | |
| The ℓ-th layer trainable weight matrix. | |
| The embedding matrix of the nodes in . | |
| The row vector of feature representations of the sampled sample learned by the ℓ-th implicit layer. | |
| The embedding vector of . | |
| The embedding vector of . | |
| The weight matrix of the comparison recognition module. | |
| The prediction score of . | |
| The contamination value of the sampled sample . | |
| R | The number of sampling rounds. |
| a | The number of nodes in adjacent sample groups. |
| b | The dimensionality of embedding. |
| W | The lowest contamination value of the unqualified samples. |
| U | The more obvious boundary value between the contaminated sample and the negligible contamination sample (default 0.5). |
| Z | Set the number of edges when structuring. |
| Item | Requirements | Testing Method | |
|---|---|---|---|
| physicochemical index | Lactose/(g/100 g) | ⩽2.0 | GB 5009.8-2016 |
| Protein/(g/100 g) | ⩾3.1 | GB 5009.5-2010 | |
| Acidity/(°T) | 11∼16 | GB 5413.34-2010 | |
| Fat/(g/100 g) | ⩾3.7 | GB 5413.3-2010 | |
| Nonfat Milk Solids/(g/100 g) | ⩾8.5 | GB 5413.39-2010 | |
| mycotoxin index | Aflatoxin M/(g/kg) | ⩽0.5 | GB 2761-2017 |
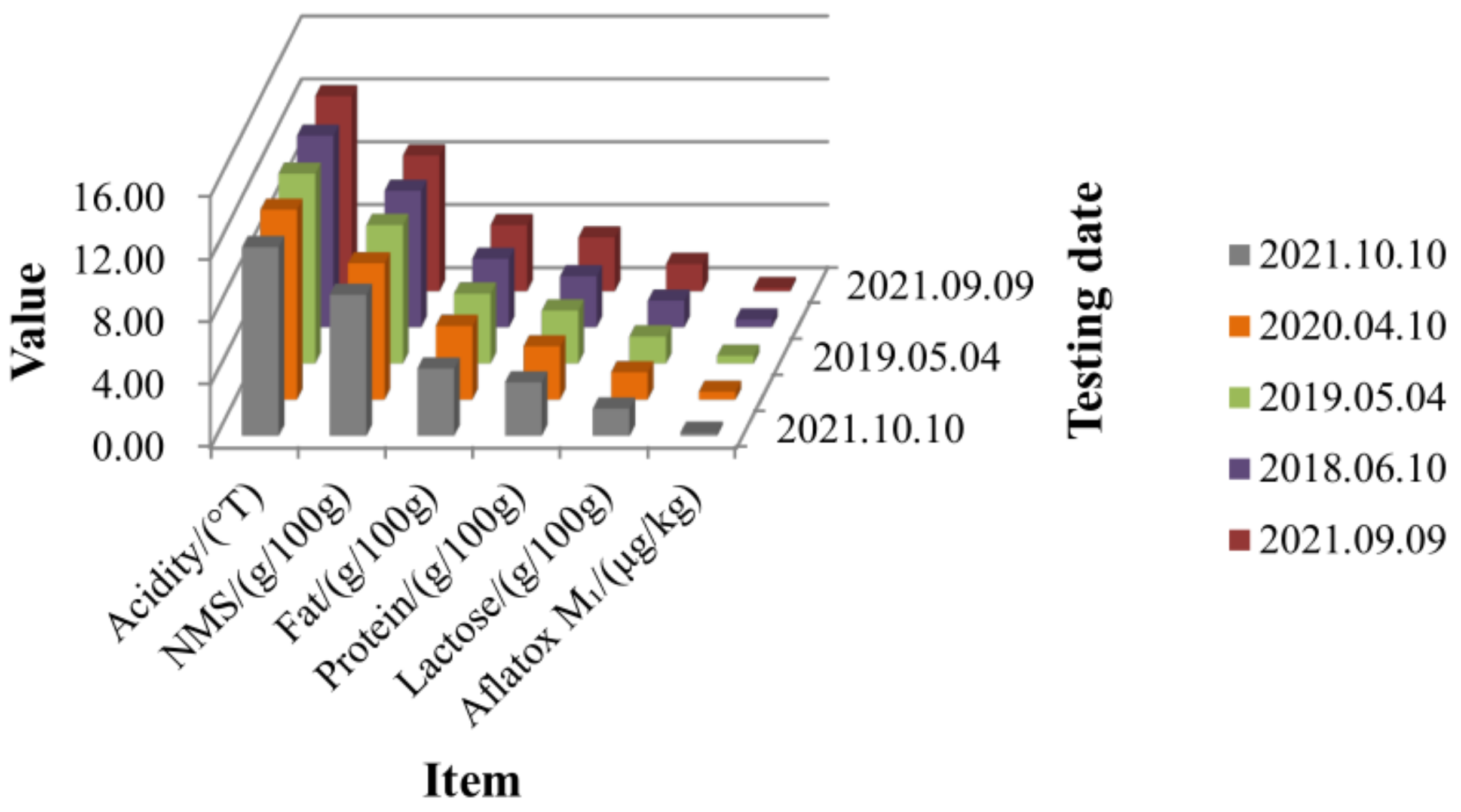
| Sample Number | Testing Date | Item | |||||
|---|---|---|---|---|---|---|---|
| Lactose | Nonfat Milk Solids | Protein | Acidity | Aflatoxin M | Fat | ||
| 20211010-578 | 10 October 2021 | 1.73 | 8.97 | 3.40 | 12.00 | 0.2 | 4.28 |
| 20200410-525 | 10 April 2020 | 1.74 | 8.67 | 3.39 | 12.08 | 0.5 | 4.69 |
| 20190504-166 | 4 May 2019 | 1.72 | 8.81 | 3.37 | 12.07 | 0.5 | 4.44 |
| 20180610-453 | 10 June 2018 | 1.70 | 8.68 | 3.25 | 12.19 | 0.5 | 4.36 |
| 20210909-512 | 9 September 2021 | 1.73 | 8.62 | 3.42 | 12.40 | 0.2 | 4.20 |
| Categories | Item | Requirements |
|---|---|---|
| Positive indicators | Aflatoxin M/(g/kg) | ⩽0.5 |
| Lactose/(g/100 g) | ⩽2.0 | |
| Inverse indicators | Protein/(g/100 g) | ⩾3.1 |
| Fat/(g/100 g) | ⩾3.7 | |
| Nonfat Milk Solids/(g/100 g) | ⩾8.5 | |
| Oscillatory indicators | Acidity/(°T) | 11∼16 |
| Real Label | |||
|---|---|---|---|
| 1 | 0 | ||
| Predicted Label | 1 | True Positive (TP) | False Positive (FP) |
| 0 | False Negative (FN) | True Negative (TN) | |
| Input | Models | AUC | Precision | Precision of Qualified Samples | Recall of Unqualified Samples | FAR |
|---|---|---|---|---|---|---|
| (X,Y) | NNLM | 0.7602 | 0.9668 | 0.9904 | 0.7059 | 0.2941 * |
| CNN | 0.6765 | 0.9833 | 0.9829 | 0.3529 | 0.6470 | |
| GCN | 0.9988 | 0.9979 | 1.0000 | 1.0000 | 0.0024 | |
| (X) | LOF | 0.9150 | 0.9787 | 0.9787 | 0.8823 | 0.0523 |
| GAN | 0.5804 | 0.9546 | 0.9782 | 0.2353 | 0.7647 | |
| CSGNN | 0.9140 | 0.9829 | 1.0000 | 1.0000 | 0.1719 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Li, H.; Zuo, E.; Li, T.; Chen, C.; Chen, C.; Lv, X. CSGNN: Contamination Warning and Control of Food Quality via Contrastive Self-Supervised Learning-Based Graph Neural Network. Foods 2023, 12, 1048. https://doi.org/10.3390/foods12051048
Yan J, Li H, Zuo E, Li T, Chen C, Chen C, Lv X. CSGNN: Contamination Warning and Control of Food Quality via Contrastive Self-Supervised Learning-Based Graph Neural Network. Foods. 2023; 12(5):1048. https://doi.org/10.3390/foods12051048
Chicago/Turabian StyleYan, Junyi, Hongyi Li, Enguang Zuo, Tianle Li, Chen Chen, Cheng Chen, and Xiaoyi Lv. 2023. "CSGNN: Contamination Warning and Control of Food Quality via Contrastive Self-Supervised Learning-Based Graph Neural Network" Foods 12, no. 5: 1048. https://doi.org/10.3390/foods12051048
APA StyleYan, J., Li, H., Zuo, E., Li, T., Chen, C., Chen, C., & Lv, X. (2023). CSGNN: Contamination Warning and Control of Food Quality via Contrastive Self-Supervised Learning-Based Graph Neural Network. Foods, 12(5), 1048. https://doi.org/10.3390/foods12051048




