Role of PI3K-AKT Pathway in Ultraviolet Ray and Hydrogen Peroxide-Induced Oxidative Damage and Its Repair by Grain Ferments
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of S. commune Fermented Broths
2.3. Cell Culture and the Cell Viability Measurement
2.4. UVA/H2O2-Induced Model Establishment
2.5. Transcriptome Sequencing
2.6. Screening for Differential Expressed Genes (DEGs) and Functional Analysis
2.7. RT-qPCR
2.8. Statistics
3. Results
3.1. The Establishment of UVA/H2O2-Induced Models and Identification of DEGs
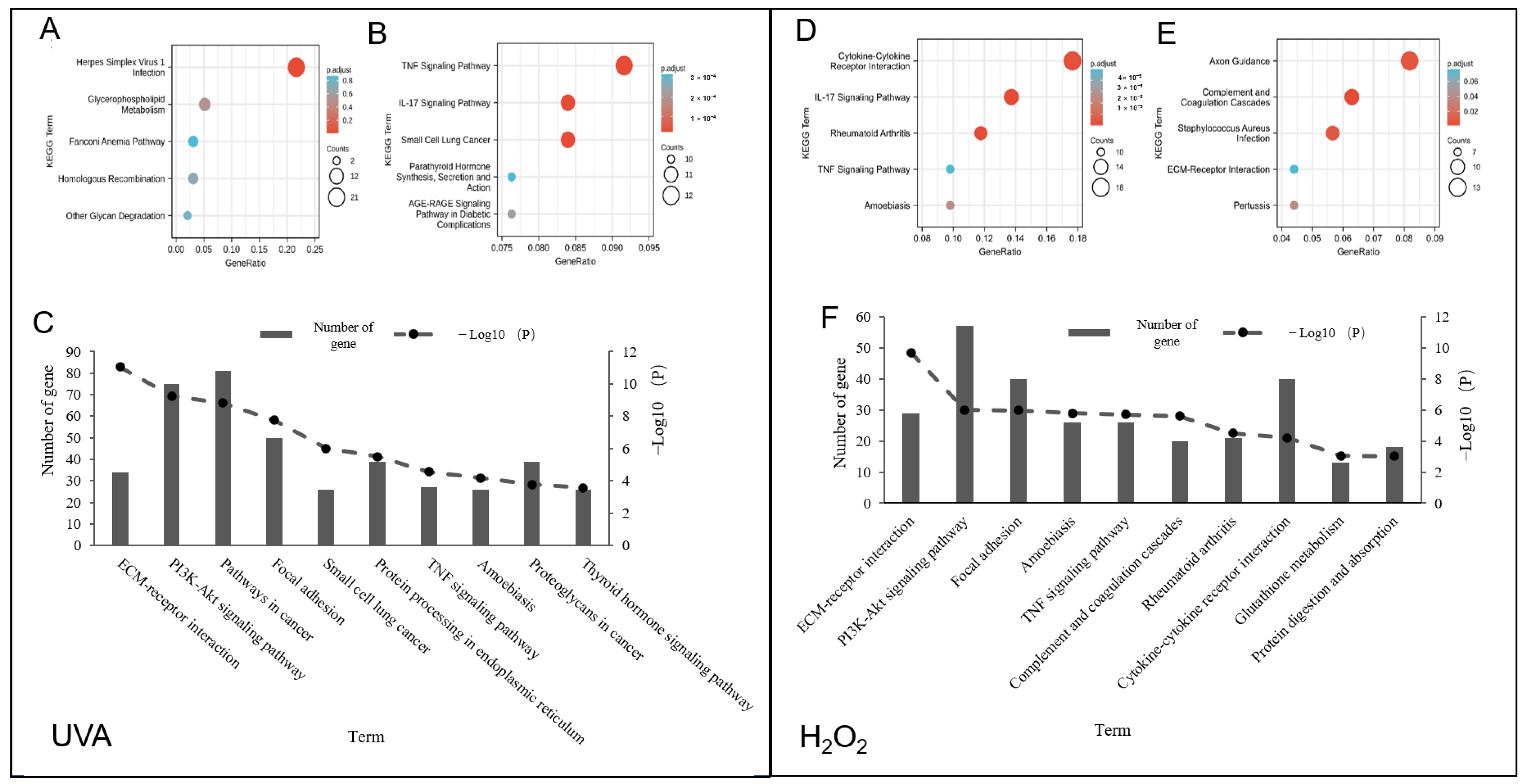
3.2. Gene Ontology (GO) and KEGG Pathway Analysis
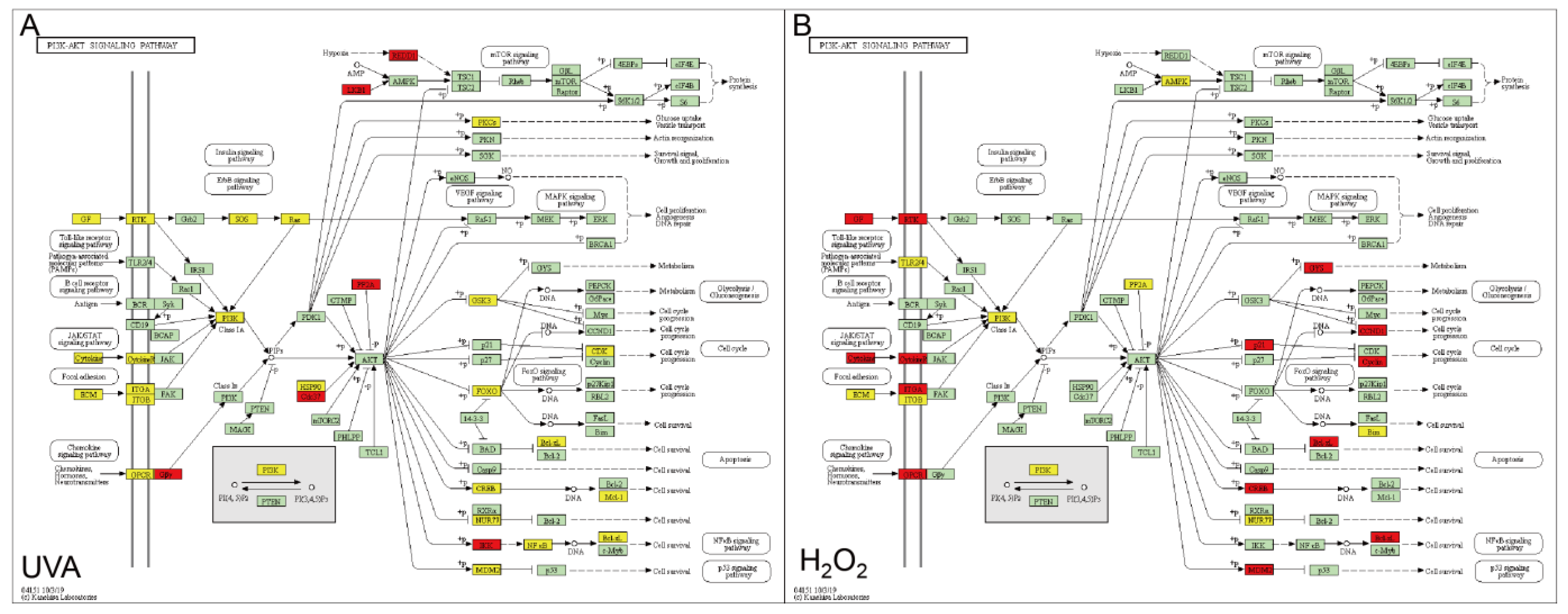
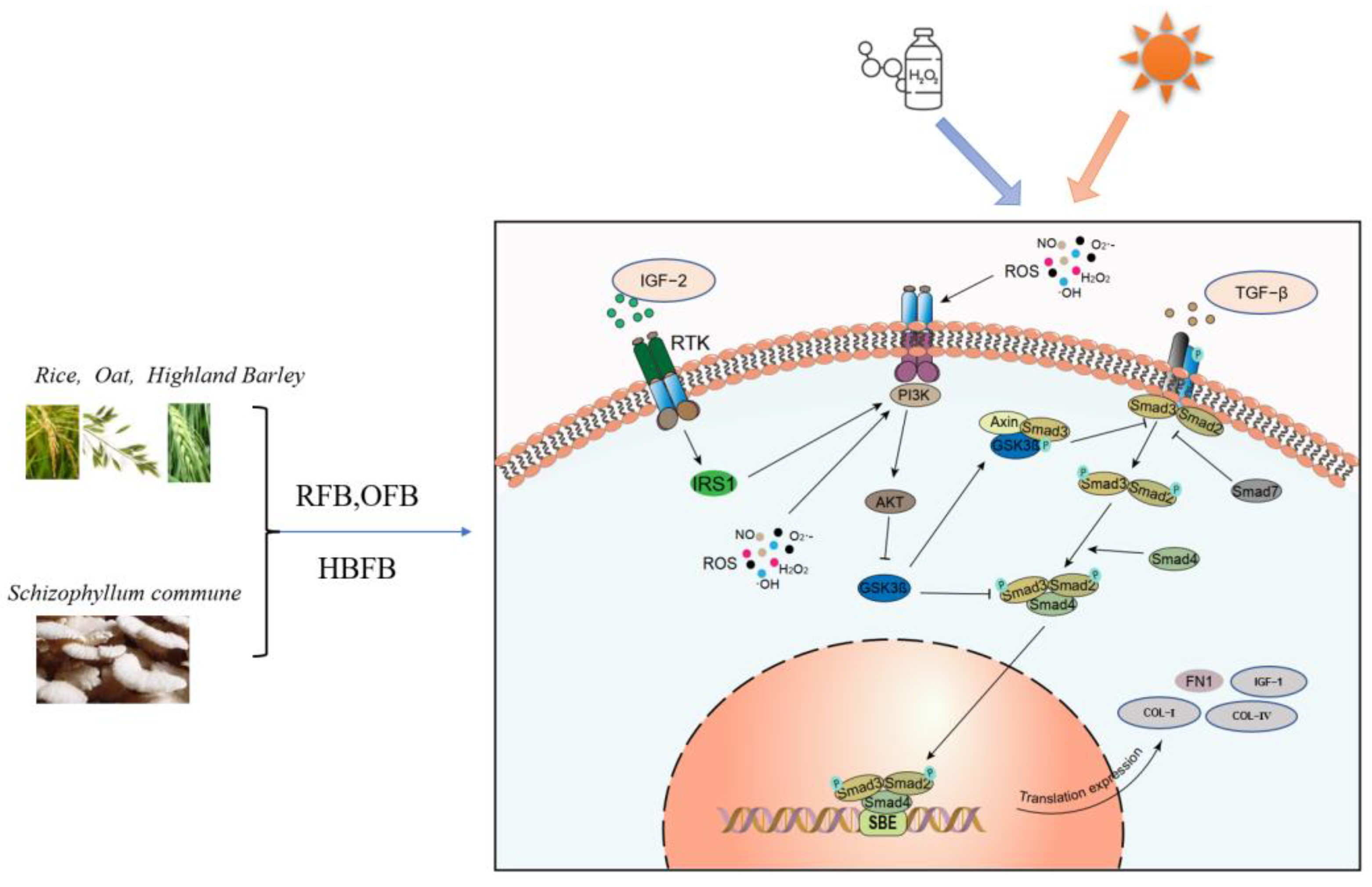
3.3. Key DEGs Functioned in PI3K-AKT Pathway
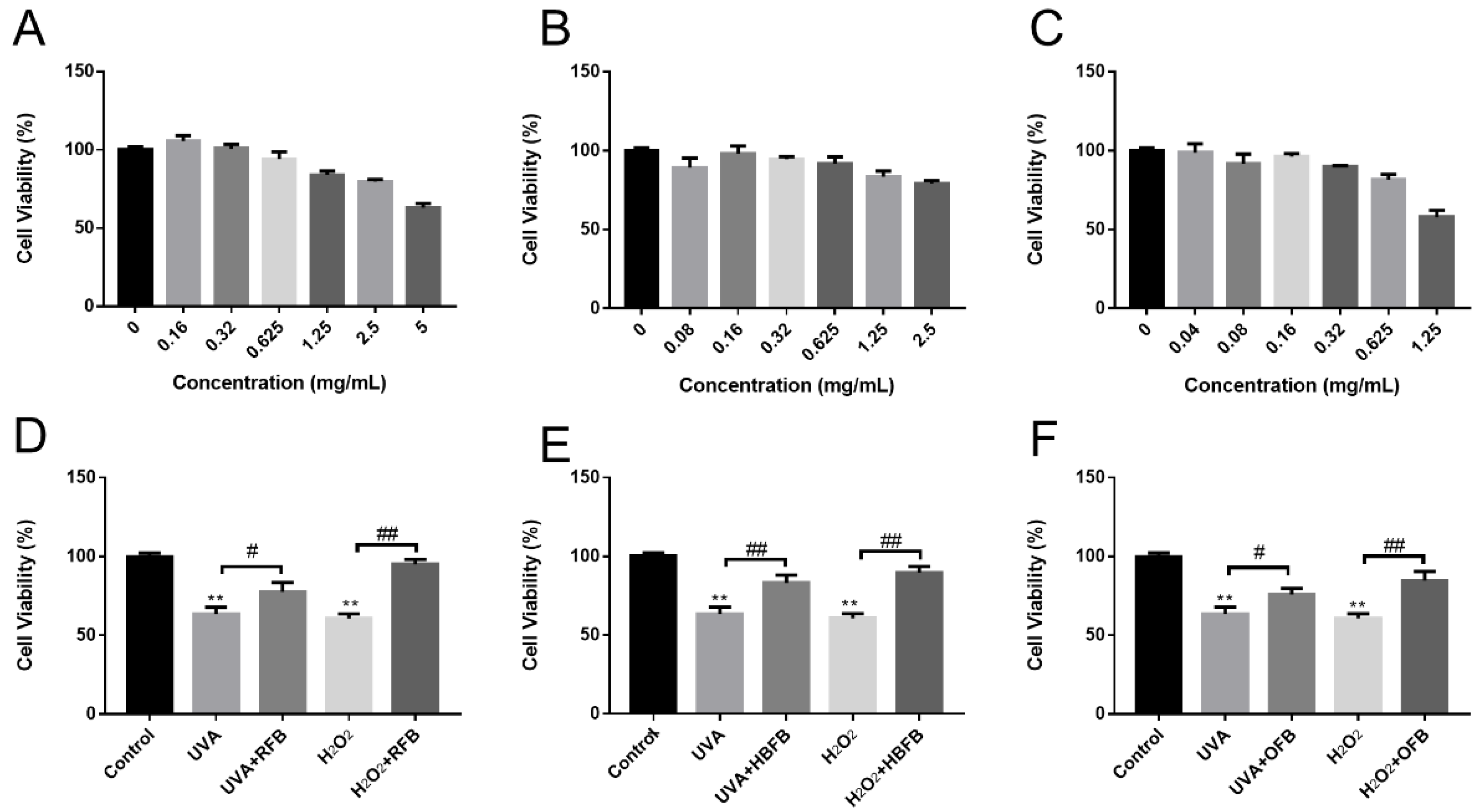
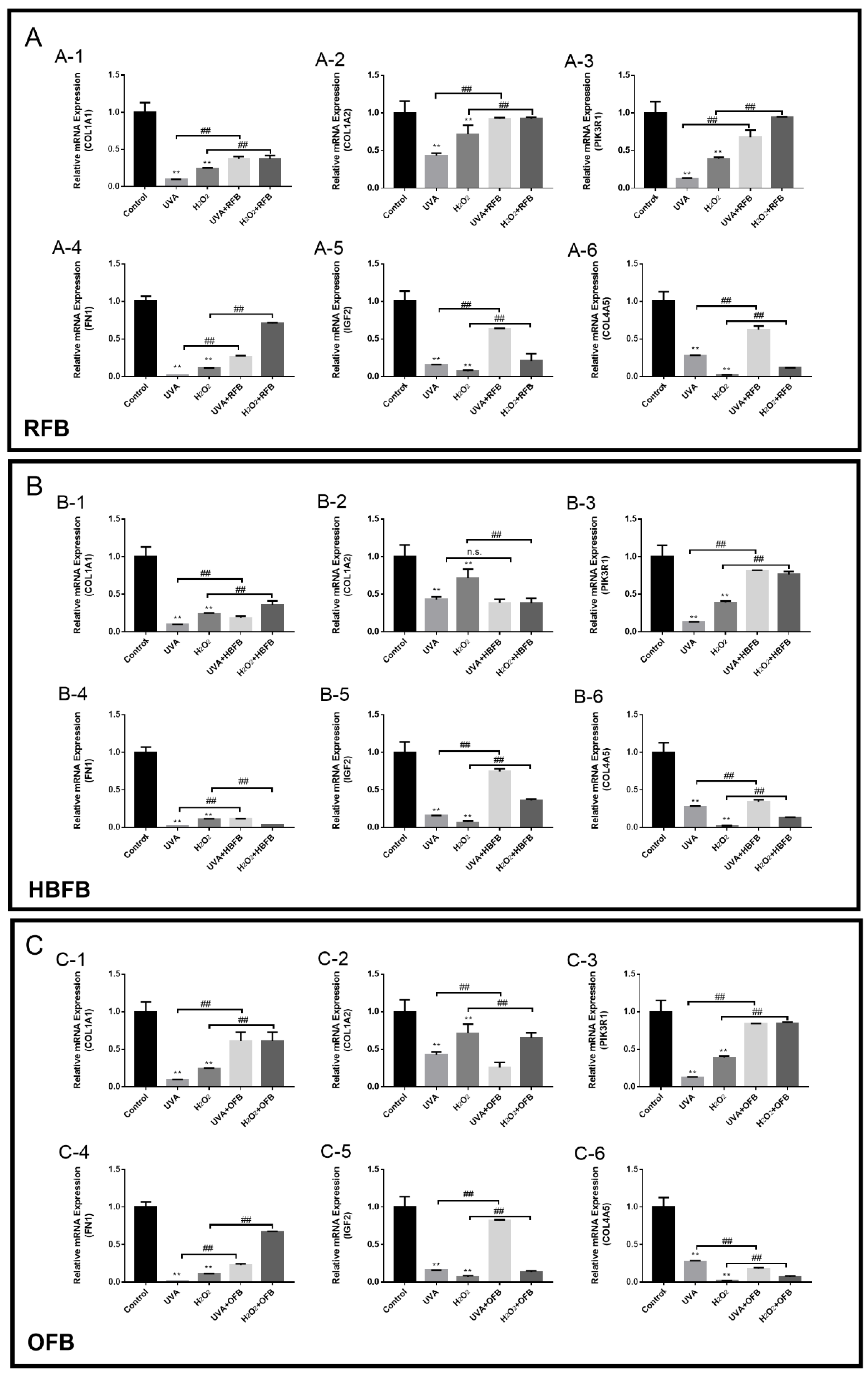
3.4. Composition Analysis and Protective Effects of RFB, HBFB and OFB
3.5. Validation of Key DEGs by RT-qPCR
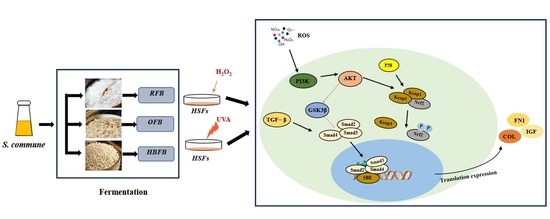
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pollman, M.J.; Hall, J.L.; Gibbons, G.H. Determinants of vascular smooth muscle cell apoptosis after balloon angioplasty injury. Influence of redox state and cell phenotype. Circ. Res. 1999, 84, 113–121. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, Y.; An, Q.; Wang, D.; You, S.; Zhao, D.; Zhang, J.; Wang, C.; Li, M. Anti-Photoaging effect of rhodiola rosea fermented by lactobacillus plantarum on UVA-damaged fibroblasts. Nutrients 2022, 14, 2324. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Wang, Z.H.; Zhang, R.; Piao, M.J.; Kim, K.C.; Kang, S.S.; Kim, Y.W.; Lee, J.; Park, D.; Hyun, J.W. Myricetin protects cells against oxidative stress-induced apoptosis via regulation of PI3K/AKT and MAPK Signaling Pathway. Int. J. Mol. Sci. 2010, 11, 4348–4360. [Google Scholar] [CrossRef] [PubMed]
- Morry, J.; Ngamcherdtrakul, W.; Yantasee, W. Oxidative stress in cancer and fibrosis: Opportunity for therapeutic intervention with antioxidant compounds, enzymes, and nanoparticles. Redox Biol. 2017, 11, 240–253. [Google Scholar] [CrossRef]
- Yang, S.S.; Zhou, L.; Nephrology, D.O. Resveratrol inhibits oxidative stress-mediated apoptosis in renal tubular epithelial cells by activating PI3K/AKT pathway. Mod. Chin. Med. 2019, 21, 913–919. [Google Scholar]
- Cheng, W.; An, Q.; Zhang, J.; Shi, X.; Wang, C.; Li, M.; Zhao, D. Protective effect of ectoin on UVA/H2O2-induced oxidative damage in human skin fibroblast cells. Appl. Sci. 2022, 12, 8531. [Google Scholar] [CrossRef]
- Abadie, S.; Bedos, P.; Rouquette, J. A human skin model to evaluate the protective effect of compounds against UVA damage. Int. J. Cosmet. Sci. 2019, 41, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Yaar, M.; Gilchrest, B.A. Photoageing: Mechanism, prevention and therapy. Br. J. Dermatol. 2007, 157, 874–887. [Google Scholar] [CrossRef]
- Chen, T.; Guo, Z.P.; Jiao, X.Y.; Zhang, Y.H.; Li, J.Y.; Liu, H.J. Protective effects of peoniflorin against hydrogen peroxide-induced oxidative stress in human umbilical vein endothelial cells. Can. J. Physiol. Pharmacol. 2011, 89, 445–453. [Google Scholar] [CrossRef]
- Park, K.J.; Kim, Y.J.; Kim, J.; Kim, S.; Lee, S.Y.; Bae, J.W. Protective effects of peroxiredoxin on hydrogen peroxide induced oxidative stress and apoptosis in cardiomyocytes. Korean Circ. J. 2012, 42, 23–32. [Google Scholar] [CrossRef]
- Zhu, L.; Chang, R.Z.; Qiu, L.J. Application of serial analysis of gene expression in the analysis of gene expression in plant. Sci. Agric. Sin. 2003, 36, 1233–1240. [Google Scholar]
- Su, Z.Q.; Fang, H.; Hong, H.X.; Shi, L.M.; Zhang, W.Q.; Zhang, Y.Y. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014, 15, 523. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.B.; Lu, W.H.; Lin, Y.; Luo, J.Z. Development and application of transcriptome sequencing. Environ. Sci. Technol. 2018, 35, 20–26. [Google Scholar]
- Qi, Y.X.; Liu, Y.B.; Rong, W.H. RNA-Seq and its applications: A new technology for transcriptomics. Hereditas 2011, 33, 1191–1202. [Google Scholar] [CrossRef]
- Bhatt, G.; Gupta, A.; Rangan, L.; Limaye, A.M. Global transcriptome analysis reveals partial estrogen-like effects of karanjin in MCF-7 breast cancer cells. Gene 2022, 830, 146507. [Google Scholar] [CrossRef]
- Deng, Y.; Huang, Q.; Hu, L.; Liu, T.; Zheng, B.; Lu, D.; Guo, C.; Zhou, L. Enhanced exopolysaccharide yield and antioxidant activities of Schizophyllum Sommune fermented products by the addition of radix puerariae. RSC Adv. 2021, 11, 38219–38234. [Google Scholar] [CrossRef]
- Caseiro, C.; Ribeiro Dias, J.N.; Godinho de Andrade Fontes, C.M.; Bule, P. From cancer therapy to winemaking: The molecular structure and applications of β-glucans and β-1,3-glucanases. Int. J. Mol. Sci. 2022, 23, 3156. [Google Scholar] [CrossRef]
- Badalyan, S.M.; Barkhudaryan, A.; Rapior, S. Medicinal macrofungi as cosmeceuticals: A review. Int. J. Med. Mushrooms 2022, 24, 1–13. [Google Scholar] [CrossRef]
- Vuong, V.; Muthuramalingam, K.; Singh, V.; Choi, C.; Kim, Y.M.; Unno, T.; Cho, M. Schizophyllum commune-derived β-glucan improves intestinal health demonstrating protective effects against constipation and common metabolic disorders. Appl. Biol. Chem. 2022, 65, 9. [Google Scholar]
- Shen, Y.; Hu, C.; Zhang, H.; Jiang, H. Characteristics of three typical chinese highland barley varieties: Phenolic compounds and antioxidant activities. J. Food Biochem. 2018, 42, e12488. [Google Scholar] [CrossRef]
- Okonogi, S.; Kaewpinta, A.; Junmahasathien, T.; Yotsawimonwat, S. Effect of rice variety and modification on antioxidant and anti-inflammatory activities. Drug Discov. Ther. 2018, 12, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, T.; Fu, X.; Brennan, M.; Abbasi, A.M.; Zheng, B.; Liu, R.H. The use of an enzymatic extraction procedure for the enhancement of highland barley (Hordeum vulgare L.) phenolic and antioxidant compounds. Int. J. Food Sci. Tech. 2016, 51, 1916–1924. [Google Scholar] [CrossRef]
- Chang, Y.W.; Alli, I.; Konishi, Y.; Ziomek, E. Characterization of protein fractions from Chickpea (Cicer Arietinum L.) and Oat (Avena Sativa L.) seeds using proteomic techniques. Food Res. Int. 2011, 44, 3094–3104. [Google Scholar] [CrossRef]
- Abd Razak, D.L.; Abd Rashid, N.Y.; Jamaluddin, A.; Sharifudin, S.A.; Long, K. Enhancement of phenolic acid content and antioxidant activity of rice bran fermented with rhizopus oligosporus and monascus purpureus. Biocatal. Agric. Biotechnol. 2015, 4, 33–38. [Google Scholar] [CrossRef]
- Wu, H.; Liu, H.N.; Ma, A.M.; Zhou, J.Z.; Xia, X.D. Synergetic effects of lactobacillus plantarum and rhizopus oryzae on physicochemical, nutritional and antioxidant properties of whole-grain Oats (Avena Sativa L.) during solid-state fermentation. LWT 2022, 154, 112687. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, H.; Huang, Q.; Tu, L.; Hu, L.; Zheng, B.; Sun, H.; Lu, D.; Guo, C.; Zhou, L. Mechanism of longevity extension of caenorhabditis elegans induced by Schizophyllum Commune fermented supernatant with added radix puerariae. Front. Nutr. 2022, 9, 847064. [Google Scholar] [CrossRef]
- Liu, W.; Chen, Y.; Wang, Q. Attenuated and synergistic effects of liquid fermentation polysaccharide of Schizophyllum on tumor-bearing mice treated with chemotherapy. Lishizhen Med. Mater. Med. Res. 2012, 23, 321–322. [Google Scholar]
- Song, Y.R.; Han, A.R.; Park, S.G. Effect of enzyme-assisted extraction on the physicochemical properties and bioactive potential of lotus leaf polysaccharides. Int. J. Biol. Macromol. 2020, 153, 169–179. [Google Scholar] [CrossRef]
- You, C.H.; Heng, W.L.; Mallikarjuna, K. Dermato-protective properties of ergothioneine through induction of Nrf2/ARE-mediated antioxidant genes in UVA-irradiated Human keratinocytes. Free Radical Bio. Med. 2015, 86, 102–117. [Google Scholar]
- Li, Q.; Bai, D.; Qin, L.; Shao, M.; Zhang, S.; Yan, C.; Yu, G.; Hao, J. Protective effect of D-tetramannuronic acid tetrasodium salt on UVA-induced photo-aging in HaCaT cells. Biomed. Pharmacother. 2020, 126, 110094. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.X.; Rosales-Corral, S.; Galano, A.; Zhou, X.J.; Xu, B. Mitochondria: Central Organelles for Melatonin’s Antioxidant and Anti-Aging Actions. Molecules 2018, 23, 509. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Fang, J.; Wang, Z.; Song, Z.; Geng, J.; Wang, D.; Wang, C.; Li, M. Two Laminaria japonica Fermentation Broths Alleviate Oxidative Stress and Inflammatory Response Caused by UVB Damage: Photoprotective and Reparative Effects. Mar. Drugs 2022, 20, 650. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Kim, J.W.; Jo, E.H.; Moon, J.E.; Cha, H.; Chang, M.H.; Cho, H.T. In vitro and in vivo inhibitory effect of citrus junos tanaka peel extract against oxidative stress-induced apoptotic death of Lung cells. Antioxidants 2020, 9, 1231. [Google Scholar] [CrossRef]
- Zhou, F.; Huang, X.; Pan, Y.; Cao, D.; Liu, C.; Liu, Y.; Chen, A. Resveratrol protects HaCaT cells from ultraviolet B-induced photoaging via upregulation of HSP27 and modulation of mitochondrial caspase-dependent apoptotic pathway. Biochem. Biophys. Res. Commun. 2018, 499, 662–668. [Google Scholar] [CrossRef]
- Sies, H. Role of metabolic H2O2 generation: Redox signaling and oxidative stress. J. Biol. Chem. 2014, 289, 8735–8741. [Google Scholar] [CrossRef]
- Feehan, R.P.; Coleman, C.S.; Ebanks, S.; Lang, C.H.; Shantz, L.M. REDD1 interacts with AIF and regulates mitochondrial reactive oxygen species generation in the keratinocyte response to UVB. Biochem. Bioph. Res. Commun. 2022, 616, 56–62. [Google Scholar] [CrossRef]
- Baumann, L. Skin ageing and its treatment. J. Pathol. 2007, 211, 241–251. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, Y.; Fu, H.; Yao, F.; Liu, P.; Mo, Q.; Wang, D.; Zhao, D.; Wang, C.; Li, M. Physicochemical and anti-UVB-induced skin inflammatory properties of Lacticaseibacillus paracasei Subsp. paracasei SS-01 strain exopolysaccharide. Fermentation 2022, 8, 198. [Google Scholar]
- Alafiatayo, A.A.; Lai, K.S.; Ahmad, S.; Mahmood, M.; Shaharuddin, N.A. RNA-seq analysis revealed genes associated with UV-induced cell necrosis through MAPK/TNF-α pathways in human dermal fibroblast cells as an inducer of premature photoaging. Genomics 2020, 112, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xu, Q.; Chen, H. Transcriptome analysis of ultraviolet A-induced photoaging cells with deep sequencing. J. Dermatol. 2018, 45, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Barandalla, M.; Shi, H.; Xiao, H.; Colleoni, S.; Galli, C.; Lio, P.; Trotter, M.; Lazzari, G. Global gene expression profiling and senescence biomarker analysis of hESC exposed to H2O2 induced non-cytotoxic oxidative stress. Stem Cell Res. Ther. 2017, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Song, Y.; Zhang, H.; Ji, J.; Yang, J. IL-17 inhibits apoptosis of Hep-2 cells by regulating PI3K/AKT/FAS/FASL signaling pathway. Acta Univ. Med. Anhui 2018, 53, 1681–1684. [Google Scholar]
- Zha, L.; He, L.; Liang, Y.; Qin, H.; Yu, B.; Chang, L.; Xue, L. TNF-Alpha Contributes to Postmenopausal Osteoporosis by Synergistically Promoting RANKL-Induced Osteoclast Formation. Biomed. Pharmacother. 2018, 102, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.R.; Long, X.; Zhao, Q.; Zheng, Y.D.; Song, M.S.; Ma, S. A Single-Cell Transcriptomic Atlas of Human Skin Aging. Dev. Cell 2020, 3, 383. [Google Scholar] [CrossRef]
- Hao, D.; Wen, X.; Liu, L.; Wang, L.; Zhou, X.; Li, Y. Sanshool improves UVB-induced skin photodamage by targeting JAK2/STAT3-dependent autophagy. Cell Death Dis. 2019, 10, 485–494. [Google Scholar] [CrossRef]
- Ma, C.; Jia, X. Research progress on the role of extracellular matrix in skin cryopreservation. Acad. J. PLA Postgrad. Med. Sch. 2006, 27, 398–400. [Google Scholar]
- Miller, L.S.; Cho, J.S. Immunity against staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 2011, 11, 505–518. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, J.; Zhou, Z.; Wu, P.; Huo, R.; Wang, B.; Shen, Z.; Li, H.; Zhai, T.; Shen, B.; et al. CCN1, a pro-inflammatory factor, aggravates psoriasis skin lesions by promoting keratinocyte activation. J. Investig. Dermatol. 2015, 135, 2666–2675. [Google Scholar] [CrossRef]
- Kalutharage, N.K.; Rathnasinghe, D.L. A Study of Chitosan and Glucosamine Isolated from Sri Lankan Local Mushroom Schizophyllum Commune and Oyster Mushroom (Pleurotus Ostreatus). Mater. Today Proc. 2020, 23, 119–122. [Google Scholar] [CrossRef]
- Kumar, A.; Bharti, A.K.; Bezie, Y. Schizophyllum Commune: A Fungal Cell-Factory for Production of Valuable Metabolites and Enzymes. Bioresources 2022, 17, 5420–5436. [Google Scholar] [CrossRef]
- Patel, S.; Goyal, A. Recent developments in mushrooms as anti-cancer therapeutics: A review. 3 Biotech 2011, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yin, C.; Fan, X.; Ma, K.; Yao, F.; Zhou, R.; Shi, D.; Cheng, W.; Gao, H. Characterization of physicochemical and biological properties of Schizophyllum Commune polysaccharide extracted with different methods. Int. J. Biol. Macromol. 2020, 156, 1425–1434. [Google Scholar] [CrossRef]
- Meade, B.; Thome, K. International Food Security Assessment, 2017–2027; U.S. Department of Agriculture: Washington, DC, USA, 2017; pp. 26–37. [Google Scholar]
- Zhang, F.; Jiang, G.; Li, W.; Qin, S.; Li, H.; Gao, Z.; Cai, G.; Lin, T. MaxENT modeling for predicting the spatial distribution of three reptors in the Sanjiangyuan National Park. China Ecol. Evol. 2019, 9, 6643–6654. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, Q.; Li, J.; Zhao, S.; Qie, M.; Wu, X.; Bai, Y.; Zhao, Y. Study on the origin traceability of tibet highland barley (Hordeum Vulgare L.) based on its nutrients and mineral elements. Food Chem. 2021, 346, 128928. [Google Scholar] [CrossRef]
- Obadi, M.; Sun, J.; Xu, B. Highland barley: Chemical composition, bioactive compounds, health effects, and applications. Food Res. Int. 2021, 140, 110065. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Yan, J.; Dong, R.; Hu, X. Research progress on oat nutrients, functional properties and related products. Sci. Technol. Food Ind. 2022, 5, 1–14. [Google Scholar]
- Yang, C.; Chen, M.; Dai, T.; Chen, J. Research advances in functional properties and application of Oat β-glucan. J. Chin. Inst. Food Sci. Technol. 2021, 6, 301–311. [Google Scholar]

| Term | Sample | Raw Reads | Clean Reads | Total Reads | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|---|---|
| H2O2 | control1 | 52,006,260 | 51,489,806 | 51,489,806 | 1,585,446 (3.08%) | 48,550,893 (94.29%) |
| control2 | 51,434,366 | 51,007,382 | 51,007,382 | 1,656,100 (3.25%) | 48,073,211(94.25%) | |
| contro13 | 47,992,480 | 47,570,278 | 47,570,278 | 1,767,599 (3.72%) | 44,528,485 (93.61%) | |
| model1 | 42,375,544 | 41,851,304 | 41,851,304 | 2,523,225 (6.03%) | 37,561,116 (89.75%) | |
| model2 | 55,834,672 | 55,256,324 | 55,256,324 | 1,946,513 (3.52%) | 51,879,913 (93.89%) | |
| model3 | 53,168,944 | 52,654,864 | 52,654,864 | 1,852,768 (3.52%) | 49,508,846 (94.03%) | |
| UVA | control1 | 203,492,014 | 201,140,460 | 201,140,460 | 9,220,602 (4.58%) | 108,745,918 (54.06%) |
| control2 | 214,473,458 | 212,140,016 | 212,140,016 | 10,108,543 (4.77%) | 114,327,168 (53.89%) | |
| control3 | 201,264,476 | 199,240,206 | 199,240,206 | 8,290,552 (4.16%) | 103,655,047 (52.03%) | |
| model1 | 183,171,608 | 180,873,264 | 180,873,264 | 10,508,759 (5.81%) | 117542871 (64.99%) | |
| model2 | 219,499,948 | 216,959,018 | 216,959,018 | 12,593,146 (5.8%) | 137,233,440 (63.25%) | |
| model3 | 207,223,010 | 204,370,646 | 204,370,646 | 12,256,006 (6.0%) | 131,192,428 (64.19%) |
| Gene | Direction | Primer Pair Sequence (5′→3′) |
|---|---|---|
| GAPDH | F | TCAGACACCATGGGGAAGGT |
| R | TCCCGTTCTCAGCCATGTAG | |
| COL4A5 | F | CAAGGTCTACCAGGTCCAGAA |
| R | TCATTCCATTGAGACCCGGC | |
| FN1 | F | CCCAATTGAGTGCTTCATGCC |
| R | CCTCCAGAGCAAAGGGCTTA | |
| IGF2 | F | TCCTGTGAAAGAGACTTCCAG |
| R | GTCTCACTGGGGCGGTAAG | |
| COL1A1 | F | GAGGGCCAAGACGAAGACATC |
| R | CAGATCACGTCATCGCACAAC | |
| COL1A2 | F | GTTGCTGCTTGCAGTAACCTT |
| R | AGGGCCAAGTCCAACTCCTT | |
| PIK3R1 | F | ACCACTACCGGAATGAATCTCT |
| R | GGGATGTGCGGGTATATTCTTC |
| Gene_Id | Gene Name | Gene Description | Log2FC | P-Adjust | Function |
|---|---|---|---|---|---|
| ENSG00000187961 | KLHL17 | kelch like family member 17 | 0.457 | 8.97 × 10−5 | Cytoskeleton organization |
| ENSG00000125817 | CENPB | centromere protein B | 0.342 | 1.87 × 10−10 | Cytoskeleton organization |
| ENSG00000011426 | ANLN | anillin actin binding protein | −0.299 | 3.31 × 10−2 | Cytoskeleton organization |
| ENSG00000123384 | LRP1 | LDL receptor related protein 1 | −0.435 | 2.99 × 10−16 | Cytoskeleton organization |
| ENSG00000288380 | CRIPAK | cysteine rich PAK1 inhibitor | −0.401 | 1.97 × 10−2 | Cytoskeleton organization |
| ENSG00000275993 | SIK1B | salt inducible kinase 1B (putative) | −2.211 | 1.70 × 10−22 | Cytoskeleton organization |
| ENSG00000147202 | DIAPH2 | diaphanous related formin 2 | −0.334 | 1.34 × 10−8 | Cytoskeleton organization |
| ENSG00000160551 | TAOK1 | TAO kinase 1 | −0.264 | 4.71 × 10−9 | Cytoskeleton organization |
| ENSG00000075826 | SEC31B | SEC31 homolog B, COPII coat complex component | 0.370 | 6.71 × 10−5 | Transport |
| ENSG00000137700 | SLC37A4 | solute carrier family 37 member 4 | 0.485 | 1.26 × 10−3 | Transport |
| ENSG00000225697 | SLC26A6 | solute carrier family 26 member 6 | 0.448 | 3.28 × 10−12 | Transport |
| ENSG00000155287 | SLC25A28 | solute carrier family 25 member28 | 0.403 | 3.14 × 10−6 | Transport |
| ENSG00000153291 | SLC25A27 | solute carrier family 25 member 27 | 0.278 | 6.30 × 10−4 | Transport |
| ENSG00000213901 | SLC23A3 | solute carrier family 23 member 3 | 0.340 | 3.43 × 10−2 | Transport |
| ENSG00000197208 | SLC22A4 | solute carrier family 22 member4 | 0.292 | 2.76 × 10−5 | Transport |
| ENSG00000101194 | SLC17A9 | solute carrier family 17 member 9 | 0.421 | 2.03 × 10−5 | Transport |
| ENSG00000105643 | ARRDC2 | arrestin domain containing 2 | 0.276 | 4.49 × 10−2 | Transport |
| ENSG00000205593 | DENND6B | DENN domain containing 6B | 0.293 | 4.04 × 10−2 | Transport |
| ENSG00000157514 | TSC22D3 | TSC22 domain family member 3 | 0.285 | 7.88 × 10−4 | Transport |
| ENSG00000140104 | CLBA1 | clathrin binding box of aftiphilin containing 1 | 0.328 | 2.62 × 10−2 | Transport |
| ENSG00000106266 | SNX8 | sorting nexin 8 | 0.294 | 2.33 × 10−4 | Transport |
| ENSG00000274512 | TBC1D3L | TBC1 domain family member 3L | 0.377 | 2.85 × 10−3 | Transport |
| ENSG00000104886 | PLEKHJ1 | pleckstrin homology domain containing J1 | 0.311 | 9.15 × 10−3 | Transport |
| ENSG00000177096 | PHETA2 | PH domain containing endocytic trafficking adaptor 2 | 0.305 | 1.35 × 10−2 | Transport |
| ENSG00000257390 | AC023055.1 | novel protein | 0.467 | 1.20 × 10−3 | Transport |
| ENSG00000254852 | NPIPA2 | nuclear pore complex interacting protein family member A2 | 0.279 | 4.98 × 10−3 | Transport |
| ENSG00000095066 | HOOK2 | hook microtubule tethering protein 2 | 0.297 | 1.92 × 10−2 | Transport |
| ENSG00000101199 | ARFGAP1 | ADP ribosylation factor GTPase activating protein 1 | 0.280 | 9.43 × 10−7 | Transport |
| ENSG00000213983 | AP1G2 | adaptor related protein complex 1 subunit gamma 2 | 0.318 | 1.78 × 10−2 | Transport |
| ENSG00000181404 | WASHC1 | WASH complex subunit 1 | 0.450 | 6.60 × 10−18 | Transport |
| ENSG00000139190 | VAMP1 | vesicle associated membrane protein 1 | 0.601 | 5.83 × 10−6 | Transport |
| ENSG00000162341 | TPCN2 | two pore segment channel 2 | 0.292 | 1.43 × 10−2 | Transport |
| ENSG00000114268 | PFKFB4 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | 0.632 | 1.33 × 10−3 | Transport |
| ENSG00000196655 | TRAPPC4 | trafficking protein particle complex 4 | 0.275 | 4.52 × 10−2 | Transport |
| ENSG00000102287 | GABRE | gamma-aminobutyric acid type A receptor epsilon subunit | 0.317 | 4.13 × 10−4 | Transport |
| ENSG00000225663 | MCRIP1 | MAPK regulated corepressor interacting protein 1 | 0.270 | 4.63 × 10−3 | Transport |
| ENSG00000081692 | JMJD4 | jumonji domain containing 4 | 0.353 | 2.60 × 10−2 | Transport |
| ENSG00000168026 | TTC21A | tetratricopeptide repeat domain 21A | 0.383 | 4.65 × 10−2 | Transport |
| ENSG00000108932 | SLC16A6 | solute carrier family 16 member 6 | −0.416 | 1.95 × 10−4 | Transport |
| ENSG00000117479 | SLC19A2 | solute carrier family 19 member 2 | −1.360 | 6.72 × 10−42 | Transport |
| ENSG00000140199 | SLC12A6 | solute carrier family 12 member6 | −0.279 | 2.21 × 10−5 | Transport |
| ENSG00000171488 | LRRC8C | leucine rich repeat containing 8 VRAC subunit C | −0.269 | 2.37 × 10−2 | Transport |
| ENSG00000169446 | MMGT1 | membrane magnesium transporter 1 | −0.306 | 4.40 × 10−4 | Transport |
| ENSG00000228253 | MT-ATP8 | mitochondrially encoded ATP synthase membrane subunit 8 | −0.567 | 6.75 × 10−4 | Transport |
| ENSG00000198899 | MT-ATP6 | mitochondrially encoded ATP synthase membrane subunit 6 | −0.457 | 2.44 × 10−3 | Transport |
| ENSG00000165714 | BORCS5 | BLOC-1 related complex subunit 5 | −0.285 | 4.97 × 10−3 | Transport |
| ENSG00000181333 | HEPHL1 | hephaestin like 1 | −0.285 | 3.27 × 10−3 | Transport |
| ENSG00000107771 | CCSER2 | coiled-coil serine rich protein 2 | −0.272 | 1.36 × 10−6 | Transport |
| ENSG00000115657 | ABCB6 | ATP binding cassette subfamily B member 6 (Langereis blood group) | 0.303 | 7.37 × 10−4 | Cellular and extracellular matrix |
| ENSG00000117425 | PTCH2 | patched 2 | 0.285 | 1.16 × 10−2 | Cellular and extracellular matrix |
| ENSG00000125551 | PLGLB2 | plasminogen like B2 | 0.383 | 1.82 × 10−2 | Cellular and extracellular matrix |
| ENSG00000164877 | MICALL2 | MICAL like 2 | 0.285 | 9.72 × 10−3 | Cellular and extracellular matrix |
| ENSG00000156535 | CD109 | CD109 molecule | −0.399 | 1.50 × 10−25 | Cellular and extracellular matrix |
| ENSG00000185022 | MAFF | MAF bZIP transcription factor F | −0.968 | 5.78 × 10−80 | Cellular and extracellular matrix |
| ENSG00000166147 | FBN1 | fibrillin 1 | −0.421 | 4.89 × 10−30 | Cellular and extracellular matrix |
| ENSG00000165240 | ATP7A | ATPase copper transporting alpha | −0.277 | 2.43 × 10−4 | Cellular and extracellular matrix |
| ENSG00000142871 | CCN1 | cellular communication network factor 1 | −0.410 | 4.51 × 10−25 | Cellular and extracellular matrix |
| ENSG00000038427 | VCAN | versican | −0.492 | 2.64 × 10−13 | Cellular and extracellular matrix |
| ENSG00000116962 | NID1 | nidogen 1 | −0.356 | 1.04 × 10−21 | Cellular and extracellular matrix |
| ENSG00000187955 | COL14A1 | collagen type XIV alpha 1 chain | −0.303 | 1.32 × 10−3 | Cellular and extracellular matrix |
| ENSG00000196569 | LAMA2 | laminin subunit alpha 2 | −0.407 | 5.66 × 10−28 | Cellular and extracellular matrix |
| ENSG00000142798 | HSPG2 | heparan sulfate proteoglycan 2 | −0.440 | 8.61 × 10−14 | Cellular and extracellular matrix |
| ENSG00000123500 | COL10A1 | collagen type X alpha 1 chain | −0.282 | 2.54 × 10−2 | Cellular and extracellular matrix |
| ENSG00000187498 | COL4A1 | collagen type IV alpha 1 chain | −0.265 | 7.12 × 10−12 | Cellular and extracellular matrix |
| ENSG00000115414 | FN1 | fibronectin 1 | −0.446 | 1.23 × 10−34 | Cellular and extracellular matrix |
| ENSG00000163359 | COL6A3 | collagen type VI alpha 3 chain | −0.412 | 3.41 × 10−38 | Cellular and extracellular matrix |
| ENSG00000103196 | CRISPLD2 | cysteine rich secretory protein LCCL domain containing 2 | −0.321 | 6.09 × 10−4 | Cellular and extracellular matrix |
| ENSG00000111799 | COL12A1 | collagen type XII alpha 1 chain | −0.388 | 7.55 × 10−28 | Cellular and extracellular matrix |
| ENSG00000080561 | MID2 | midline 2 | −0.275 | 8.28 × 10−4 | Cellular and extracellular matrix |
| ENSG00000171223 | JUNB | JunB proto-oncogene, AP-1 transcription factor subunit | −1.704 | 9.13 × 10−173 | Cellular and extracellular matrix |
| ENSG00000101825 | MXRA5 | matrix remodeling associated 5 | −0.387 | 8.91 × 10−32 | Cellular and extracellular matrix |
| ENSG00000113369 | ARRDC3 | arrestin domain containing 3 | −0.289 | 6.30 × 10−4 | Cellular and extracellular matrix |
| ENSG00000138759 | FRAS1 | Fraser extracellular matrix complex subunit 1 | −0.447 | 4.82 × 10−14 | Cellular and extracellular matrix |
| ENSG00000106780 | MEGF9 | multiple EGF like domains 9 | −0.284 | 3.72 × 10−3 | Cellular and extracellular matrix |
| ENSG00000119681 | LTBP2 | latent transforming growth factor beta binding protein 2 | −0.270 | 3.80 × 10−10 | Cellular and extracellular matrix |
| ENSG00000182179 | UBA7 | ubiquitin like modifier activating enzyme 7 | 0.278 | 6.06 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000148399 | DPH7 | diphthamide biosynthesis 7 | 0.345 | 5.72 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167733 | HSD11B1L | hydroxysteroid 11-beta dehydrogenase 1 like | 0.284 | 4.11 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000104852 | SNRNP70 | small nuclear ribonucleoprotein U1 subunit 70 | 0.340 | 6.65 × 10−17 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000125901 | MRPS26 | mitochondrial ribosomal protein S26 | 0.289 | 3.13 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000183513 | COA5 | cytochrome c oxidase assembly factor 5 | 0.273 | 4.15 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000165792 | METTL17 | methyltransferase like 17 | 0.401 | 5.91 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000100429 | HDAC10 | histone deacetylase 10 | 0.274 | 1.11 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000141519 | CCDC40 | coiled-coil domain containing 40 | 0.383 | 3.27 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000172828 | CES3 | carboxylesterase 3 | 0.506 | 2.88 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139631 | CSAD | cysteine sulfinic acid decarboxylase | 0.311 | 8.25 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000112578 | BYSL | bystin like | 0.331 | 1.17 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000213398 | LCAT | lecithin-cholesterol acyltransferase | 0.488 | 3.70 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000232653 | GOLGA8N | golgin A8 family member N | 0.348 | 2.84 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000145020 | AMT | aminomethyltransferase | 0.388 | 4.57 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000178038 | ALS2CL | ALS2 C-terminal like | 0.409 | 1.90 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000179886 | TIGD5 | tigger transposable element derived 5 | 0.313 | 4.31 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128710 | HOXD10 | homeobox D10 | 0.319 | 3.71 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000108641 | B9D1 | B9 domain containing 1 | 0.286 | 3.29 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197774 | EME2 | essential meiotic structure-specific endonuclease subunit 2 | 0.385 | 8.64 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000172732 | MUS81 | MUS81 structure-specific endonuclease subunit | 0.276 | 7.00 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000039650 | PNKP | polynucleotide kinase 3′-phosphatase | 0.284 | 1.49 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000129250 | KIF1C | kinesin family member 1C | 0.366 | 1.72 × 10−17 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000010292 | NCAPD2 | non-SMC condensin I complex subunit D2 | 0.328 | 2.36 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000146063 | TRIM41 | tripartite motif containing 41 | 0.332 | 1.10 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000140983 | RHOT2 | ras homolog family member T2 | 0.328 | 5.44 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000163482 | STK36 | serine/threonine kinase 36 | 0.273 | 4.31 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000138834 | MAPK8IP3 | mitogen-activated protein kinase 8 interacting protein 3 | 0.309 | 1.58 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000148120 | AOPEP | aminopeptidase O (putative) | 0.469 | 1.77 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167100 | SAMD14 | sterile alpha motif domain containing 14 | 0.270 | 4.18 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000110455 | ACCS | 1-aminocyclopropane-1-carboxylate synthase homolog (inactive) | 0.357 | 3.47 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000158062 | UBXN11 | UBX domain protein 11 | 0.303 | 1.60 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000107829 | FBXW4 | F-box and WD repeat domain containing 4 | 0.347 | 1.00 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000138835 | RGS3 | regulator of G protein signaling 3 | 0.343 | 9.63 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000232119 | MCTS1 | MCTS1 re-initiation and release factor | 0.278 | 1.81 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167280 | ENGASE | endo-beta-N-acetylglucosaminidase | 0.409 | 1.64 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000149716 | LTO1 | LTO1 maturation factor of ABCE1 | 0.307 | 4.33 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000250151 | ARPC4-TTLL3 | ARPC4-TTLL3 readthrough | 0.354 | 1.51 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000073605 | GSDMB | gasdermin B | 0.448 | 1.17 × 10−8 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000148824 | MTG1 | mitochondrial ribosome associated GTPase 1 | 0.303 | 1.28 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137504 | CREBZF | CREB/ATF bZIP transcription factor | 0.287 | 3.31 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000180902 | D2HGDH | D-2-hydroxyglutarate dehydrogenase | 0.274 | 3.10 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000042429 | MED17 | mediator complex subunit 17 | 0.408 | 1.86 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000149930 | TAOK2 | TAO kinase 2 | 0.456 | 8.73 × 10−17 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139546 | TARBP2 | TARBP2 subunit of RISC loading complex | 0.310 | 9.44 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000116001 | TIA1 | TIA1 cytotoxic granule associated RNA binding protein | 0.332 | 8.51 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115053 | NCL | nucleolin | 0.275 | 1.29 × 10−12 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000173559 | NABP1 | nucleic acid binding protein 1 | 0.285 | 4.36 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000198585 | NUDT16 | nudix hydrolase 16 | 0.317 | 4.87 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167393 | PPP2R3B | protein phosphatase 2 regulatory subunit B’’beta | 0.331 | 2.50 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000141456 | PELP1 | proline, glutamate and leucine rich protein 1 | 0.337 | 3.84 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128159 | TUBGCP6 | tubulin gamma complex associated protein 6 | 0.283 | 1.47 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000078399 | HOXA9 | homeobox A9 | 0.355 | 4.10 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000213339 | QTRT1 | queuine tRNA-ribosyltransferase catalytic subunit 1 | 0.292 | 4.27 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000177192 | PUS1 | pseudouridine synthase 1 | 0.275 | 4.58 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000144785 | AC073896.1 | novel protein | 0.382 | 4.61 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000185024 | BRF1 | BRF1 RNA polymerase III transcription initiation factor subunit | 0.301 | 7.22 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000228049 | POLR2J2 | RNA polymerase II subunit J2 | 0.339 | 3.50 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197782 | ZNF780A | zinc finger protein 780A | 0.281 | 1.47 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000147789 | ZNF7 | zinc finger protein 7 | 0.282 | 1.60 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000171163 | ZNF692 | zinc finger protein 692 | 0.390 | 1.49 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000176024 | ZNF613 | zinc finger protein 613 | 0.405 | 3.87 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000180626 | ZNF594 | zinc finger protein 594 | 0.552 | 8.53 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000161551 | ZNF577 | zinc finger protein 577 | 0.309 | 9.79 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167785 | ZNF558 | zinc finger protein 558 | 0.330 | 6.43 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000152433 | ZNF547 | zinc finger protein 547 | 0.503 | 2.07 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000144026 | ZNF514 | zinc finger protein 514 | 0.270 | 4.19 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000196653 | ZNF502 | zinc finger protein 502 | 0.320 | 4.78 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000083817 | ZNF416 | zinc finger protein 416 | 0.337 | 4.62 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000175213 | ZNF408 | zinc finger protein 408 | 0.394 | 1.40 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000130684 | ZNF337 | zinc finger protein 337 | 0.340 | 6.05 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000083812 | ZNF324 | zinc finger protein 324 | 0.378 | 1.21 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000182986 | ZNF320 | zinc finger protein 320 | 0.406 | 6.47 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000205903 | ZNF316 | zinc finger protein 316 | 0.313 | 1.86 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000174652 | ZNF266 | zinc finger protein 266 | 0.433 | 1.55 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167380 | ZNF226 | zinc finger protein 226 | 0.337 | 1.45 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197841 | ZNF181 | zinc finger protein 181 | 0.298 | 3.56 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000154957 | ZNF18 | zinc finger protein 18 | 0.339 | 2.32 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000179909 | ZNF154 | zinc finger protein 154 | 0.341 | 3.55 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000213762 | ZNF134 | zinc finger protein 134 | 0.280 | 2.08 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000125846 | ZNF133 | zinc finger protein 133 | 0.325 | 1.48 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000204946 | ZNF783 | zinc finger family member 783 | 0.265 | 1.58 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197114 | ZGPAT | zinc finger CCCH-type and G-patch domain containing | 0.270 | 1.89 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000140987 | ZSCAN32 | zinc finger and SCAN domain containing 32 | 0.486 | 6.71 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000114853 | ZBTB47 | zinc finger and BTB domain containing 47 | 0.372 | 4.54 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000099899 | TRMT2A | tRNA methyltransferase 2 homolog A | 0.293 | 7.14 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000100038 | TOP3B | DNA topoisomerase III beta | 0.295 | 1.09 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000198056 | PRIM1 | DNA primase subunit 1 | 0.461 | 1.32 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000178028 | DMAP1 | DNA methyltransferase 1 associated protein 1 | 0.311 | 4.49 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000168209 | DDIT4 | DNA damage inducible transcript 4 | 0.278 | 7.35 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000221978 | CCNL2 | cyclin L2 | 0.429 | 2.31 × 10−23 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000250506 | CDK3 | cyclin dependent kinase 3 | 0.535 | 6.59 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000156345 | CDK20 | cyclin dependent kinase 20 | 0.287 | 3.81 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000248333 | CDK11B | cyclin dependent kinase 11B | 0.299 | 8.71 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000185324 | CDK10 | cyclin dependent kinase 10 | 0.344 | 1.84 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000130305 | NSUN5 | NOP2/Sun RNA methyltransferase 5 | 0.264 | 4.48 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000134186 | PRPF38B | pre-mRNA processing factor 38B | 0.269 | 6.68 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000103168 | TAF1C | TATA-box binding protein associated factor, RNA polymerase I subunit C | 0.324 | 7.71 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000178718 | RPP25 | ribonuclease P and MRP subunit p25 | 0.280 | 3.45 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000264668 | AC138696.1 | novel protein | 0.384 | 3.75 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000041988 | THAP3 | THAP domain containing 3 | 0.347 | 3.09 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000104129 | DNAJC17 | DnaJ heat shock protein family (Hsp40) member C17 | 0.364 | 3.96 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000187531 | SIRT7 | sirtuin 7 | 0.386 | 1.73 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000108479 | GALK1 | galactokinase 1 | 0.311 | 9.61 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000140400 | MAN2C1 | mannosidase alpha class 2C member 1 | 0.384 | 3.49 × 10−12 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000142102 | PGGHG | protein-glucosylgalactosylhydroxylysine glucosidase | 0.388 | 6.40 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000181274 | FRAT2 | FRAT regulator of WNT signaling pathway 2 | 0.331 | 4.77 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000215788 | TNFRSF25 | TNF receptor superfamily member 25 | 0.558 | 1.63 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000153179 | RASSF3 | Ras association domain family member 3 | 0.326 | 5.71 × 10−8 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115875 | SRSF7 | serine and arginine rich splicing factor 7 | 0.293 | 1.90 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000162910 | MRPL55 | mitochondrial ribosomal protein L55 | 0.295 | 4.41 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000172586 | CHCHD1 | coiled-coil-helix-coiled-coil-helix domain containing 1 | 0.415 | 2.99 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000136938 | ANP32B | acidic nuclear phosphoprotein 32 family member B | 0.309 | 1.46 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000177595 | PIDD1 | p53-induced death domain protein 1 | 0.300 | 2.42 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000129473 | BCL2L2 | BCL2 like 2 | 0.273 | 1.67 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000184207 | PGP | phosphoglycolate phosphatase | 0.409 | 5.30 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000136271 | DDX56 | DEAD-box helicase 56 | 0.292 | 5.37 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000106404 | CLDN15 | claudin 15 | 0.436 | 2.44 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000130734 | ATG4D | autophagy related 4D cysteine peptidase | 0.360 | 4.83 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000256825 | AC026786.1 | novel protein | 0.466 | 1.84 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000113240 | CLK4 | CDC like kinase 4 | 0.348 | 8.35 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000176444 | CLK2 | CDC like kinase 2 | 0.319 | 4.36 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000114735 | HEMK1 | HemK methyltransferase family member 1 | 0.313 | 9.02 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000135414 | GDF11 | growth differentiation factor 11 | 0.306 | 2.35 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000141013 | GAS8 | growth arrest specific 8 | 0.395 | 1.02 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000065268 | WDR18 | WD repeat domain 18 | 0.321 | 9.31 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000071246 | VASH1 | vasohibin 1 | 0.374 | 2.50 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000005007 | UPF1 | UPF1 RNA helicase and ATPase | 0.275 | 4.60 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000126790 | L3HYPDH | trans-L-3-hydroxyproline dehydratase | 0.376 | 9.63 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000102871 | TRADD | TNFRSF1A associated via death domain | 0.399 | 2.50 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000146109 | ABT1 | activator of basal transcription 1 | 0.263 | 1.99 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000264343 | NOTCH2NLA | notch 2 N-terminal like A | 0.540 | 1.22 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000258674 | AC011448.1 | novel protein | 0.432 | 3.06 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000149451 | ADAM33 | ADAM metallopeptidase domain 33 | 0.382 | 5.58 × 10−20 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000215041 | NEURL4 | neuralized E3 ubiquitin protein ligase 4 | 0.308 | 1.02 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000150401 | DCUN1D2 | defective in cullin neddylation 1 domain containing 2 | 0.282 | 1.23 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000251287 | ALG1L2 | ALG1 chitobiosyldiphosphodolichol beta-mannosyltransferase like 2 | 0.356 | 6.01 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000073169 | SELENOO | selenoprotein O | 0.364 | 1.01 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000234616 | JRK | Jrk helix-turn-helix protein | 0.374 | 3.11 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000105559 | PLEKHA4 | pleckstrin homology domain containing A4 | 0.442 | 5.49 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000007392 | LUC7L | LUC7 like | 0.322 | 8.83 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000131584 | ACAP3 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 | 0.334 | 3.35 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000175137 | SH3BP5L | SH3 binding domain protein 5 like | 0.422 | 5.89 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000159692 | CTBP1 | C-terminal binding protein 1 | 0.342 | 2.13 × 10−13 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000173706 | HEG1 | heart development protein with EGF like domains 1 | −0.277 | 5.78 × 10−14 | Metablism/proliferation/regulation |
| ENSG00000155008 | APOOL | apolipoprotein O like | −0.267 | 3.99 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000178385 | PLEKHM3 | pleckstrin homology domain containing M3 | −0.264 | 1.23 × 10−3 | Metablism/proliferation/regulation |
| ENSG0000017207 | EIF2AK3 | eukaryotic translation initiation factor 2 alpha kinase 3 | −0.307 | 2.27 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000106392 | C1GALT1 | core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1 | −0.268 | 1.02 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000165195 | PIGA | phosphatidylinositol glycan anchor biosynthesis class A | −0.726 | 1.08 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000155090 | KLF10 | Kruppel like factor 10 | −1.136 | 1.11 × 10−43 | Metablism/proliferation/regulation |
| ENSG00000144655 | CSRNP1 | cysteine and serine rich nuclear protein 1 | −1.225 | 4.13 × 10−35 | Metablism/proliferation/regulation |
| ENSG00000255112 | CHMP1B | charged multivesicular body protein 1B | −0.526 | 1.54 × 10−16 | Metablism/proliferation/regulation |
| ENSG00000179241 | LDLRAD3 | low density lipoprotein receptor class A domain containing 3 | −0.272 | 1.15 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000108582 | CPD | carboxypeptidase D | −0.308 | 1.62 × 10−15 | Metablism/proliferation/regulation |
| ENSG00000176641 | RNF152 | ring finger protein 152 | −0.326 | 7.13 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000136542 | GALNT5 | polypeptide N-acetylgalactosaminyltransferase 5 | −0.264 | 7.28 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000203814 | HIST2H2BF | histone cluster 2 H2B family member f | −0.332 | 3.21 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000162924 | REL | REL proto-oncogene, NF-kB subunit | −0.591 | 5.43 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000160888 | IER2 | immediate early response 2 | −1.137 | 6.07 × 10−73 | Metablism/proliferation/regulation |
| ENSG00000087074 | PPP1R15A | protein phosphatase 1 regulatory subunit 15A | −0.598 | 1.91 × 10−38 | Metablism/proliferation/regulation |
| ENSG00000136158 | SPRY2 | sprouty RTK signaling antagonist 2 | −0.356 | 8.30 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000137075 | RNF38 | ring finger protein 38 | −0.266 | 3.06 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000166340 | TPP1 | tripeptidyl peptidase 1 | −0.341 | 1.59 × 10−9 | Metablism/proliferation/regulation |
| ENSG00000130164 | LDLR | low density lipoprotein receptor | −0.282 | 9.25 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000112245 | PTP4A1 | protein tyrosine phosphatase 4A1 | −0.303 | 1.01 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000119138 | KLF9 | Kruppel like factor 9 | −0.409 | 7.12 × 10−12 | Metablism/proliferation/regulation |
| ENSG00000134107 | BHLHE40 | basic helix-loop-helix family member e40 | −0.597 | 7.53 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000115520 | COQ10B | coenzyme Q10B | −0.414 | 2.48 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000049323 | LTBP1 | latent transforming growth factor beta binding protein 1 | −0.308 | 1.58 × 10−15 | Metablism/proliferation/regulation |
| ENSG00000138166 | DUSP5 | dual specificity phosphatase 5 | −0.738 | 6.55 × 10−23 | Metablism/proliferation/regulation |
| ENSG00000130513 | GDF15 | growth differentiation factor 15 | −0.915 | 1.32 × 10−35 | Metablism/proliferation/regulation |
| ENSG00000119986 | AVPI1 | arginine vasopressin induced 1 | −0.734 | 1.12 × 10−21 | Metablism/proliferation/regulation |
| ENSG00000157168 | NRG1 | neuregulin 1 | −0.295 | 2.04 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000166225 | FRS2 | fibroblast growth factor receptor substrate 2 | −0.292 | 7.11 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000197081 | IGF2R | insulin like growth factor 2 receptor | −0.264 | 3.17 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000136527 | TRA2B | transformer 2 beta homolog | −0.421 | 2.33 × 10−14 | Metablism/proliferation/regulation |
| ENSG00000283782 | AC116366.3 | novel protein | −0.435 | 5.49 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000164220 | F2RL2 | coagulation factor II thrombin receptor like 2 | −0.303 | 1.82 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000185483 | ROR1 | receptor tyrosine kinase like orphan receptor 1 | −0.272 | 2.88 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000080200 | CRYBG3 | crystallin beta-gamma domain containing 3 | −0.468 | 3.60 × 10−16 | Metablism/proliferation/regulation |
| ENSG00000162433 | AK4 | adenylate kinase 4 | −0.266 | 1.58 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000178607 | ERN1 | endoplasmic reticulum to nucleus signaling 1 | −0.334 | 4.90 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000004799 | PDK4 | pyruvate dehydrogenase kinase 4 | −1.212 | 9.25 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000155816 | FMN2 | formin 2 | −0.275 | 3.38 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000132475 | H3F3B | H3 histone family member 3B | −0.400 | 6.12 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000184260 | HIST2H2AC | histone cluster 2 H2A family member c | −0.362 | 6.93 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000272196 | HIST2H2AA4 | histone cluster 2 H2A family member a4 | −0.754 | 4.41 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000105835 | NAMPT | nicotinamide phosphoribosyltransferase | −0.700 | 1.23 × 10−34 | Metablism/proliferation/regulation |
| ENSG00000143384 | MCL1 | MCL1 apoptosis regulator, BCL2 family member | −0.436 | 1.54 × 10−22 | Metablism/proliferation/regulation |
| ENSG00000067082 | KLF6 | Kruppel like factor 6 | −0.645 | 2.26 × 10−44 | Metablism/proliferation/regulation |
| ENSG00000177606 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | −1.114 | 2.64 × 10−153 | Metablism/proliferation/regulation |
| ENSG00000153936 | HS2ST1 | heparan sulfate 2-O-sulfotransferase 1 | −0.264 | 4.02 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000168621 | GDNF | glial cell derived neurotrophic factor | −0.518 | 3.74 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000175592 | FOSL1 | FOS like 1, AP-1 transcription factor subunit | −0.568 | 2.02 × 10−19 | Metablism/proliferation/regulation |
| ENSG00000108306 | FBXL20 | F-box and leucine rich repeat protein 20 | −0.264 | 6.71 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000174010 | KLHL15 | kelch like family member 15 | −0.339 | 8.75 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000118263 | KLF7 | Kruppel like factor 7 | −0.415 | 2.68 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000144959 | NCEH1 | neutral cholesterol ester hydrolase 1 | −0.401 | 1.07 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000167470 | MIDN | midnolin | −0.473 | 1.19 × 10−15 | Metablism/proliferation/regulation |
| ENSG00000069667 | RORA | RAR related orphan receptor A | −0.292 | 1.71 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000075213 | SEMA3A | semaphorin 3A | −0.321 | 2.38 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000176542 | USF3 | upstream transcription factor family member 3 | −0.359 | 2.91 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000100354 | TNRC6B | trinucleotide repeat containing adaptor 6B | −0.265 | 3.40 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000185650 | ZFP36L1 | ZFP36 ring finger protein like 1 | −0.377 | 3.07 × 10−20 | Metablism/proliferation/regulation |
| ENSG00000122641 | INHBA | inhibin subunit beta A | −0.301 | 9.16 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000092969 | TGFB2 | transforming growth factor beta 2 | −0.309 | 7.30 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000165997 | ARL5B | ADP ribosylation factor like GTPase 5B | −0.828 | 2.28 × 10−25 | Metablism/proliferation/regulation |
| ENSG00000221869 | CEBPD | CCAAT enhancer binding protein delta | −0.326 | 1.21 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000136731 | UGGT1 | UDP-glucose glycoprotein glucosyltransferase 1 | −0.366 | 1.03 × 10−17 | Metablism/proliferation/regulation |
| ENSG00000148737 | TCF7L2 | transcription factor 7 like 2 | −0.339 | 2.90 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000152377 | SPOCK1 | SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 | −0.323 | 7.57 × 10−14 | Metablism/proliferation/regulation |
| ENSG00000124813 | RUNX2 | RUNX family transcription factor 2 | −0.313 | 1.99 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000143190 | POU2F1 | POU class 2 homeobox 1 | −0.331 | 1.05 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000184384 | MAML2 | mastermind like transcriptional coactivator 2 | −0.314 | 3.02 × 10−12 | Metablism/proliferation/regulation |
| ENSG00000128342 | LIF | LIF interleukin 6 family cytokine | −0.763 | 2.73 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000120616 | EPC1 | enhancer of polycomb homolog 1 | −0.321 | 9.31 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000175197 | DDIT3 | DNA damage inducible transcript 3 | −0.427 | 6.45 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000171681 | ATF7IP | activating transcription factor 7 interacting protein | −0.277 | 9.25 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000162772 | ATF3 | activating transcription factor 3 | −2.877 | 3.76 × 10−175 | Metablism/proliferation/regulation |
| ENSG00000003989 | SLC7A2 | solute carrier family 7 member2 | −0.359 | 4.70 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000131389 | SLC6A6 | solute carrier family 6 member6 | −0.269 | 2.71 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000059804 | SLC2A3 | solute carrier family 2 member3 | −1.000 | 6.82 × 10−60 | Metablism/proliferation/regulation |
| ENSG00000155850 | SLC26A2 | solute carrier family 26 member2 | −0.266 | 1.54 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000118596 | SLC16A7 | solute carrier family 16 member7 | −0.389 | 1.14 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000163660 | CCNL1 | cyclin L1 | −1.027 | 4.70 × 10−78 | Metablism/proliferation/regulation |
| ENSG00000182263 | FIGN | fidgetin, microtubule severing factor | −0.360 | 2.43 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000137331 | IER3 | immediate early response 3 | −0.884 | 1.62 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000143878 | RHOB | ras homolog family member B | −1.048 | 2.20 × 10−87 | Metablism/proliferation/regulation |
| ENSG00000123975 | CKS2 | CDC28 protein kinase regulatory subunit 2 | −0.448 | 1.76 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000102781 | KATNAL1 | katanin catalytic subunit A1 like 1 | −0.274 | 3.80 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000185621 | LMLN | leishmanolysin like peptidase | −0.293 | 4.91 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000204131 | NHSL2 | NHS like 2 | −0.292 | 1.07 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000134954 | ETS1 | ETS proto-oncogene 1, transcription factor | −0.298 | 8.39 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000105810 | CDK6 | cyclin dependent kinase 6 | −0.306 | 2.07 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000204524 | ZNF805 | zinc finger protein 805 | −0.310 | 4.10 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000197483 | ZNF628 | zinc finger protein 628 | −0.439 | 1.94 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000197714 | ZNF460 | zinc finger protein 460 | −0.305 | 3.43 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000130844 | ZNF331 | zinc finger protein 331 | −0.277 | 5.71 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000285253 | AC090517.4 | zinc finger protein 280D | −0.444 | 2.01 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000185947 | ZNF267 | zinc finger protein 267 | −0.284 | 3.90 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000091656 | ZFHX4 | zinc finger homeobox 4 | −0.313 | 1.06 × 10−9 | Metablism/proliferation/regulation |
| ENSG00000180776 | ZDHHC20 | zinc finger DHHC-type containing 20 | −0.287 | 5.38 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000163874 | ZC3H12A | zinc finger CCCH-type containing 12A | −0.274 | 3.54 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000173276 | ZBTB21 | zinc finger and BTB domain containing 21 | −0.288 | 1.05 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000030419 | IKZF2 | IKAROS family zinc finger 2 | −0.412 | 4.37 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000164122 | ASB5 | ankyrin repeat and SOCS box containing 5 | −0.269 | 6.59 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000113448 | PDE4D | phosphodiesterase 4D | −0.374 | 1.92 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000184588 | PDE4B | phosphodiesterase 4B | −0.474 | 2.45 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000142892 | PIGK | phosphatidylinositol glycan anchor biosynthesis class K | −0.264 | 9.96 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000118523 | CCN2 | cellular communication network factor 2 | −0.313 | 3.41 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000112419 | PHACTR2 | phosphatase and actin regulator 2 | −0.281 | 1.95 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000179094 | PER1 | period circadian regulator 1 | −0.443 | 1.38 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000106460 | TMEM106B | transmembrane protein 106B | −0.311 | 8.77 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000182752 | PAPPA | pappalysin 1 | −0.354 | 1.08 × 10−26 | Metablism/proliferation/regulation |
| ENSG00000139496 | NUP58 | nucleoporin 58 | −0.328 | 2.66 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000119508 | NR4A3 | nuclear receptor subfamily 4 group A member 3 | −3.256 | 9.06 × 10−169 | Metablism/proliferation/regulation |
| ENSG00000153234 | NR4A2 | nuclear receptor subfamily 4 group A member 2 | −3.941 | 1.48 × 10−78 | Metablism/proliferation/regulation |
| ENSG00000123358 | NR4A1 | nuclear receptor subfamily 4 group A member 1 | −3.834 | 4.26 × 10−130 | Metablism/proliferation/regulation |
| ENSG00000165030 | NFIL3 | nuclear factor, interleukin 3 regulated | −0.605 | 9.27 × 10−18 | Metablism/proliferation/regulation |
| ENSG00000102908 | NFAT5 | nuclear factor of activated T cells 5 | −0.312 | 1.58 × 10−15 | Metablism/proliferation/regulation |
| ENSG00000162599 | NFIA | nuclear factor I A | −0.312 | 1.38 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000141458 | NPC1 | NPC intracellular cholesterol transporter 1 | −0.649 | 6.52 × 10−53 | Metablism/proliferation/regulation |
| ENSG00000284057 | AP001273.2 | novel protein, C11orf54-MED17 readthrough | −0.356 | 2.99 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000138336 | TET1 | tet methylcytosine dioxygenase 1 | −0.348 | 3.53 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000163960 | UBXN7 | UBX domain protein 7 | −0.265 | 1.61 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000172059 | KLF11 | Kruppel like factor 11 | −0.437 | 8.40 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000196233 | LCOR | ligand dependent nuclear receptor corepressor | −0.382 | 2.07 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000004776 | HSPB6 | heat shock protein family B (small) member 6 | −0.286 | 1.62 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000177570 | SAMD12 | sterile alpha motif domain containing 12 | −0.334 | 7.44 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000197620 | CXorf40A | chromosome X open reading frame 40A | −0.320 | 8.98 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000198142 | SOWAHC | sosondowah ankyrin repeat domain family member C | −0.427 | 6.40 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000154175 | ABI3BP | ABI family member 3 binding protein | −0.352 | 8.25 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000280987 | MATR3 | matrin 3 | −0.323 | 4.21 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000164296 | TIGD6 | tigger transposable element derived 6 | −0.272 | 2.37 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000205189 | ZBTB10 | zinc finger and BTB domain containing 10 | −0.266 | 6.94 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000151967 | SCHIP1 | schwannomin interacting protein 1 | −0.628 | 3.95 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000130962 | PRRG1 | proline rich and Gla domain 1 | −0.307 | 1.52 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000169908 | TM4SF1 | transmembrane 4 L six family member 1 | −0.945 | 7.67 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000236383 | CCDC200 | coiled-coil domain containing 200 | −0.411 | 1.06 × 10−16 | Metablism/proliferation/regulation |
| ENSG00000219481 | NBPF1 | NBPF member 1 | −0.850 | 4.05 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000041982 | TNC | tenascin C | −0.466 | 1.86 × 10−26 | Metablism/proliferation/regulation |
| ENSG00000130702 | LAMA5 | laminin subunit alpha 5 | −0.285 | 6.65 × 10−5 | Metablism/proliferation/regulation |
| ENSG00000150907 | FOXO1 | forkhead box O1 | −0.283 | 2.58 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000135842 | NIBAN1 | niban apoptosis regulator 1 | −0.310 | 2.05 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000181827 | RFX7 | regulatory factor X7 | −0.264 | 2.26 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000156030 | ELMSAN1 | ELM2 and Myb/SANT domain containing 1 | −0.295 | 6.51 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000167244 | IGF2 | insulin like growth factor 2 | −0.409 | 6.22 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000152409 | JMY | junction mediating and regulatory protein, p53 cofactor | −0.265 | 4.94 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000059728 | MXD1 | MAX dimerization protein 1 | −0.392 | 1.74 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000170653 | ATF7 | activating transcription factor 7 | −0.340 | 1.15 × 10−8 | Metablism/proliferation/regulation |
| ENSG00000118922 | KLF12 | Kruppel like factor 12 | −0.272 | 3.37 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000158711 | ELK4 | ETS transcription factor ELK4 | −0.266 | 1.36 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000095951 | HIVEP1 | human immunodeficiency virus type I enhancer binding protein 1 | −0.305 | 1.59 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000119314 | PTBP3 | polypyrimidine tract binding protein 3 | −0.309 | 1.49 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000173889 | PHC3 | polyhomeotic homolog 3 | −0.265 | 3.70 × 10−10 | Metablism/proliferation/regulation |
| ENSG00000099250 | NRP1 | neuropilin 1 | −0.302 | 1.84 × 10−12 | Metablism/proliferation/regulation |
| ENSG00000091409 | ITGA6 | integrin subunit alpha 6 | −0.277 | 4.73 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000148841 | ITPRIP | inositol 1,4,5-trisphosphate receptor interacting protein | −0.961 | 1.51 × 10−44 | Metablism/proliferation/regulation |
| ENSG00000118503 | TNFAIP3 | TNF alpha induced protein 3 | −0.658 | 3.99 × 10−25 | Metablism/proliferation/regulation |
| ENSG00000277075 | HIST1H2AE | histone cluster 1 H2A family member e | −0.403 | 2.84 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000173611 | SCAI | suppressor of cancer cell invasion | −0.281 | 3.22 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000197594 | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | −0.293 | 1.62 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000213064 | SFT2D2 | SFT2 domain containing 2 | −0.363 | 3.39 × 10−11 | Metablism/proliferation/regulation |
| ENSG00000184897 | H1FX | H1 histone family member X | −0.307 | 1.90 × 10−4 | Metablism/proliferation/regulation |
| ENSG00000168298 | HIST1H1E | histone cluster 1 H1 family member e | −0.314 | 2.88 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000163125 | RPRD2 | regulation of nuclear pre-mRNA domain containing 2 | −0.265 | 1.20 × 10−6 | Metablism/proliferation/regulation |
| ENSG00000148773 | MKI67 | marker of proliferation Ki-67 | −0.339 | 6.61 × 10−3 | Metablism/proliferation/regulation |
| ENSG00000164307 | ERAP1 | endoplasmic reticulum aminopeptidase 1 | −0.280 | 9.38 × 10−7 | Metablism/proliferation/regulation |
| ENSG00000076770 | MBNL3 | muscleblind like splicing regulator 3 | −0.269 | 1.35 × 10−2 | Metablism/proliferation/regulation |
| ENSG00000139636 | LMBR1L | limb development membrane protein 1 like | 0.349 | 3.97 × 10−5 | Immune response |
| ENSG00000107281 | NPDC1 | neural proliferation, differentiation and control 1 | 0.279 | 3.67 × 10−4 | Immune response |
| ENSG00000121716 | PILRB | paired immunoglobin like type 2 receptor beta | 0.512 | 3.18 × 10−3 | Immune response |
| ENSG00000160360 | GPSM1 | G protein signaling modulator 1 | 0.316 | 1.77 × 10−9 | Immune response |
| ENSG00000104522 | TSTA3 | tissue specific transplantation antigen P35B | 0.289 | 1.26 × 10−2 | Immune response |
| ENSG00000214279 | SCART1 | scavenger receptor family member expressed on T cells 1 | 0.588 | 1.35 × 10−7 | Immune response |
| ENSG00000169228 | RAB24 | RAB24, member RAS oncogene family | 0.286 | 3.64 × 10−3 | Immune response |
| ENSG0000010877 | DHX58 | DExH-box helicase 58 | 0.361 | 1.47 × 10−3 | Immune response |
| ENSG00000160072 | ATAD3B | ATPase family AAA domain containing 3B | 0.276 | 4.14 × 10−3 | Immune response |
| ENSG00000112715 | VEGFA | vascular endothelial growth factor A | 0.263 | 3.33 × 10−4 | Immune response |
| ENSG00000110719 | TCIRG1 | T cell immune regulator 1, ATPase H+ transporting V0 subunit a3 | 0.278 | 2.19 × 10−6 | Immune response |
| ENSG00000204104 | TRAF3IP1 | TRAF3 interacting protein 1 | 0.313 | 2.78 × 10−4 | Immune response |
| ENSG00000173531 | MST1 | macrophage stimulating 1 | 0.575 | 2.35 × 10−7 | Immune response |
| ENSG00000273802 | HIST1H2BG | histone cluster 1 H2B family member g | −0.339 | 5.97 × 10−4 | Immune response |
| ENSG00000277224 | HIST1H2BF | histone cluster 1 H2B family member f | −0.332 | 2.65 × 10−2 | Immune response |
| ENSG00000135870 | RC3H1 | ring finger and CCCH-type domains 1 | −0.270 | 4.05 × 10−7 | Immune response |
| ENSG00000144802 | NFKBIZ | NFKB inhibitor zeta | −1.500 | 1.51 × 10−32 | Immune response |
| ENSG00000125730 | C3 | complement C3 | −0.321 | 2.55 × 10−17 | Immune response |
| ENSG00000184678 | HIST2H2BE | histone cluster 2 H2B family member e | −0.615 | 1.72 × 10−20 | Immune response |
| ENSG00000164949 | GEM | GTP binding protein overexpressed in skeletal muscle | −1.068 | 2.18 × 10−56 | Immune response |
| ENSG00000113494 | PRLR | prolactin receptor | −0.372 | 3.94 × 10−3 | Immune response |
| ENSG00000069702 | TGFBR3 | transforming growth factor beta receptor 3 | −0.279 | 2.29 × 10−11 | Immune response |
| ENSG00000121210 | TMEM131L | transmembrane 131 like | −0.281 | 1.44 × 10−2 | Immune response |
| ENSG00000120738 | EGR1 | early growth response 1 | −2.195 | 0.00 | Immune response |
| ENSG00000181722 | ZBTB20 | zinc finger and BTB domain containing 20 | −0.511 | 4.80 × 10−22 | Immune response |
| ENSG00000067992 | PDK3 | pyruvate dehydrogenase kinase 3 | −0.294 | 2.41 × 10−3 | Immune response |
| ENSG00000157764 | BRAF | B-Raf proto-oncogene, serine/threonine kinase | −0.277 | 1.91 × 10−5 | Immune response |
| ENSG00000249437 | NAIP | NLR family apoptosis inhibitory protein | −0.318 | 7.20 × 10−3 | Immune response |
| ENSG00000131016 | AKAP12 | A-kinase anchoring protein 12 | −0.494 | 1.87 × 10−20 | Immune response |
| ENSG00000106089 | STX1A | syntaxin 1A | 0.438 | 2.44 × 10−2 | inflammatory response |
| ENSG00000185338 | SOCS1 | suppressor of cytokine signaling 1 | 0.351 | 6.41 × 10−4 | inflammatory response |
| ENSG00000136244 | IL6 | interleukin 6 | −2.301 | 4.19 × 10−65 | inflammatory response |
| ENSG00000172292 | CERS6 | ceramide synthase 6 | −0.286 | 5.45 × 10−5 | inflammatory response |
| ENSG00000170345 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit | −2.530 | 1.24 × 10−62 | inflammatory response |
| ENSG00000101966 | XIAP | X-linked inhibitor of apoptosis | −0.276 | 3.96 × 10−6 | inflammatory response |
| ENSG00000169429 | CXCL8 | C-X-C motif chemokine ligand 8 | −0.572 | 3.49 × 10−4 | inflammatory response |
| ENSG00000081041 | CXCL2 | C-X-C motif chemokine ligand 2 | −1.212 | 3.32 × 10−29 | inflammatory response |
| ENSG00000128016 | ZFP36 | ZFP36 ring finger protein | −1.515 | 1.97 × 10−82 | inflammatory response |
| ENSG00000179954 | SSC5D | scavenger receptor cysteine rich family member with 5 domains | −0.307 | 3.19 × 10−7 | inflammatory response |
| ENSG00000100906 | NFKBIA | NFKB inhibitor alpha | −0.406 | 3.02 × 10−14 | inflammatory response |
| ENSG00000034152 | MAP2K3 | mitogen-activated protein kinase kinase 3 | −0.402 | 3.85 × 10−12 | inflammatory response |
| ENSG00000172817 | CYP7B1 | cytochrome P450 family 7 subfamily B member 1 | −0.377 | 7.82 × 10−3 | inflammatory response |
| ENSG00000173992 | CCS | copper chaperone for superoxide dismutase | 0.283 | 5.84 × 10−3 | Response to oxidative stress |
| ENSG00000002016 | RAD52 | RAD52 homolog, DNA repair protein | 0.297 | 1.52 × 10−2 | Response to oxidative stress |
| ENSG00000172780 | RAB43 | RAB43, member RAS oncogene family | 0.311 | 5.51 × 10−3 | Response to oxidative stress |
| ENSG00000103245 | CIAO3 | cytosolic iron-sulfur assembly component 3 | 0.324 | 2.22 × 10−2 | Response to oxidative stress |
| ENSG00000163516 | ANKZF1 | ankyrin repeat and zinc finger peptidyl tRNA hydrolase 1 | 0.292 | 1.87 × 10−4 | Response to oxidative stress |
| ENSG00000140398 | NEIL1 | nei like DNA glycosylase 1 | 0.459 | 4.10 × 10−5 | Response to oxidative stress |
| ENSG00000159363 | ATP13A2 | ATPase cation transporting 13A2 | 0.284 | 3.52 × 10−2 | Response to oxidative stress |
| ENSG00000151012 | SLC7A11 | solute carrier family 7 member11 | −0.327 | 2.98 × 10−4 | Response to oxidative stress |
| ENSG00000172115 | CYCS | cytochrome c, somatic | −0.323 | 3.86 × 10−4 | Response to oxidative stress |
| ENSG00000146648 | EGFR | epidermal growth factor receptor | −0.303 | 3.91 × 10−19 | Response to oxidative stress |
| ENSG00000198840 | MT-ND3 | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 3 | −0.477 | 6.34 × 10−14 | Response to oxidative stress |
| ENSG00000158615 | PPP1R15B | protein phosphatase 1 regulatory subunit 15B | −0.631 | 1.15 × 10−36 | Response to oxidative stress |
| ENSG00000109819 | PPARGC1A | PPARG coactivator 1 alpha | −0.285 | 1.53 × 10−2 | Response to oxidative stress |
| ENSG00000137449 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 | −0.313 | 9.29 × 10−6 | Response to oxidative stress |
| ENSG00000198763 | MT-ND2 | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 2 | −0.463 | 3.74 × 10−18 | Response to oxidative stress |
| ENSG00000198786 | MT-ND5 | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 5 | −0.493 | 3.60 × 10−4 | Response to oxidative stress |
| ENSG00000198886 | MT-ND4 | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 4 | −0.556 | 4.78 × 10−4 | Response to oxidative stress |
| ENSG00000198888 | MT-ND1 | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 1 | −0.511 | 1.15 × 10−5 | Response to oxidative stress |
| ENSG00000212907 | MT-ND4L | mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 4L | −0.467 | 2.65 × 10−3 | Response to oxidative stress |
| ENSG00000132510 | KDM6B | lysine demethylase 6B | −0.543 | 1.21 × 10−18 | Response to oxidative stress |
| ENSG00000198712 | MT-CO2 | mitochondrially encoded cytochrome c oxidase II | −0.397 | 2.79 × 10−3 | Response to oxidative stress |
| ENSG00000143322 | ABL2 | ABL proto-oncogene 2, non-receptor tyrosine kinase | −0.359 | 9.88 × 10−14 | Response to oxidative stress |
| ENSG00000127528 | KLF2 | Kruppel like factor 2 | −1.221 | 4.51 × 10−14 | Response to oxidative stress |
| ENSG00000286268 | AF196969.1 | novel protein | 0.712 | 9.99 × 10−3 | Lipids |
| ENSG00000100288 | CHKB | choline kinase beta | 0.359 | 6.43 × 10−5 | Lipids |
| ENSG00000132793 | LPIN3 | lipin 3 | 0.342 | 2.57 × 10−8 | Lipids |
| ENSG00000243708 | PLA2G4B | phospholipase A2 group IVB | 0.565 | 1.56 × 10−2 | Lipids |
| ENSG00000240303 | ACAD11 | acyl-CoA dehydrogenase family member 11 | 0.284 | 9.86 × 10−3 | Lipids |
| ENSG00000072778 | ACADVL | acyl-CoA dehydrogenase very long chain | 0.268 | 7.46 × 10−10 | Lipids |
| ENSG00000197943 | PLCG2 | phospholipase C gamma 2 | 0.310 | 5.49 × 10−3 | Lipids |
| ENSG00000102125 | TAZ | tafazzin | 0.408 | 5.69 × 10−4 | Lipids |
| ENSG00000111664 | GNB3 | G protein subunit beta 3 | 0.506 | 3.20 × 10−2 | Lipids |
| ENSG00000123689 | G0S2 | G0/G1 switch 2 | −0.470 | 4.89 × 10−2 | Lipids |
| ENSG00000073756 | PTGS2 | prostaglandin-endoperoxide synthase 2 | −2.220 | 1.77 × 10−13 | Lipids |
| ENSG00000151176 | PLBD2 | phospholipase B domain containing 2 | −0.446 | 2.90 × 10−18 | Lipids |
| ENSG00000137642 | SORL1 | sortilin related receptor 1 | −0.407 | 3.23 × 10−2 | Lipids |
| ENSG00000172954 | LCLAT1 | lysocardiolipin acyltransferase 1 | −0.319 | 9.16 × 10−5 | Lipids |
| ENSG00000171132 | PRKCE | protein kinase C epsilon | −0.267 | 4.13 × 10−4 | Lipids |
| ENSG00000165029 | ABCA1 | ATP binding cassette subfamily A member 1 | −0.299 | 1.82 × 10−13 | Lipids |
| ENSG00000117600 | PLPPR4 | phospholipid phosphatase related 4 | −0.311 | 1.43 × 10−3 | Lipids |
| ENSG00000147872 | PLIN2 | perilipin 2 | −0.340 | 8.19 × 10−15 | Lipids |
| ENSG00000140474 | ULK3 | unc-51 like kinase 3 | 0.302 | 4.43 × 10−3 | Communication |
| ENSG00000204084 | INPP5B | inositol polyphosphate-5-phosphatase B | 0.295 | 3.88 × 10−5 | Communication |
| ENSG00000170153 | RNF150 | ring finger protein 150 | −0.340 | 4.94 × 10−9 | Communication |
| ENSG00000198753 | PLXNB3 | plexin B3 | −0.372 | 1.56 × 10−3 | Communication |
| ENSG00000184226 | PCDH9 | protocadherin 9 | −0.439 | 2.47 × 10−4 | Communication |
| ENSG00000253537 | PCDHGA7 | protocadherin gamma subfamily A, 7 | −0.264 | 4.29 × 10−3 | Communication |
| ENSG00000253873 | PCDHGA11 | protocadherin gamma subfamily A, 11 | −0.302 | 9.94 × 10−4 | Communication |
| ENSG00000189152 | GRAPL | GRB2 related adaptor protein like | −0.270 | 1.68 × 10−5 | Communication |
| ENSG00000187678 | SPRY4 | sprouty RTK signaling antagonist 4 | −0.310 | 1.38 × 10−4 | Communication |
| ENSG00000075568 | TMEM131 | transmembrane protein 131 | −0.268 | 1.03 × 10−8 | Communication |
| ENSG00000013588 | GPRC5A | G protein-coupled receptor class C group 5 member A | −0.422 | 1.06 × 10−10 | Communication |
| ENSG00000113441 | LNPEP | leucyl and cystinyl aminopeptidase | −0.336 | 1.61 × 10−9 | Communication |
| ENSG00000158258 | CLSTN2 | calsyntenin 2 | −0.388 | 9.23 × 10−10 | Communication |
| ENSG00000091129 | NRCAM | neuronal cell adhesion molecule | −0.346 | 1.21 × 10−3 | Communication |
| ENSG00000083857 | FAT1 | FAT atypical cadherin 1 | −0.387 | 4.48 × 10−33 | Communication |
| ENSG00000070018 | LRP6 | LDL receptor related protein 6 | −0.284 | 1.70 × 10−8 | Communication |
| ENSG00000253953 | PCDHGB4 | protocadherin gamma subfamily B, 4 | −0.269 | 5.57 × 10−4 | Communication |
| ENSG00000134982 | APC | APC regulator of WNT signaling pathway | −0.282 | 2.50 × 10−7 | Communication |
| ENSG00000112902 | SEMA5A | semaphorin 5A | −0.341 | 1.31 × 10−25 | Communication |
| ENSG00000218336 | TENM3 | teneurin transmembrane protein 3 | −0.304 | 9.06 × 10−4 | Communication |
| ENSG00000165124 | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | −0.481 | 7.92 × 10−36 | Communication |
| ENSG00000123096 | SSPN | sarcospan | −0.315 | 1.10 × 10−4 | Communication |
| ENSG00000145012 | LPP | LIM domain containing preferred translocation partner in lipoma | −0.319 | 2.31 × 10−15 | Communication |
| ENSG00000067141 | NEO1 | neogenin 1 | −0.272 | 2.21 × 10−7 | Communication |
| ENSG00000196159 | FAT4 | FAT atypical cadherin 4 | −0.318 | 2.88 × 10−12 | Communication |
| ENSG00000143341 | HMCN1 | hemicentin 1 | −0.421 | 2.95 × 10−4 | Communication |
| ENSG00000078401 | EDN1 | endothelin 1 | −0.429 | 3.86 × 10−3 | Response to stimulus |
| ENSG00000135999 | EPC2 | enhancer of polycomb homolog 2 | −0.284 | 7.13 × 10−4 | Response to stimulus |
| ENSG00000283321 | AC019117.3 | novel protein | −0.337 | 5.32 × 10−3 | Response to stimulus |
| ENSG00000110395 | CBL | Cbl proto-oncogene | −0.303 | 2.57 × 10−8 | Response to stimulus |
| ENSG00000114933 | INO80D | INO80 complex subunit D | −0.303 | 6.43 × 10−7 | Response to stimulus |
| ENSG00000130522 | JUND | JunD proto-oncogene, AP-1 transcription factor subunit | −0.681 | 5.90 × 10−30 | Response to stimulus |
| ENSG00000120129 | DUSP1 | dual specificity phosphatase 1 | −2.123 | 1.05 × 10−267 | Response to stimulus |
| ENSG00000148339 | SLC25A25 | solute carrier family 25 member25 | −0.303 | 6.18 × 10−4 | Response to stimulus |
| ENSG00000125740 | FOSB | FosB proto-oncogene, AP-1 transcription factor subunit | −4.090 | 0.00 | Response to stimulus |
| ENSG00000198727 | MT-CYB | mitochondrially encoded cytochrome b | −0.320 | 3.34 × 10−8 | Response to stimulus |
| ENSG00000177694 | NAALADL2 | N-acetylated alpha-linked acidic dipeptidase like 2 | −0.320 | 1.80 × 10−4 | Response to stimulus |
| ENSG00000184557 | SOCS3 | suppressor of cytokine signaling 3 | −0.506 | 6.46 × 10−7 | Response to stimulus |
| ENSG00000159167 | STC1 | stanniocalcin 1 | −0.534 | 1.49 × 10−4 | Response to stimulus |
| ENSG00000164603 | BMT2 | base methyltransferase of 25S rRNA 2 homolog | −0.323 | 1.14 × 10−2 | Response to stimulus |
| ENSG00000181072 | CHRM2 | cholinergic receptor muscarinic 2 | −0.273 | 2.59 × 10−3 | Response to stimulus |
| ENSG00000132635 | PCED1A | PC-esterase domain containing 1A | 0.280 | 1.88 × 10−3 | Unknown |
| ENSG00000186166 | CCDC84 | coiled-coil domain containing 84 | 0.282 | 7.88 × 10−7 | Unknown |
| ENSG00000117616 | RSRP1 | arginine and serine rich protein 1 | 0.347 | 1.04 × 10−9 | Unknown |
| ENSG00000214193 | SH3D21 | SH3 domain containing 21 | 0.284 | 1.03 × 10−2 | Unknown |
| ENSG00000134884 | ARGLU1 | arginine and glutamate rich 1 | 0.289 | 9.33 × 10−6 | Unknown |
| ENSG00000268350 | FAM156A | family with sequence similarity 156 member A | 0.449 | 9.92 × 10−5 | Unknown |
| ENSG00000182700 | IGIP | IgA inducing protein | 0.400 | 3.33 × 10−4 | Unknown |
| ENSG00000140365 | COMMD4 | COMM domain containing 4 | 0.319 | 1.83 × 10−4 | Unknown |
| ENSG00000146067 | FAM193B | family with sequence similarity 193 member B | 0.285 | 7.51 × 10−3 | Unknown |
| ENSG00000014914 | MTMR11 | myotubularin related protein 11 | 0.286 | 1.90 × 10−2 | Unknown |
| ENSG00000148341 | SH3GLB2 | SH3 domain containing GRB2 like, endophilin B2 | 0.278 | 1.62 × 10−4 | Unknown |
| ENSG00000188483 | IER5L | immediate early response 5 like | 0.325 | 1.54 × 10−2 | Unknown |
| ENSG00000135637 | CCDC142 | coiled-coil domain containing 142 | 0.407 | 1.10 × 10−4 | Unknown |
| ENSG00000213563 | C8orf82 | chromosome 8 open reading frame 82 | 0.275 | 5.43 × 10−3 | Unknown |
| ENSG00000143469 | SYT14 | synaptotagmin 14 | −0.272 | 8.77 × 10−4 | Unknown |
| ENSG00000215472 | RPL17-C18orf32 | RPL17-C18orf32 readthrough | −0.298 | 6.96 × 10−3 | Unknown |
| Gene_Id | Gene Name | Gene Description | Log2FC | P-Adjust | Function |
|---|---|---|---|---|---|
| ENSG00000113494 | PRLR | prolactin receptor | 1.509 | 2.51 × 10−4 | immune response |
| ENSG00000124882 | EREG | epiregulin | 1.029 | 1.75 × 10−10 | immune response |
| ENSG00000128271 | ADORA2A | adenosine A2a receptor | 1.938 | 1.64 × 10−10 | immune response |
| ENSG00000160224 | AIRE | autoimmune regulator | 1.490 | 1.30 × 10−2 | immune response |
| ENSG00000169508 | GPR183 | G protein-coupled receptor 183 | 2.593 | 2.80 × 10−3 | immune response |
| ENSG00000198805 | PNP | purine nucleoside phosphorylase | 1.397 | 1.63 × 10−4 | immune response |
| ENSG00000105088 | OLFM2 | olfactomedin 2 | 1.794 | 6.44 × 10−9 | immune response |
| ENSG00000243509 | TNFRSF6B | TNF receptor superfamily member 6b | 1.875 | 3.16 × 10−5 | immune response |
| ENSG00000113070 | HBEGF | heparin binding EGF like growth factor | 1.237 | 4.11 × 10−2 | immune response |
| ENSG00000122861 | PLAU | plasminogen activator, urokinase | 1.872 | 5.61 × 10−35 | immune response |
| ENSG00000284337 | AC013271.1 | LIM and senescent cell antigen-like-containing domain protein 3 | 1.835 | 6.10 × 10−4 | immune response |
| ENSG00000198535 | C2CD4A | C2 calcium dependent domain containing 4A | 2.260 | 2.90 × 10−7 | immune response |
| ENSG00000237264 | FTH1P11 | ferritin heavy chain 1 pseudogene 11 | 4.468 | 4.70 × 10−2 | immune response |
| ENSG00000219507 | FTH1P8 | ferritin heavy chain 1 pseudogene 8 | 6.317 | 1.68 × 10−2 | immune response |
| ENSG00000160223 | ICOSLG | inducible T-cell costimulator ligand | 1.226 | 1.85 × 10−2 | immune response |
| ENSG00000211772 | TRBC2 | T-cell receptor beta constant 2 | 1.185 | 1.65 × 10−2 | immune response |
| ENSG00000110031 | LPXN | leupaxin | 2.068 | 9.53 × 10−12 | immune response |
| ENSG00000028277 | POU2F2 | POU class 2 homeobox 2 | 1.447 | 7.46 × 10−21 | immune response |
| ENSG00000076641 | PAG1 | phosphoprotein membrane anchor with glycosphingolipid microdomains 1 | 1.396 | 8.91 × 10−7 | immune response |
| ENSG00000173457 | PPP1R14B | protein phosphatase 1 regulatory inhibitor subunit 14B | 1.476 | 2.28 × 10−2 | immune response |
| ENSG00000131187 | F12 | coagulation factor XII | 1.808 | 4.49 × 10−2 | immune response |
| ENSG00000131015 | ULBP2 | UL16 binding protein 2 | 1.169 | 3.47 × 10−2 | immune response |
| ENSG00000069702 | TGFBR3 | transforming growth factor beta receptor 3 | −1.074 | 4.18 × 10−3 | Immune response |
| ENSG00000138735 | PDE5A | phosphodiesterase 5A | −1.754 | 3.76 × 10−13 | Immune response |
| ENSG00000164764 | SBSPON | somatomedin B and thrombospondin type 1 domain containing | −1.973 | 3.06 × 10−3 | Immune response |
| ENSG00000089127 | OAS1 | 2′-5′-oligoadenylate synthetase 1 | −1.399 | 4.55 × 10−4 | Immune response |
| ENSG00000066735 | KIF26A | kinesin family member 26A | −1.743 | 1.91 × 10−4 | Immune response |
| ENSG00000120885 | CLU | clusterin | −1.346 | 1.07 × 10−11 | Immune response |
| ENSG00000136869 | TLR4 | toll like receptor 4 | −1.989 | 7.89 × 10−6 | Immune response |
| ENSG00000164342 | TLR3 | toll like receptor 3 | −2.064 | 2.85 × 10−4 | Immune response |
| ENSG00000159403 | C1R | complement C1r | −1.476 | 6.54 × 10−10 | Immune response |
| ENSG00000182326 | C1S | complement C1s | −1.331 | 6.39 × 10−14 | Immune response |
| ENSG00000120899 | PTK2B | protein tyrosine kinase 2 beta | −2.970 | 6.43 × 10−31 | Immune response |
| ENSG00000130303 | BST2 | bone marrow stromal cell antigen 2 | −1.112 | 7.88 × 10−3 | Immune response |
| ENSG00000158270 | COLEC12 | collectin subfamily member 12 | −1.382 | 2.11 × 10−7 | Immune response |
| ENSG00000244731 | C4A | complement C4A (Rodgers blood group) | −2.248 | 3.26 × 10−6 | Immune response |
| ENSG00000224389 | C4B | complement C4B (Chido blood group) | −1.643 | 2.40 × 10−3 | Immune response |
| ENSG00000205403 | CFI | complement factor I | −1.319 | 7.62 × 10−5 | Immune response |
| ENSG00000243649 | CFB | complement factor B | −2.221 | 1.40 × 10−26 | Immune response |
| ENSG00000197766 | CFD | complement factor D | −1.657 | 2.00 × 10−2 | Immune response |
| ENSG00000000971 | CFH | complement factor H | −1.106 | 1.44 × 10−12 | Immune response |
| ENSG00000125730 | C3 | complement C3 | −1.190 | 3.38 × 10−4 | Immune response |
| ENSG00000185745 | IFIT1 | interferon induced protein with tetratricopeptide repeats 1 | −1.393 | 2.81 × 10−7 | Immune response |
| ENSG00000091972 | CD200 | CD200 molecule | −1.694 | 2.28 × 10−3 | Immune response |
| ENSG00000111331 | OAS3 | 2′-5′-oligoadenylate synthetase 3 | −1.246 | 3.53 × 10−2 | Immune response |
| ENSG00000101347 | SAMHD1 | SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 | −1.048 | 1.57 × 10−2 | Immune response |
| ENSG00000154188 | ANGPT1 | angiopoietin 1 | −1.648 | 7.33 × 10−9 | Immune response |
| ENSG00000107562 | CXCL12 | C-X-C motif chemokine ligand 12 | −2.798 | 5.70 × 10−42 | Immune response |
| ENSG00000142910 | TINAGL1 | tubulointerstitial nephritis antigen like 1 | −3.217 | 7.01 × 10−25 | Immune response |
| ENSG00000106537 | TSPAN13 | tetraspanin 13 | 1.228 | 3.73 × 10−2 | Transport |
| ENSG00000108932 | SLC16A6 | solute carrier family 16 member 6 | 1.447 | 1.76 × 10−8 | Transport |
| ENSG00000134955 | SLC37A2 | solute carrier family 37 member 2 | 1.488 | 6.86 × 10−5 | Transport |
| ENSG00000139508 | SLC46A3 | solute carrier family 46 member3 | 1.007 | 3.41 × 10−5 | Transport |
| ENSG00000101187 | SLCO4A1 | solute carrier organic anion transporter family member 4A1 | 1.354 | 1.02 × 10−7 | Transport |
| ENSG00000188643 | S100A16 | S100 calcium binding protein A16 | 1.023 | 6.96 × 10−3 | Transport |
| ENSG00000157445 | CACNA2D3 | calcium voltage-gated channel auxiliary subunit alpha2delta 3 | 1.659 | 1.37 × 10−2 | Transport |
| ENSG00000069849 | ATP1B3 | ATPase Na+/K+ transporting subunit beta 3 | 1.100 | 3.43 × 10−2 | Transport |
| ENSG00000166592 | RRAD | RRAD, Ras related glycolysis inhibitor and calcium channel regulator | 1.262 | 2.60 × 10−3 | Transport |
| ENSG00000159199 | ATP5G1 | ATP synthase, H+ transporting, mitochondrial Fo complex subunit C1 (subunit 9) | 1.329 | 4.87 × 10−2 | Transport |
| ENSG00000166510 | CCDC68 | coiled-coil domain containing 68 | 1.072 | 1.66 × 10−4 | Transport |
| ENSG00000157551 | KCNJ15 | potassium voltage-gated channel subfamily J member 15 | 1.993 | 1.26 × 10−3 | Transport |
| ENSG00000157542 | KCNJ6 | potassium voltage-gated channel subfamily J member 6 | 2.391 | 3.83 × 10−5 | Transport |
| ENSG00000173114 | LRRN3 | leucine rich repeat neuronal 3 | 1.515 | 7.22 × 10−8 | Transport |
| ENSG00000171246 | NPTX1 | neuronal pentraxin 1 | 2.029 | 1.12 × 10−7 | Transport |
| ENSG00000259863 | SH3RF3-AS1 | SH3RF3 antisense RNA 1 | 2.013 | 6.87 × 10−7 | Transport |
| ENSG00000102287 | GABRE | gamma-aminobutyric acid type A receptor epsilon subunit | −1.296 | 2.76 × 10−6 | Transport |
| ENSG00000198691 | ABCA4 | ATP binding cassette subfamily A member 4 | −1.740 | 1.07 × 10−11 | Transport |
| ENSG00000221955 | SLC12A8 | solute carrier family 12 member 8 | −1.424 | 5.81 × 10−7 | Transport |
| ENSG00000162383 | SLC1A7 | solute carrier family 1 member 7 | −1.588 | 1.17 × 10−3 | Transport |
| ENSG00000146411 | SLC2A12 | solute carrier family 2 member 12 | −1.807 | 1.49 × 10−6 | Transport |
| ENSG00000169507 | SLC38A11 | solute carrier family 38 member 11 | −2.235 | 7.62 × 10−10 | Transport |
| ENSG00000139209 | SLC38A4 | solute carrier family 38 member 4 | −1.638 | 1.79 × 10−4 | Transport |
| ENSG00000138821 | SLC39A8 | solute carrier family 39 member 8 | −1.053 | 6.51 × 10−7 | Transport |
| ENSG00000138449 | SLC40A1 | solute carrier family 40 member 1 | −1.479 | 7.54 × 10−8 | Transport |
| ENSG00000013293 | SLC7A14 | solute carrier family 7 member 14 | −1.849 | 2.46 × 10−6 | Transport |
| ENSG00000181804 | SLC9A9 | solute carrier family 9 member A9 | −1.018 | 4.45 × 10−3 | Transport |
| ENSG00000168356 | SCN11A | sodium voltage-gated channel alpha subunit 11 | −1.637 | 4.59 × 10−2 | Transport |
| ENSG00000144285 | SCN1A | sodium voltage-gated channel alpha subunit 1 | −4.979 | 1.95 × 10−3 | Transport |
| ENSG00000151572 | ANO4 | anoctamin 4 | −1.308 | 5.69 × 10−11 | Transport |
| ENSG00000145362 | ANK2 | ankyrin 2 | −2.002 | 3.71 × 10−6 | Transport |
| ENSG00000111859 | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 | −1.118 | 5.58 × 10−6 | Transport |
| ENSG00000144645 | OSBPL10 | oxysterol binding protein like 10 | −1.097 | 1.63 × 10−4 | Transport |
| ENSG00000162687 | KCNT2 | potassium sodium-activated channel subfamily T member 2 | −2.348 | 1.96 × 10−3 | Transport |
| ENSG00000135643 | KCNMB4 | potassium calcium-activated channel subfamily M regulatory beta subunit 4 | −1.242 | 1.75 × 10−4 | Transport |
| ENSG00000143028 | SYPL2 | synaptophysin like 2 | −1.197 | 2.42 × 10−5 | Transport |
| ENSG00000089472 | HEPH | hephaestin | −1.250 | 2.79 × 10−6 | Transport |
| ENSG00000166473 | PKD1L2 | polycystin 1 like 2 (gene/pseudogene) | −1.796 | 1.07 × 10−11 | Transport |
| ENSG00000143416 | SELENBP1 | selenium binding protein 1 | −1.411 | 1.75 × 10−19 | Transport |
| ENSG00000175538 | KCNE3 | potassium voltage-gated channel subfamily E regulatory subunit 3 | −2.405 | 1.46 × 10−5 | Transport |
| ENSG00000055732 | MCOLN3 | mucolipin 3 | −1.027 | 1.32 × 10−3 | Transport |
| ENSG00000172548 | NIPAL4 | NIPA like domain containing 4 | −1.999 | 4.27 × 10−2 | Transport |
| ENSG00000003137 | CYP26B1 | cytochrome P450 family 26 subfamily B member 1 | 1.217 | 9.43 × 10−5 | Intracellular and extracellular matrix |
| ENSG00000058085 | LAMC2 | laminin subunit gamma 2 | 1.484 | 1.02 × 10−4 | Intracellular and extracellular matrix |
| ENSG00000138316 | ADAMTS14 | ADAM metallopeptidase with thrombospondin type 1 motif 14 | 1.501 | 3.47 × 10−5 | Intracellular and extracellular matrix |
| ENSG00000135346 | CGA | glycoprotein hormones, alpha polypeptide | 3.014 | 2.16 × 10−5 | Intracellular and extracellular matrix |
| ENSG00000163347 | CLDN1 | claudin 1 | 1.065 | 1.97 × 10−6 | Intracellular and extracellular matrix |
| ENSG00000123500 | COL10A1 | collagen type X alpha 1 chain | 1.206 | 4.22 × 10−2 | Intracellular and extracellular matrix |
| ENSG00000197467 | COL13A1 | collagen type XIII alpha 1 chain | 1.299 | 2.71 × 10−7 | Intracellular and extracellular matrix |
| ENSG00000080573 | COL5A3 | collagen type V alpha 3 chain | 1.535 | 3.48 × 10−7 | Intracellular and extracellular matrix |
| ENSG00000143127 | ITGA10 | integrin subunit alpha 10 | 1.596 | 9.82 × 10−5 | Intracellular and extracellular matrix |
| ENSG00000137809 | ITGA11 | integrin subunit alpha 11 | 1.284 | 2.46 × 10−3 | Intracellular and extracellular matrix |
| ENSG00000164171 | ITGA2 | integrin subunit alpha 2 | 1.034 | 9.75 × 10−3 | Intracellular and extracellular matrix |
| ENSG00000005884 | ITGA3 | integrin subunit alpha 3 | 1.065 | 3.37 × 10−6 | Intracellular and extracellular matrix |
| ENSG00000196611 | MMP1 | matrix metallopeptidase 1 | 2.713 | 3.61 × 10−68 | Intracellular and extracellular matrix |
| ENSG00000166670 | MMP10 | matrix metallopeptidase 10 | 3.191 | 2.09 × 10−4 | Intracellular and extracellular matrix |
| ENSG00000156103 | MMP16 | matrix metallopeptidase 16 | 1.229 | 8.60 × 10−6 | Intracellular and extracellular matrix |
| ENSG00000149968 | MMP3 | matrix metallopeptidase 3 | 3.076 | 9.37 × 10−44 | Intracellular and extracellular matrix |
| ENSG00000104368 | PLAT | plasminogen activator, tissue type | 1.692 | 1.03 × 10−32 | Intracellular and extracellular matrix |
| ENSG00000196581 | AJAP1 | adherens junctions associated protein 1 | 1.032 | 8.97 × 10−5 | Intracellular and extracellular matrix |
| ENSG00000182667 | NTM | neurotrimin | 2.160 | 2.03 × 10−25 | Intracellular and extracellular matrix |
| ENSG00000261371 | PECAM1 | platelet and endothelial cell adhesion molecule 1 | 2.965 | 1.05 × 10−7 | Intracellular and extracellular matrix |
| ENSG00000235649 | MXRA5Y | matrix remodeling associated 5, Y-linked (pseudogene) | −1.849 | 3.32 × 10−2 | Cellular and extracellular matrix |
| ENSG00000124749 | COL21A1 | collagen type XXI alpha 1 chain | −3.204 | 7.20 × 10−22 | Cellular and extracellular matrix |
| ENSG00000049089 | COL9A2 | collagen type IX alpha 2 chain | −1.606 | 1.01 × 10−3 | Cellular and extracellular matrix |
| ENSG00000060718 | COL11A1 | collagen type XI alpha 1 chain | −1.577 | 1.44 × 10−4 | Cellular and extracellular matrix |
| ENSG00000187955 | COL14A1 | collagen type XIV alpha 1 chain | −2.288 | 8.17 × 10−16 | Cellular and extracellular matrix |
| ENSG00000168077 | SCARA3 | scavenger receptor class A member 3 | −1.422 | 1.51 × 10−18 | Cellular and extracellular matrix |
| ENSG00000106823 | ECM2 | extracellular matrix protein 2 | −2.612 | 3.11 × 10−3 | Cellular and extracellular matrix |
| ENSG00000242600 | MBL1P | mannose binding lectin 1, pseudogene | −1.941 | 1.06 × 10−2 | Cellular and extracellular matrix |
| ENSG00000166482 | MFAP4 | microfibrillar associated protein 4 | −1.966 | 1.51 × 10−26 | Cellular and extracellular matrix |
| ENSG00000165272 | AQP3 | aquaporin 3 (Gill blood group) | −1.789 | 4.63 × 10−11 | Cellular and extracellular matrix |
| ENSG00000138829 | FBN2 | fibrillin 2 | −1.036 | 1.55 × 10−2 | Cellular and extracellular matrix |
| ENSG00000123243 | ITIH5 | inter-alpha-trypsin inhibitor heavy chain family member 5 | −1.312 | 7.45 × 10−4 | Cellular and extracellular matrix |
| ENSG00000049540 | ELN | elastin | −1.271 | 2.11 × 10−4 | Cellular and extracellular matrix |
| ENSG00000183798 | EMILIN3 | elastin microfibril interfacer 3 | −1.626 | 8.81 × 10−4 | Cellular and extracellular matrix |
| ENSG00000091986 | CCDC80 | coiled-coil domain containing 80 | −1.606 | 9.31 × 10−7 | Cellular and extracellular matrix |
| ENSG00000135424 | ITGA7 | integrin subunit alpha 7 | −1.024 | 1.29 × 10−4 | Cellular and extracellular matrix |
| ENSG00000196569 | LAMA2 | laminin subunit alpha 2 | −1.840 | 1.50 × 10−5 | Cellular and extracellular matrix |
| ENSG00000132470 | ITGB4 | integrin subunit beta 4 | −1.792 | 1.98 × 10−11 | Cellular and extracellular matrix |
| ENSG00000105855 | ITGB8 | integrin subunit beta 8 | −1.390 | 1.53 × 10−6 | Cellular and extracellular matrix |
| ENSG00000205221 | VIT | vitrin | −2.675 | 4.97 × 10−3 | Cellular and extracellular matrix |
| ENSG00000122707 | RECK | reversion inducing cysteine rich protein with kazal motifs | −1.066 | 7.99 × 10−6 | Cellular and extracellular matrix |
| ENSG00000169908 | TM4SF1 | transmembrane 4 L six family member 1 | 1.509 | 4.14 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000157064 | NMNAT2 | nicotinamide nucleotide adenylyltransferase 2 | 1.159 | 5.85 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000165891 | E2F7 | E2F transcription factor 7 | 1.276 | 4.50 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128965 | CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | 1.667 | 1.34 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000185697 | MYBL1 | MYB proto-oncogene like 1 | 1.393 | 9.20 × 10−12 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000168405 | CMAHP | cytidine monophospho-N-acetylneuraminic acid hydroxylase, pseudogene | 1.120 | 1.46 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137331 | IER3 | immediate early response 3 | 1.390 | 4.21 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000124802 | EEF1E1 | eukaryotic translation elongation factor 1 epsilon 1 | 1.325 | 2.52 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170498 | KISS1 | KiSS-1 metastasis-suppressor | 2.954 | 1.23 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000118523 | CTGF | connective tissue growth factor | 1.010 | 6.38 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000140379 | BCL2A1 | BCL2 related protein A1 | 1.898 | 2.86 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000169372 | CRADD | CASP2 and RIPK1 domain containing adaptor with death domain | 1.186 | 1.22 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000160013 | PTGIR | prostaglandin I2 (prostacyclin) receptor (IP) | 1.162 | 3.31 × 10−18 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000183691 | NOG | noggin | 2.225 | 9.39 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000124762 | CDKN1A | cyclin dependent kinase inhibitor 1A | 1.073 | 8.29 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000232977 | LINC00327 | long intergenic non-protein coding RNA 327 | 1.356 | 6.49 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000255073 | ZFP91-CNTF | ZFP91-CNTF readthrough (NMD candidate) | 1.284 | 3.17 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000179862 | CITED4 | Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 4 | 2.519 | 6.13 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128917 | DLL4 | delta like canonical Notch ligand 4 | 1.356 | 3.41 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000205683 | DPF3 | double PHD fingers 3 | 1.292 | 1.03 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000006468 | ETV1 | ETS variant 1 | 1.229 | 8.84 × 10−14 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000175592 | FOSL1 | FOS like 1, AP-1 transcription factor subunit | 1.145 | 5.17 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000164379 | FOXQ1 | forkhead box Q1 | 1.395 | 1.63 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000134363 | FST | follistatin | 1.174 | 3.03 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000177283 | FZD8 | frizzled class receptor 8 | 1.495 | 7.35 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000114315 | HES1 | hes family bHLH transcription factor 1 | 2.075 | 1.27 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000188290 | HES4 | hes family bHLH transcription factor 4 | 1.563 | 1.16 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000164683 | HEY1 | hes related family bHLH transcription factor with YRPW motif 1 | 1.461 | 5.80 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000163909 | HEYL | hes related family bHLH transcription factor with YRPW motif-like | 1.649 | 5.79 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000187837 | HIST1H1C | histone cluster 1 H1 family member c | 1.075 | 2.17 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137309 | HMGA1 | high mobility group AT-hook 1 | 1.299 | 2.87 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115274 | INO80B | INO80 complex subunit B | 1.318 | 3.48 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000108551 | RASD1 | ras related dexamethasone induced 1 | 1.168 | 4.82 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000196460 | RFX8 | RFX family member 8, lacking RFX DNA binding domain | 1.059 | 3.06 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000147509 | RGS20 | regulator of G protein signaling 20 | 1.409 | 2.83 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000095752 | IL11 | interleukin 11 | 1.511 | 1.46 × 10−12 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000133169 | BEX1 | brain expressed X-linked 1 | 1.579 | 1.14 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000168621 | GDNF | glial cell derived neurotrophic factor | 1.029 | 1.69 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000122641 | INHBA | inhibin beta A subunit | 1.188 | 6.30 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000254858 | MPV17L2 | MPV17 mitochondrial inner membrane protein like 2 | 1.060 | 4.39 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000169994 | MYO7B | myosin VIIB | 2.121 | 2.99 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000239672 | NME1 | NME/NM23 nucleoside diphosphate kinase 1 | 1.300 | 2.18 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139289 | PHLDA1 | pleckstrin homology like domain family A member 1 | 1.113 | 4.84 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000181649 | PHLDA2 | pleckstrin homology like domain family A member 2 | 1.679 | 1.79 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000176641 | RNF152 | ring finger protein 152 | 1.800 | 7.61 × 10−16 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000154133 | ROBO4 | roundabout guidance receptor 4 | 2.382 | 2.98 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197632 | SERPINB2 | serpin family B member 2 | 1.263 | 3.41 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000006327 | TNFRSF12A | TNF receptor superfamily member 12A | 1.168 | 5.04 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000130513 | GDF15 | growth differentiation factor 15 | 1.930 | 5.16 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000092345 | DAZL | deleted in azoospermia like | 1.516 | 2.85 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000268439 | EMG1 | EMG1, N1-specific pseudouridine methyltransferase | 1.048 | 2.25 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000003249 | DBNDD1 | dysbindin domain containing 1 | 1.267 | 1.40 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000077348 | EXOSC5 | exosome component 5 | 1.275 | 2.99 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000162669 | HFM1 | HFM1, ATP dependent DNA helicase homolog | 2.020 | 9.62 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000156265 | MAP3K7CL | MAP3K7 C-terminal like | 1.040 | 3.11 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000158747 | NBL1 | neuroblastoma 1, DAN family BMP antagonist | 1.065 | 1.69 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000135919 | SERPINE2 | serpin family E member 2 | 1.245 | 7.42 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128805 | ARHGAP22 | Rho GTPase activating protein 22 | 1.187 | 3.55 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000130702 | LAMA5 | laminin subunit alpha 5 | −1.325 | 6.48 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000110455 | ACCS | 1-aminocyclopropane-1-carboxylate synthase homolog (inactive) | −1.435 | 2.85 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000138772 | ANXA3 | annexin A3 | −1.922 | 8.29 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000151632 | AKR1C2 | aldo-keto reductase family 1 member C2 | −1.608 | 1.64 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170962 | PDGFD | platelet derived growth factor D | −1.790 | 1.64 × 10−43 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128606 | LRRC17 | leucine rich repeat containing 17 | −1.332 | 5.81 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000102466 | FGF14 | fibroblast growth factor 14 | −1.333 | 3.65 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000109339 | MAPK10 | mitogen-activated protein kinase 10 | −1.329 | 1.44 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000108984 | MAP2K6 | mitogen-activated protein kinase kinase 6 | −1.954 | 1.33 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000064687 | ABCA7 | ATP binding cassette subfamily A member 7 | −1.603 | 1.39 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000173706 | HEG1 | heart development protein with EGF like domains 1 | −1.524 | 1.95 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000174348 | PODN | podocan | −1.891 | 4.35 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000003096 | KLHL13 | kelch like family member 13 | −2.855 | 4.79 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000146021 | KLHL3 | kelch like family member 3 | −1.014 | 2.99 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000145819 | ARHGAP26 | Rho GTPase activating protein 26 | −2.155 | 2.00 × 10−13 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139354 | GAS2L3 | growth arrest specific 2 like 3 | −1.578 | 5.53 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000198796 | ALPK2 | alpha kinase 2 | −1.018 | 2.28 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000082438 | COBLL1 | cordon-bleu WH2 repeat protein like 1 | −1.576 | 1.97 × 10−8 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000176971 | FIBIN | fin bud initiation factor homolog (zebrafish) | −1.382 | 9.25 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000183098 | GPC6 | glypican 6 | −1.037 | 3.16 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139263 | LRIG3 | leucine rich repeats and immunoglobulin like domains 3 | −1.084 | 2.12 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170500 | LONRF2 | LON peptidase N-terminal domain and ring finger 2 | −2.063 | 3.13 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000163017 | ACTG2 | actin, gamma 2, smooth muscle, enteric | −1.699 | 5.71 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000111684 | LPCAT3 | lysophosphatidylcholine acyltransferase 3 | −1.021 | 1.61 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000182575 | NXPH3 | neurexophilin 3 | −1.451 | 7.04 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000243970 | PPIEL | peptidylprolyl isomerase E like pseudogene | −1.019 | 1.09 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000113231 | PDE8B | phosphodiesterase 8B | −1.113 | 6.47 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000185483 | ROR1 | receptor tyrosine kinase like orphan receptor 1 | −1.344 | 6.02 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000187164 | SHTN1 | shootin 1 | −1.080 | 2.96 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000177409 | SAMD9L | sterile alpha motif domain containing 9 like | −1.003 | 1.80 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000196562 | SULF2 | sulfatase 2 | −1.218 | 2.03 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000186854 | TRABD2A | TraB domain containing 2A | −1.386 | 9.04 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000182179 | UBA7 | ubiquitin like modifier activating enzyme 7 | −1.234 | 7.71 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000112303 | VNN2 | vanin 2 | −3.082 | 2.31 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115339 | GALNT3 | polypeptide N-acetylgalactosaminyltransferase 3 | −1.284 | 2.38 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000167311 | ART5 | ADP-ribosyltransferase 5 | −1.826 | 3.84 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000088882 | CPXM1 | carboxypeptidase X, M14 family member 1 | −1.278 | 1.34 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000138180 | CEP55 | centrosomal protein 55 | −1.272 | 1.45 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000064886 | CHI3L2 | chitinase 3 like 2 | −1.354 | 2.34 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115468 | EFHD1 | EF-hand domain family member D1 | −1.752 | 5.70 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000106565 | TMEM176B | transmembrane protein 176B | −1.559 | 4.58 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000143869 | GDF7 | growth differentiation factor 7 | −2.047 | 2.63 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139269 | INHBE | inhibin beta E subunit | −2.466 | 7.03 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000105989 | WNT2 | Wnt family member 2 | −3.133 | 2.67 × 10−32 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139304 | PTPRQ | protein tyrosine phosphatase, receptor type Q | −2.790 | 5.05 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128045 | RASL11B | RAS like family 11 member B | −2.577 | 1.04 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000138615 | CILP | cartilage intermediate layer protein | −1.854 | 3.03 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000002933 | TMEM176A | transmembrane protein 176A | −1.660 | 4.32 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000121858 | TNFSF10 | TNF superfamily member 10 | −1.790 | 9.42 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000171462 | DLK2 | delta like non-canonical Notch ligand 2 | −1.329 | 1.62 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000178662 | CSRNP3 | cysteine and serine rich nuclear protein 3 | −2.392 | 8.91 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000090006 | LTBP4 | latent transforming growth factor beta binding protein 4 | −1.122 | 1.82 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000100626 | GALNT16 | polypeptide N-acetylgalactosaminyltransferase 16 | −1.780 | 2.60 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000168453 | HR | HR, lysine demethylase and nuclear receptor corepressor | −2.686 | 1.55 × 10−18 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000054654 | SYNE2 | spectrin repeat containing nuclear envelope protein 2 | −1.497 | 9.55 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000064692 | SNCAIP | synuclein alpha interacting protein | −1.993 | 3.08 × 10−20 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000101938 | CHRDL1 | chordin like 1 | −1.622 | 1.48 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000132846 | ZBED3 | zinc finger BED-type containing 3 | −1.501 | 2.00 × 10−13 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000187764 | SEMA4D | semaphorin 4D | −1.142 | 1.53 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000104081 | BMF | Bcl2 modifying factor | −1.067 | 7.29 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170214 | ADRA1B | adrenoceptor alpha 1B | −3.324 | 4.07 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000151692 | RNF144A | ring finger protein 144A | −1.343 | 1.09 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000111885 | MAN1A1 | mannosidase alpha class 1A member 1 | −1.117 | 1.09 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000048740 | CELF2 | CUGBP Elav-like family member 2 | −1.545 | 2.49 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000134245 | WNT2B | Wnt family member 2B | −1.467 | 5.53 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000136859 | ANGPTL2 | angiopoietin like 2 | −1.707 | 9.06 × 10−22 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170624 | SGCD | sarcoglycan delta | −1.300 | 8.69 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000135472 | FAIM2 | Fas apoptotic inhibitory molecule 2 | −1.531 | 2.10 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000065717 | TLE2 | transducin like enhancer of split 2 | −1.275 | 2.37 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000064205 | WISP2 | WNT1 inducible signaling pathway protein 2 | −2.029 | 5.00 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000184347 | SLIT3 | slit guidance ligand 3 | −1.835 | 7.13 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137834 | SMAD6 | SMAD family member 6 | −1.545 | 9.50 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000178573 | MAF | MAF bZIP transcription factor | −1.077 | 2.32 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000248746 | ACTN3 | actinin alpha 3 (gene/pseudogene) | −1.987 | 1.15 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000154734 | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | −1.667 | 4.48 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000166106 | ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif 15 | −1.395 | 2.83 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000158555 | GDPD5 | glycerophosphodiester phosphodiesterase domain containing 5 | −2.275 | 1.82 × 10−8 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000173599 | PC | pyruvate carboxylase | −1.123 | 4.07 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000122877 | EGR2 | early growth response 2 | −2.430 | 3.34 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000179388 | EGR3 | early growth response 3 | −1.946 | 1.08 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000052850 | ALX4 | ALX homeobox 4 | −1.188 | 4.54 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000168502 | MTCL1 | microtubule crosslinking factor 1 | −1.144 | 3.19 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000276600 | RAB7B | RAB7B, member RAS oncogene family | −1.749 | 1.91 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000118849 | RARRES1 | retinoic acid receptor responder 1 | −1.041 | 7.15 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000213626 | LBH | limb bud and heart development | −1.890 | 1.54 × 10−25 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000169744 | LDB2 | LIM domain binding 2 | −1.115 | 1.09 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000071282 | LMCD1 | LIM and cysteine rich domains 1 | −1.747 | 1.80 × 10−22 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000163431 | LMOD1 | leiomodin 1 | −1.048 | 1.94 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000170312 | CDK1 | cyclin dependent kinase 1 | −1.104 | 4.72 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000123080 | CDKN2C | cyclin dependent kinase inhibitor 2C | −1.247 | 1.27 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000066279 | ASPM | abnormal spindle microtubule assembly | −1.597 | 7.20 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000116741 | RGS2 | regulator of G protein signaling 2 | −1.023 | 7.35 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000128482 | RNF112 | ring finger protein 112 | −1.258 | 5.99 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137193 | PIM1 | Pim-1 proto-oncogene, serine/threonine kinase | −1.080 | 6.87 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000116133 | DHCR24 | 24-dehydrocholesterol reductase | −1.042 | 2.13 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000148773 | MKI67 | marker of proliferation Ki-67 | −1.255 | 4.78 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000092621 | PHGDH | phosphoglycerate dehydrogenase | −1.408 | 5.84 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000108387 | SEPT4 | septin 4 | −1.274 | 3.41 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000125354 | SEPT6 | septin 6 | −1.003 | 4.95 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000136531 | SCN2A | sodium voltage-gated channel alpha subunit 2 | −1.828 | 1.68 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000139211 | AMIGO2 | adhesion molecule with Ig like domain 2 | −1.127 | 1.26 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000120738 | EGR1 | early growth response 1 | −1.344 | 4.50 × 10−16 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000165959 | CLMN | calmin | −2.182 | 1.51 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000090539 | CHRD | chordin | −1.001 | 4.24 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000106003 | LFNG | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | −1.029 | 3.52 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000164920 | OSR2 | odd-skipped related transciption factor 2 | −1.249 | 3.36 × 10−8 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000115252 | PDE1A | phosphodiesterase 1A | −2.989 | 1.15 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000111341 | MGP | matrix Gla protein | −1.673 | 2.70 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000187098 | MITF | melanogenesis associated transcription factor | −1.741 | 1.10 × 10−16 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000169184 | MN1 | MN1 proto-oncogene, transcriptional regulator | −1.857 | 3.34 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000120278 | PLEKHG1 | pleckstrin homology and RhoGEF domain containing G1 | −1.308 | 2.49 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000240694 | PNMA2 | paraneoplastic Ma antigen 2 | −1.284 | 1.77 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000143125 | PROK1 | prokineticin 1 | −1.352 | 1.04 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000106772 | PRUNE2 | prune homolog 2 | −1.792 | 1.75 × 10−10 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000160801 | PTH1R | parathyroid hormone 1 receptor | −1.442 | 3.49 × 10−7 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000107551 | RASSF4 | Ras association domain family member 4 | −1.457 | 6.36 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000141068 | KSR1 | kinase suppressor of ras 1 | −1.157 | 2.05 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000117155 | SSX2IP | SSX family member 2 interacting protein | −1.190 | 2.62 × 10−16 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000162595 | DIRAS3 | DIRAS family GTPase 3 | −1.371 | 6.86 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000179981 | TSHZ1 | teashirt zinc finger homeobox 1 | −1.094 | 3.79 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000088756 | ARHGAP28 | Rho GTPase activating protein 28 | −1.425 | 3.47 × 10−23 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000137727 | ARHGAP20 | Rho GTPase activating protein 20 | −2.101 | 1.84 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000172346 | CSDC2 | cold shock domain containing C2 | −2.139 | 2.20 × 10−14 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000181444 | ZNF467 | zinc finger protein 467 | −1.210 | 2.94 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000206538 | VGLL3 | vestigial like family member 3 | −1.052 | 9.47 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000112246 | SIM1 | single-minded family bHLH transcription factor 1 | −1.095 | 1.58 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000173227 | SYT12 | synaptotagmin 12 | −2.160 | 1.04 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000258818 | RNASE4 | ribonuclease A family member 4 | −1.050 | 4.07 × 10−3 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000153993 | SEMA3D | semaphorin 3D | −1.488 | 9.82 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000012171 | SEMA3B | semaphorin 3B | −1.515 | 5.39 × 10−11 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000182175 | RGMA | repulsive guidance molecule family member a | −1.225 | 8.69 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000120693 | SMAD9 | SMAD family member 9 | −1.359 | 8.99 × 10−5 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000112984 | KIF20A | kinesin family member 20A | −1.607 | 1.70 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000211448 | DIO2 | iodothyronine deiodinase 2 | −1.249 | 7.48 × 10−6 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000197406 | DIO3 | iodothyronine deiodinase 3 | −1.593 | 3.51 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000010932 | FMO1 | flavin containing monooxygenase 1 | −3.430 | 1.99 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000076258 | FMO4 | flavin containing monooxygenase 4 | −1.825 | 1.53 × 10−9 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000131781 | FMO5 | flavin containing monooxygenase 5 | −1.218 | 4.49 × 10−2 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000162496 | DHRS3 | dehydrogenase/reductase 3 | −1.489 | 1.03 × 10−4 | Metabolism/Cell Proliferation/Regulation |
| ENSG00000159167 | STC1 | stanniocalcin 1 | 1.249 | 7.50 × 10−13 | Response to stimulus |
| ENSG00000133805 | AMPD3 | adenosine monophosphate deaminase 3 | 1.087 | 8.42 × 10−9 | Response to stimulus |
| ENSG00000143786 | CNIH3 | cornichon family AMPA receptor auxiliary protein 3 | 1.380 | 4.64 × 10−7 | Response to stimulus |
| ENSG00000170373 | CST1 | cystatin SN | 2.721 | 3.26 × 10−6 | Response to stimulus |
| ENSG00000134769 | DTNA | dystrobrevin alpha | 1.118 | 4.90 × 10−7 | Response to stimulus |
| ENSG00000120875 | DUSP4 | dual specificity phosphatase 4 | 1.040 | 3.70 × 10−11 | Response to stimulus |
| ENSG00000060558 | GNA15 | G protein subunit alpha 15 | 1.725 | 2.90 × 10−2 | Response to stimulus |
| ENSG00000181773 | GPR3 | G protein-coupled receptor 3 | 1.345 | 4.88 × 10−4 | Response to stimulus |
| ENSG00000180875 | GREM2 | gremlin 2, DAN family BMP antagonist | 1.351 | 8.05 × 10−7 | Response to stimulus |
| ENSG00000148834 | GSTO1 | glutathione S-transferase omega 1 | 1.653 | 1.22 × 10−3 | Response to stimulus |
| ENSG00000128285 | MCHR1 | melanin concentrating hormone receptor 1 | 2.294 | 3.65 × 10−2 | Response to stimulus |
| ENSG00000169715 | MT1E | metallothionein 1E | 1.731 | 6.29 × 10−3 | Response to stimulus |
| ENSG00000104490 | NCALD | neurocalcin delta | 1.428 | 1.58 × 10−2 | Response to stimulus |
| ENSG00000171208 | NETO2 | neuropilin and tolloid like 2 | 1.272 | 2.96 × 10−8 | Response to stimulus |
| ENSG00000154217 | PITPNC1 | phosphatidylinositol transfer protein, cytoplasmic 1 | 1.257 | 2.90 × 10−5 | Response to stimulus |
| ENSG00000087494 | PTHLH | parathyroid hormone like hormone | 1.458 | 9.51 × 10−5 | Response to stimulus |
| ENSG00000041353 | RAB27B | RAB27B, member RAS oncogene family | 1.385 | 2.72 × 10−6 | Response to stimulus |
| ENSG00000136237 | RAPGEF5 | Rap guanine nucleotide exchange factor 5 | 1.659 | 2.41 × 10−2 | Response to stimulus |
| ENSG00000181625 | SLX1B | SLX1 homolog B, structure-specific endonuclease subunit | 1.410 | 3.68 × 10−3 | Response to stimulus |
| ENSG00000101463 | SYNDIG1 | synapse differentiation inducing 1 | 1.162 | 7.33 × 10−4 | Response to stimulus |
| ENSG00000011347 | SYT7 | synaptotagmin 7 | 1.678 | 1.56 × 10−2 | Response to stimulus |
| ENSG00000184292 | TACSTD2 | tumor associated calcium signal transducer 2 | 1.229 | 1.93 × 10−2 | Response to stimulus |
| ENSG00000105825 | TFPI2 | tissue factor pathway inhibitor 2 | 1.940 | 2.13 × 10−7 | Response to stimulus |
| ENSG00000178726 | THBD | thrombomodulin | 2.311 | 2.52 × 10−4 | Response to stimulus |
| ENSG00000147573 | TRIM55 | tripartite motif containing 55 | 2.227 | 2.68 × 10−6 | Response to stimulus |
| ENSG00000081181 | ARG2 | arginase 2 | 1.398 | 3.08 × 10−2 | Response to stimulus |
| ENSG00000169627 | BOLA2B | bolA family member 2B | 1.289 | 3.05 × 10−2 | Response to stimulus |
| ENSG00000144476 | ACKR3 | atypical chemokine receptor 3 | −2.220 | 3.17 × 10−14 | Response to stimulus |
| ENSG00000114698 | PLSCR4 | phospholipid scramblase 4 | −1.101 | 6.91 × 10−15 | Response to stimulus |
| ENSG00000137959 | IFI44L | interferon induced protein 44 like | −1.666 | 1.47 × 10−8 | Response to stimulus |
| ENSG00000069431 | ABCC9 | ATP binding cassette subfamily C member 9 | −1.279 | 7.83 × 10−6 | Response to stimulus |
| ENSG00000132561 | MATN2 | matrilin 2 | −1.966 | 5.43 × 10−21 | Response to stimulus |
| ENSG00000162551 | ALPL | alkaline phosphatase, liver/bone/kidney | −2.494 | 8.25 × 10−25 | Response to stimulus |
| ENSG00000184979 | USP18 | ubiquitin specific peptidase 18 | −1.057 | 3.81 × 10−2 | Response to stimulus |
| ENSG00000172264 | MACROD2 | MACRO domain containing 2 | −2.645 | 6.64 × 10−3 | Response to stimulus |
| ENSG00000250722 | SELENOP | selenoprotein P | −2.805 | 1.50 × 10−36 | Response to stimulus |
| ENSG00000069482 | GAL | galanin and GMAP prepropeptide | 2.335 | 3.68 × 10−3 | Inflammatory process |
| ENSG00000151651 | ADAM8 | ADAM metallopeptidase domain 8 | 1.251 | 5.04 × 10−7 | Inflammatory process |
| ENSG00000011201 | ANOS1 | anosmin 1 | 1.877 | 9.50 × 10−16 | Inflammatory process |
| ENSG00000108691 | CCL2 | C-C motif chemokine ligand 2 | 1.339 | 5.92 × 10−3 | Inflammatory process |
| ENSG00000164400 | CSF2 | colony stimulating factor 2 | 2.520 | 3.09 × 10−5 | Inflammatory process |
| ENSG00000108342 | CSF3 | colony stimulating factor 3 | 2.372 | 3.36 × 10−17 | Inflammatory process |
| ENSG00000163739 | CXCL1 | C-X-C motif chemokine ligand 1 | 1.612 | 1.83 × 10−5 | Inflammatory process |
| ENSG00000081041 | CXCL2 | C-X-C motif chemokine ligand 2 | 1.713 | 8.35 × 10−5 | Inflammatory process |
| ENSG00000163734 | CXCL3 | C-X-C motif chemokine ligand 3 | 1.149 | 7.49 × 10−6 | Inflammatory process |
| ENSG00000163735 | CXCL5 | C-X-C motif chemokine ligand 5 | 2.107 | 3.74 × 10−36 | Inflammatory process |
| ENSG00000169429 | CXCL8 | C-X-C motif chemokine ligand 8 | 2.340 | 3.78 × 10−17 | Inflammatory process |
| ENSG00000123496 | IL13RA2 | interleukin 13 receptor subunit alpha 2 | 1.324 | 9.69 × 10−6 | Inflammatory process |
| ENSG00000125538 | IL1B | interleukin 1 beta | 1.940 | 2.34 × 10−14 | Inflammatory process |
| ENSG00000162892 | IL24 | interleukin 24 | 3.105 | 2.62 × 10−16 | Inflammatory process |
| ENSG00000136244 | IL6 | interleukin 6 | 1.654 | 2.51 × 10−6 | Inflammatory process |
| ENSG00000134070 | IRAK2 | interleukin 1 receptor associated kinase 2 | 1.446 | 1.34 × 10−24 | Inflammatory process |
| ENSG00000119714 | GPR68 | G protein-coupled receptor 68 | 1.839 | 5.85 × 10−6 | Inflammatory process |
| ENSG00000175040 | CHST2 | carbohydrate sulfotransferase 2 | 1.046 | 1.22 × 10−9 | Inflammatory process |
| ENSG00000271605 | MILR1 | mast cell immunoglobulin like receptor 1 | 1.339 | 8.66 × 10−4 | Inflammatory process |
| ENSG00000130203 | APOE | apolipoprotein E | −1.092 | 4.88 × 10−9 | Inflammatory process |
| ENSG00000196639 | HRH1 | histamine receptor H1 | −1.494 | 6.54 × 10−10 | Inflammatory process |
| ENSG00000172156 | CCL11 | C-C motif chemokine ligand 11 | −1.416 | 4.72 × 10−2 | Inflammatory process |
| ENSG00000168952 | STXBP6 | syntaxin binding protein 6 | −1.283 | 3.40 × 10−3 | Inflammatory process |
| ENSG00000129009 | ISLR | immunoglobulin superfamily containing leucine rich repeat | −1.501 | 9.35 × 10−20 | Inflammatory process |
| ENSG00000170989 | S1PR1 | sphingosine-1-phosphate receptor 1 | −1.683 | 3.54 × 10−8 | Inflammatory process |
| ENSG00000235568 | NFAM1 | NFAT activating protein with ITAM motif 1 | −2.546 | 3.56 × 10−4 | Inflammatory process |
| ENSG00000124212 | PTGIS | prostaglandin I2 synthase | −2.422 | 1.80 × 10−22 | Inflammatory process |
| ENSG00000185432 | METTL7A | methyltransferase like 7A | −1.523 | 7.00 × 10−10 | Inflammatory process |
| ENSG00000169432 | SCN9A | sodium voltage-gated channel alpha subunit 9 | −1.245 | 1.42 × 10−4 | Inflammatory process |
| ENSG00000163701 | IL17RE | interleukin 17 receptor E | −1.421 | 2.73 × 10−2 | Inflammatory process |
| ENSG00000167191 | GPRC5B | G protein-coupled receptor class C group 5 member B | −1.650 | 1.14 × 10−9 | Inflammatory process |
| ENSG00000162645 | GBP2 | guanylate binding protein 2 | −2.361 | 2.44 × 10−34 | Inflammatory process |
| ENSG00000170323 | FABP4 | fatty acid binding protein 4 | −4.620 | 4.47 × 10−6 | Inflammatory process |
| ENSG00000162692 | VCAM1 | vascular cell adhesion molecule 1 | −2.990 | 7.32 × 10−9 | Inflammatory process |
| ENSG00000144730 | IL17RD | interleukin 17 receptor D | −1.157 | 4.32 × 10−2 | Inflammatory process |
| ENSG00000116106 | EPHA4 | EPH receptor A4 | −1.815 | 2.62 × 10−7 | Inflammatory process |
| ENSG00000182580 | EPHB3 | EPH receptor B3 | −1.264 | 6.89 × 10−5 | Inflammatory process |
| ENSG00000106123 | EPHB6 | EPH receptor B6 | −1.367 | 1.33 × 10−10 | Inflammatory process |
| ENSG00000238271 | IFNWP19 | interferon omega 1 pseudogene 19 | −1.612 | 4.34 × 10−2 | Inflammatory process |
| ENSG00000133056 | PIK3C2B | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta | −1.449 | 2.38 × 10−2 | Inflammatory process |
| ENSG00000145675 | PIK3R1 | phosphoinositide-3-kinase regulatory subunit 1 | −1.017 | 1.15 × 10−2 | Inflammatory process |
| ENSG00000164099 | PRSS12 | protease, serine 12 | −1.081 | 1.70 × 10−4 | Inflammatory process |
| ENSG00000090889 | KIF4A | kinesin family member 4A | −1.560 | 3.32 × 10−2 | Inflammatory process |
| ENSG00000099998 | GGT5 | gamma-glutamyltransferase 5 | 1.009 | 9.64 × 10−14 | Response to oxidative stress |
| ENSG00000148834 | GSTO1 | glutathione S-transferase omega 1 | 1.653 | 1.22 × 10−3 | Response to oxidative stress |
| ENSG00000198763 | MT-ND2 | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 | 1.118 | 3.23 × 10−2 | Response to oxidative stress |
| ENSG00000086991 | NOX4 | NADPH oxidase 4 | 1.604 | 2.24 × 10−5 | Response to oxidative stress |
| ENSG00000158125 | XDH | xanthine dehydrogenase | 1.317 | 2.20 × 10−4 | Response to oxidative stress |
| ENSG00000122378 | FAM213A | family with sequence similarity 213 member A | 1.054 | 9.19 × 10−7 | Response to oxidative stress |
| ENSG00000117592 | PRDX6 | peroxiredoxin 6 | 1.014 | 1.90 × 10−2 | Response to oxidative stress |
| ENSG00000188906 | LRRK2 | leucine rich repeat kinase 2 | −1.082 | 2.49 × 10−2 | Response to oxidative stress |
| ENSG00000196139 | AKR1C3 | aldo-keto reductase family 1 member C3 | −1.442 | 3.29 × 10−7 | Response to oxidative stress |
| ENSG00000123453 | SARDH | sarcosine dehydrogenase | −1.445 | 1.03 × 10−2 | Response to oxidative stress |
| ENSG00000196616 | ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide | −3.297 | 1.15 × 10−47 | Response to oxidative stress |
| ENSG00000165092 | ALDH1A1 | aldehyde dehydrogenase 1 family member A1 | −1.811 | 7.67 × 10−56 | Response to oxidative stress |
| ENSG00000109819 | PPARGC1A | PPARG coactivator 1 alpha | −1.062 | 2.51 × 10−6 | Response to oxidative stress |
| ENSG00000155962 | CLIC2 | chloride intracellular channel 2 | −1.595 | 2.65 × 10−2 | Response to oxidative stress |
| ENSG00000240069 | GPAA1P1 | glycosylphosphatidylinositol anchor attachment 1 pseudogene 1 | 2.788 | 5.44 × 10−4 | Communication |
| ENSG00000230596 | GPAA1P2 | glycosylphosphatidylinositol anchor attachment 1 pseudogene 2 | 2.360 | 2.70 × 10−3 | Communication |
| ENSG00000115756 | HPCAL1 | hippocalcin like 1 | 1.011 | 3.56 × 10−3 | Communication |
| ENSG00000180332 | KCTD4 | potassium channel tetramerization domain containing 4 | 2.199 | 5.29 × 10−4 | Communication |
| ENSG00000166562 | SEC11C | SEC11 homolog C, signal peptidase complex subunit | 1.536 | 1.19 × 10−3 | Communication |
| ENSG00000139364 | TMEM132B | transmembrane protein 132B | 1.465 | 1.59 × 10−7 | Communication |
| ENSG00000249992 | TMEM158 | transmembrane protein 158 (gene/pseudogene) | 3.051 | 2.25 × 10−6 | Communication |
| ENSG00000157111 | TMEM171 | transmembrane protein 171 | 1.540 | 4.72 × 10−2 | Communication |
| ENSG00000095209 | TMEM38B | transmembrane protein 38B | 1.152 | 1.87 × 10−5 | Communication |
| ENSG00000179431 | FJX1 | four jointed box 1 | 1.432 | 2.05 × 10−4 | Communication |
| ENSG00000143341 | HMCN1 | hemicentin 1 | −2.093 | 1.63 × 10−6 | Communication |
| ENSG00000144681 | STAC | SH3 and cysteine rich domain | −3.241 | 3.30 × 10−12 | Communication |
| ENSG00000171992 | SYNPO | synaptopodin | −1.400 | 2.83 × 10−3 | Communication |
| ENSG00000172403 | SYNPO2 | synaptopodin 2 | −3.016 | 9.64 × 10−14 | Communication |
| ENSG00000111452 | ADGRD1 | adhesion G protein-coupled receptor D1 | −1.390 | 1.36 × 10−3 | Communication |
| ENSG00000126016 | AMOT | angiomotin | −2.152 | 4.31 × 10−12 | Communication |
| ENSG00000113209 | PCDHB5 | protocadherin beta 5 | −1.191 | 4.99 × 10−2 | Communication |
| ENSG00000113212 | PCDHB7 | protocadherin beta 7 | −1.428 | 1.70 × 10−2 | Communication |
| ENSG00000163531 | NFASC | neurofascin | −1.183 | 1.11 × 10−2 | Communication |
| ENSG00000007944 | MYLIP | myosin regulatory light chain interacting protein | −1.078 | 1.18 × 10−6 | Communication |
| ENSG00000169760 | NLGN1 | neuroligin 1 | −1.523 | 3.37 × 10−5 | Communication |
| ENSG00000144857 | BOC | BOC cell adhesion associated, oncogene regulated | −1.222 | 6.43 × 10−5 | Communication |
| ENSG00000162849 | KIF26B | kinesin family member 26B | −1.849 | 5.18 × 10−6 | Communication |
| ENSG00000226237 | GAS1RR | GAS1 adjacent regulatory RNA | −1.923 | 4.31 × 10−3 | Communication |
| ENSG00000185565 | LSAMP | limbic system-associated membrane protein | −1.733 | 2.28 × 10−3 | Communication |
| ENSG00000158258 | CLSTN2 | calsyntenin 2 | −2.251 | 3.02 × 10−8 | Communication |
| ENSG00000163520 | FBLN2 | fibulin 2 | −1.135 | 8.04 × 10−5 | Communication |
| ENSG00000153707 | PTPRD | protein tyrosine phosphatase, receptor type D | −1.549 | 1.33 × 10−4 | Communication |
| ENSG00000175356 | SCUBE2 | signal peptide, CUB domain and EGF like domain containing 2 | −1.335 | 8.72 × 10−4 | Communication |
| ENSG00000182010 | RTKN2 | rhotekin 2 | −2.248 | 3.82 × 10−3 | Communication |
| ENSG00000068831 | RASGRP2 | RAS guanyl releasing protein 2 | −1.188 | 1.22 × 10−2 | Communication |
| ENSG00000171408 | PDE7B | phosphodiesterase 7B | −1.071 | 1.94 × 10−3 | Communication |
| ENSG00000103710 | RASL12 | RAS like family 12 | −1.189 | 2.06 × 10−6 | Communication |
| ENSG00000145934 | TENM2 | teneurin transmembrane protein 2 | −1.527 | 6.39 × 10−3 | Communication |
| ENSG00000166448 | TMEM130 | transmembrane protein 130 | −2.267 | 5.15 × 10−6 | Communication |
| ENSG00000178401 | DNAJC22 | DnaJ heat shock protein family (Hsp40) member C22 | −1.357 | 1.14 × 10−4 | Communication |
| ENSG00000185442 | FAM174B | family with sequence similarity 174 member B | −1.141 | 2.79 × 10−7 | Communication |
| ENSG00000177363 | LRRN4CL | LRRN4 C-terminal like | −1.072 | 2.24 × 10−5 | Communication |
| ENSG00000170153 | RNF150 | ring finger protein 150 | −2.208 | 3.10 × 10−13 | Communication |
| ENSG00000185561 | TLCD2 | TLC domain containing 2 | −1.134 | 2.38 × 10−5 | Communication |
| ENSG00000151690 | MFSD6 | major facilitator superfamily domain containing 6 | −1.024 | 2.12 × 10−7 | Communication |
| ENSG00000226887 | ERVMER34-1 | endogenous retrovirus group MER34 member 1, envelope | −2.372 | 5.16 × 10−3 | Communication |
| ENSG00000005379 | TSPOAP1 | TSPO associated protein 1 | −2.017 | 1.32 × 10−5 | Communication |
| ENSG00000013297 | CLDN11 | claudin 11 | −1.263 | 1.23 × 10−5 | Communication |
| ENSG00000172348 | RCAN2 | regulator of calcineurin 2 | −2.240 | 9.99 × 10−8 | Communication |
| ENSG00000198542 | ITGBL1 | integrin subunit beta like 1 | −1.045 | 1.55 × 10−8 | Communication |
| ENSG00000113805 | CNTN3 | contactin 3 | −1.285 | 1.94 × 10−4 | Communication |
| ENSG00000165124 | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | −1.546 | 1.36 × 10−4 | Communication |
| ENSG00000162804 | SNED1 | sushi, nidogen and EGF like domains 1 | −1.163 | 8.26 × 10−4 | Communication |
| ENSG00000187244 | BCAM | basal cell adhesion molecule (Lutheran blood group) | −1.088 | 3.79 × 10−5 | Communication |
| ENSG00000064309 | CDON | cell adhesion associated, oncogene regulated | −2.033 | 2.00 × 10−13 | Communication |
| ENSG00000095203 | EPB41L4B | erythrocyte membrane protein band 4.1 like 4B | −1.389 | 6.09 × 10−3 | Communication |
| ENSG00000157193 | LRP8 | LDL receptor related protein 8 | 1.448 | 9.21 × 10−4 | Lipids(barrier function) |
| ENSG00000073756 | PTGS2 | prostaglandin-endoperoxide synthase 2 | 1.274 | 1.20 × 10−8 | Lipids(barrier function) |
| ENSG00000123689 | G0S2 | G0/G1 switch 2 | 1.382 | 7.06 × 10−5 | Lipids(barrier function) |
| ENSG00000176170 | SPHK1 | sphingosine kinase 1 | 1.014 | 2.42 × 10−4 | Lipids(barrier function) |
| ENSG00000105499 | PLA2G4C | phospholipase A2 group IVC | 1.145 | 7.84 × 10−11 | Lipids(barrier function) |
| ENSG00000101188 | NTSR1 | neurotensin receptor 1 | 1.895 | 2.44 × 10−2 | Lipids(barrier function) |
| ENSG00000076555 | ACACB | acetyl-CoA carboxylase beta | −1.405 | 3.86 × 10−3 | Lipids(barrier function) |
| ENSG00000100342 | APOL1 | apolipoprotein L1 | −1.841 | 2.88 × 10−4 | Lipids(barrier function) |
| ENSG00000157399 | ARSE | arylsulfatase E (chondrodysplasia punctata 1) | −1.230 | 1.09 × 10−3 | Lipids(barrier function) |
| ENSG00000106484 | MEST | mesoderm specific transcript | −1.829 | 6.91 × 10−15 | Lipids(barrier function) |
| ENSG00000169255 | B3GALNT1 | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) | −1.071 | 9.26 × 10−4 | Lipids(barrier function) |
| ENSG00000120915 | EPHX2 | epoxide hydrolase 2 | −1.744 | 1.48 × 10−3 | Lipids(barrier function) |
| ENSG00000169174 | PCSK9 | proprotein convertase subtilisin/kexin type 9 | −2.368 | 3.78 × 10−2 | Lipids(barrier function) |
| ENSG00000162407 | PLPP3 | phospholipid phosphatase 3 | −1.408 | 1.06 × 10−15 | Lipids(barrier function) |
| ENSG00000133321 | RARRES3 | retinoic acid receptor responder 3 | −1.892 | 1.00 × 10−2 | Lipids(barrier function) |
| ENSG00000137642 | SORL1 | sortilin related receptor 1 | −1.848 | 2.32 × 10−2 | Lipids(barrier function) |
| ENSG00000172296 | SPTLC3 | serine palmitoyltransferase long chain base subunit 3 | −2.612 | 3.34 × 10−5 | Lipids(barrier function) |
| ENSG00000129951 | PLPPR3 | phospholipid phosphatase related 3 | −1.639 | 2.62 × 10−2 | Lipids(barrier function) |
| ENSG00000134343 | ANO3 | anoctamin 3 | −3.413 | 5.11 × 10−5 | Lipids(barrier function) |
| ENSG00000172594 | SMPDL3A | sphingomyelin phosphodiesterase acid like 3A | −1.331 | 9.50 × 10−5 | Lipids(barrier function) |
| ENSG00000147465 | STAR | steroidogenic acute regulatory protein | −1.107 | 4.63 × 10−2 | Lipids(barrier function) |
| ENSG00000099204 | ABLIM1 | actin binding LIM protein 1 | −1.299 | 2.25 × 10−6 | Cytoskeleton organization |
| ENSG00000167549 | CORO6 | coronin 6 | −1.256 | 4.91 × 10−4 | Cytoskeleton organization |
| ENSG00000146122 | DAAM2 | dishevelled associated activator of morphogenesis 2 | −1.158 | 3.78 × 10−2 | Cytoskeleton organization |
| ENSG00000139734 | DIAPH3 | diaphanous related formin 3 | −1.223 | 2.20 × 10−2 | Cytoskeleton organization |
| ENSG00000082397 | EPB41L3 | erythrocyte membrane protein band 4.1 like 3 | −1.296 | 2.38 × 10−8 | Cytoskeleton organization |
| ENSG00000078018 | MAP2 | microtubule associated protein 2 | −3.102 | 1.25 × 10−11 | Cytoskeleton organization |
| ENSG00000116141 | MARK1 | microtubule affinity regulating kinase 1 | −1.291 | 9.50 × 10−5 | Cytoskeleton organization |
| ENSG00000133026 | MYH10 | myosin heavy chain 10 | −1.297 | 2.58 × 10−3 | Cytoskeleton organization |
| ENSG00000099864 | PALM | paralemmin | −1.255 | 3.72 × 10−9 | Cytoskeleton organization |
| ENSG00000118898 | PPL | periplakin | −2.858 | 4.50 × 10−16 | Cytoskeleton organization |
| ENSG00000126785 | RHOJ | ras homolog family member J | −1.184 | 1.02 × 10−2 | Cytoskeleton organization |
| ENSG00000168477 | TNXB | tenascin XB | −1.431 | 1.19 × 10−3 | Cytoskeleton organization |
| ENSG00000137285 | TUBB2B | tubulin beta 2B class IIb | −1.460 | 4.75 × 10−9 | Cytoskeleton organization |
| ENSG00000148700 | ADD3 | adducin 3 | −1.009 | 2.68 × 10−6 | Cytoskeleton organization |
| ENSG00000231789 | PIK3CD-AS2 | PIK3CD antisense RNA 2 | −1.395 | 1.54 × 10−2 | Cytoskeleton organization |
| ENSG00000152527 | PLEKHH2 | pleckstrin homology, MyTH4 and FERM domain containing H2 | −1.017 | 4.84 × 10−3 | Cytoskeleton organization |
| ENSG00000188783 | PRELP | proline and arginine rich end leucine rich repeat protein | −1.594 | 2.61 × 10−29 | Cytoskeleton organization |
| ENSG00000222047 | C10orf55 | chromosome 10 open reading frame 55 | 1.712 | 4.81 × 10−3 | Unknown |
| ENSG00000163814 | CDCP1 | CUB domain containing protein 1 | 1.621 | 2.86 × 10−17 | Unknown |
| ENSG00000154319 | FAM167A | family with sequence similarity 167 member A | 1.082 | 5.30 × 10−10 | Unknown |
| ENSG00000166578 | IQCD | IQ motif containing D | 1.637 | 1.34 × 10−6 | Unknown |
| ENSG00000253522 | MIR3142HG | MIR3142 host gene | 1.555 | 1.41 × 10−2 | Unknown |
| ENSG00000164929 | BAALC | brain and acute leukemia, cytoplasmic | 1.215 | 1.50 × 10−2 | Unknown |
| ENSG00000239704 | CDRT4 | CMT1A duplicated region transcript 4 | 3.443 | 2.85 × 10−2 | Unknown |
| ENSG00000260549 | MT1L | metallothionein 1L, pseudogene | 2.582 | 6.86 × 10−5 | Unknown |
| ENSG00000187193 | MT1X | metallothionein 1X | 1.349 | 4.34 × 10−2 | Unknown |
| ENSG00000125148 | MT2A | metallothionein 2A | 2.408 | 4.51 × 10−5 | Unknown |
| ENSG00000217930 | PAM16 | presequence translocase associated motor 16 homolog | 1.400 | 3.06 × 10−2 | Unknown |
| ENSG00000241749 | RPSAP52 | ribosomal protein SA pseudogene 52 | 1.471 | 8.60 × 10−6 | Unknown |
| ENSG00000186577 | SMIM29 | small integral membrane protein 29 | 1.077 | 3.37 × 10−2 | Unknown |
| ENSG00000157111 | TMEM171 | transmembrane protein 171 | 1.540 | 4.72 × 10−2 | Unknown |
| ENSG00000185262 | UBALD2 | UBA like domain containing 2 | 1.329 | 2.38 × 10−3 | Unknown |
| ENSG00000240476 | LINC00973 | long intergenic non-protein coding RNA 973 | 2.326 | 5.72 × 10−3 | Unknown |
| ENSG00000257219 | LINC02407 | long intergenic non-protein coding RNA 2407 | 2.301 | 7.04 × 10−3 | Unknown |
| ENSG00000235385 | LINC02154 | long intergenic non-protein coding RNA 2154 | 2.177 | 2.30 × 10−3 | Unknown |
| ENSG00000164236 | ANKRD33B | ankyrin repeat domain 33B | −1.240 | 3.89 × 10−3 | Unknown |
| ENSG00000264230 | ANXA8L1 | annexin A8 like 1 | −2.813 | 4.43 × 10−4 | Unknown |
| ENSG00000198624 | CCDC69 | coiled-coil domain containing 69 | −1.320 | 6.43 × 10−13 | Unknown |
| ENSG00000055813 | CCDC85A | coiled-coil domain containing 85A | −2.721 | 1.19 × 10−2 | Unknown |
| ENSG00000105516 | DBP | D-box binding PAR bZIP transcription factor | −1.355 | 3.37 × 10−2 | Unknown |
| ENSG00000258498 | DIO3OS | DIO3 opposite strand/antisense RNA (head to head) | −1.035 | 2.02 × 10−3 | Unknown |
| ENSG00000229847 | EMX2OS | EMX2 opposite strand/antisense RNA | −1.016 | 1.90 × 10−4 | Unknown |
| ENSG00000133106 | EPSTI1 | epithelial stromal interaction 1 | −1.184 | 3.04 × 10−13 | Unknown |
| ENSG00000162636 | FAM102B | family with sequence similarity 102 member B | −1.024 | 3.21 × 10−4 | Unknown |
| ENSG00000255052 | FAM66D | family with sequence similarity 66 member D | −1.599 | 4.85 × 10−3 | Unknown |
| ENSG00000265962 | GACAT2 | gastric cancer associated transcript 2 (non-protein coding) | −1.788 | 7.03 × 10−3 | Unknown |
| ENSG00000072163 | LIMS2 | LIM zinc finger domain containing 2 | −1.501 | 3.09 × 10−17 | Unknown |
| ENSG00000225783 | MIAT | myocardial infarction associated transcript (non-protein coding) | −2.924 | 4.61 × 10−11 | Unknown |
| ENSG00000220785 | MTMR9LP | myotubularin related protein 9-like, pseudogene | −1.153 | 3.36 × 10−3 | Unknown |
| ENSG00000183486 | MX2 | MX dynamin like GTPase 2 | −1.475 | 1.38 × 10−4 | Unknown |
| ENSG00000143850 | PLEKHA6 | pleckstrin homology domain containing A6 | −1.190 | 4.27 × 10−3 | Unknown |
| ENSG00000230487 | PSMG3-AS1 | PSMG3 antisense RNA 1 (head to head) | −1.008 | 2.36 × 10−3 | Unknown |
| ENSG00000203727 | SAMD5 | sterile alpha motif domain containing 5 | −2.663 | 2.06 × 10−14 | Unknown |
| ENSG00000111907 | TPD52L1 | tumor protein D52 like 1 | −1.743 | 1.79 × 10−2 | Unknown |
| ENSG00000183801 | OLFML1 | olfactomedin like 1 | −2.388 | 6.30 × 10−18 | Unknown |
| ENSG00000116774 | OLFML3 | olfactomedin like 3 | −1.813 | 3.10 × 10−27 | Unknown |
| ENSG00000103145 | HCFC1R1 | host cell factor C1 regulator 1 | −1.254 | 9.23 × 10−4 | Unknown |
| ENSG00000156042 | CFAP70 | cilia and flagella associated protein 70 | −1.335 | 4.93 × 10−2 | Unknown |
| ENSG00000147642 | SYBU | syntabulin | −1.633 | 2.50 × 10−2 | Unknown |
| ENSG00000165507 | C10orf10 | chromosome 10 open reading frame 10 | −1.072 | 1.75 × 10−6 | Unknown |
| ENSG00000110002 | VWA5A | von Willebrand factor A domain containing 5A | −1.182 | 3.26 × 10−7 | Unknown |
| ENSG00000082497 | SERTAD4 | SERTA domain containing 4 | −1.686 | 8.66 × 10−9 | Unknown |
| ENSG00000130518 | KIAA1683 | KIAA1683 | −1.007 | 1.23 × 10−2 | Unknown |
| ENSG00000152217 | SETBP1 | SET binding protein 1 | −1.253 | 7.10 × 10−4 | Unknown |
| ENSG00000198846 | TOX | thymocyte selection associated high mobility group box | −1.792 | 1.29 × 10−4 | Unknown |
| ENSG00000139714 | MORN3 | MORN repeat containing 3 | −1.540 | 4.27 × 10−2 | Unknown |
| ENSG00000164309 | CMYA5 | cardiomyopathy associated 5 | −1.344 | 3.12 × 10−2 | Unknown |
| ENSG00000072952 | MRVI1 | murine retrovirus integration site 1 homolog | −1.251 | 7.52 × 10−5 | Unknown |
| ENSG00000204789 | ZNF204P | zinc finger protein 204, pseudogene | −2.032 | 4.81 × 10−3 | Unknown |
| ENSG00000116667 | C1orf21 | chromosome 1 open reading frame 21 | −1.219 | 3.04 × 10−12 | Unknown |
| ENSG00000120262 | CCDC170 | coiled-coil domain containing 170 | −1.921 | 8.42 × 10−9 | Unknown |
| ENSG00000204396 | VWA7 | von Willebrand factor A domain containing 7 | −2.105 | 1.73 × 10−2 | Unknown |
| Samples | RFB | HBFB | OFB | |
|---|---|---|---|---|
| Matter Content (%) | ||||
| Total sugar | 43.36 ± 0.14 | 12.16 ± 0.11 | 9.69 ± 0.17 | |
| Reducing sugar | 3.18 ± 0.01 | 0.91 ± 0.03 | 0.12 ± 0.02 | |
| β-glucan | 0.78 ± 0.05 | 0.84 ± 0.08 | 2.74 ± 0.04 | |
| Flavonoids | 1.11 ± 0.01 | 1.31 ± 0.06 | 1.17 ± 0.01 | |
| Total phenol | 1.46 ± 0.09 | 1.84 ± 0.05 | 1.75 ± 0.03 | |
| Samples | RFB | HBFB | OFB | |
|---|---|---|---|---|
| Matter Content (%) | ||||
| Fat | 0.6 | 1.1 | 0.9 | |
| Protein proportion | 7.83 | 14.9 | 27.3 | |
| Ash | 0.35 | 4.0 | 4.6 | |
| Moisture | 7.3 | 6.71 | 7.95 | |
| Carbohydrate | 83.9 | 73.3 | 59.2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, W.; Shi, X.; Zhang, J.; Li, L.; Di, F.; Li, M.; Wang, C.; An, Q.; Zhao, D. Role of PI3K-AKT Pathway in Ultraviolet Ray and Hydrogen Peroxide-Induced Oxidative Damage and Its Repair by Grain Ferments. Foods 2023, 12, 806. https://doi.org/10.3390/foods12040806
Cheng W, Shi X, Zhang J, Li L, Di F, Li M, Wang C, An Q, Zhao D. Role of PI3K-AKT Pathway in Ultraviolet Ray and Hydrogen Peroxide-Induced Oxidative Damage and Its Repair by Grain Ferments. Foods. 2023; 12(4):806. https://doi.org/10.3390/foods12040806
Chicago/Turabian StyleCheng, Wenjing, Xiuqin Shi, Jiachan Zhang, Luyao Li, Feiqian Di, Meng Li, Changtao Wang, Quan An, and Dan Zhao. 2023. "Role of PI3K-AKT Pathway in Ultraviolet Ray and Hydrogen Peroxide-Induced Oxidative Damage and Its Repair by Grain Ferments" Foods 12, no. 4: 806. https://doi.org/10.3390/foods12040806
APA StyleCheng, W., Shi, X., Zhang, J., Li, L., Di, F., Li, M., Wang, C., An, Q., & Zhao, D. (2023). Role of PI3K-AKT Pathway in Ultraviolet Ray and Hydrogen Peroxide-Induced Oxidative Damage and Its Repair by Grain Ferments. Foods, 12(4), 806. https://doi.org/10.3390/foods12040806