Identification of Milk Adulteration in Camel Milk Using FT-Mid-Infrared Spectroscopy and Machine Learning Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. FT-MIR Analysis and Data Preprocessing
2.3. Data Preprocessing and Model Building
2.4. Quality Control for the Method
3. Results and Discussion
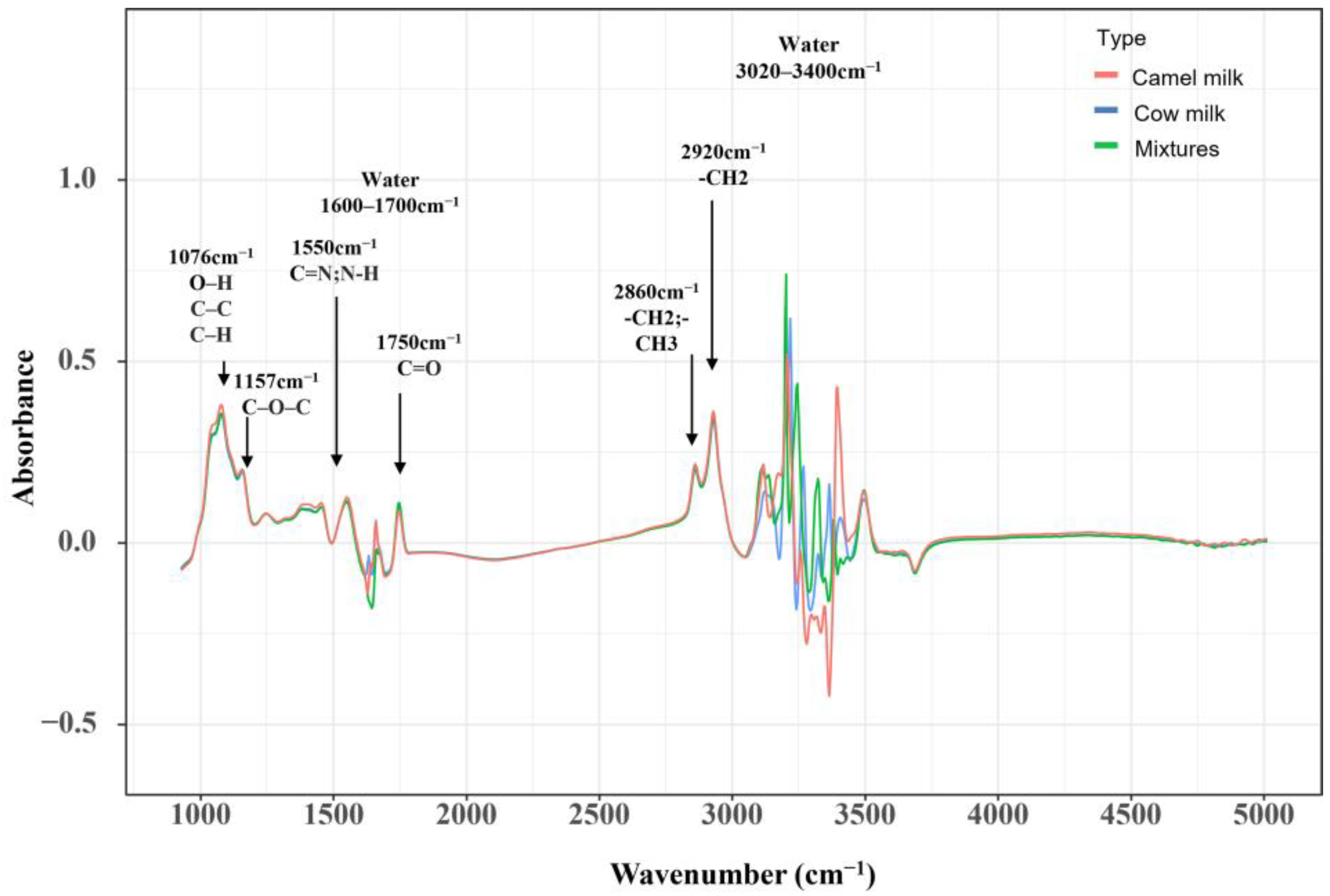
3.1. FT-MIR of Camel Milk, Cow’s Milk and Adulterated Milk
3.2. Identification of Adulteration in Camel Milk with Cow Milk
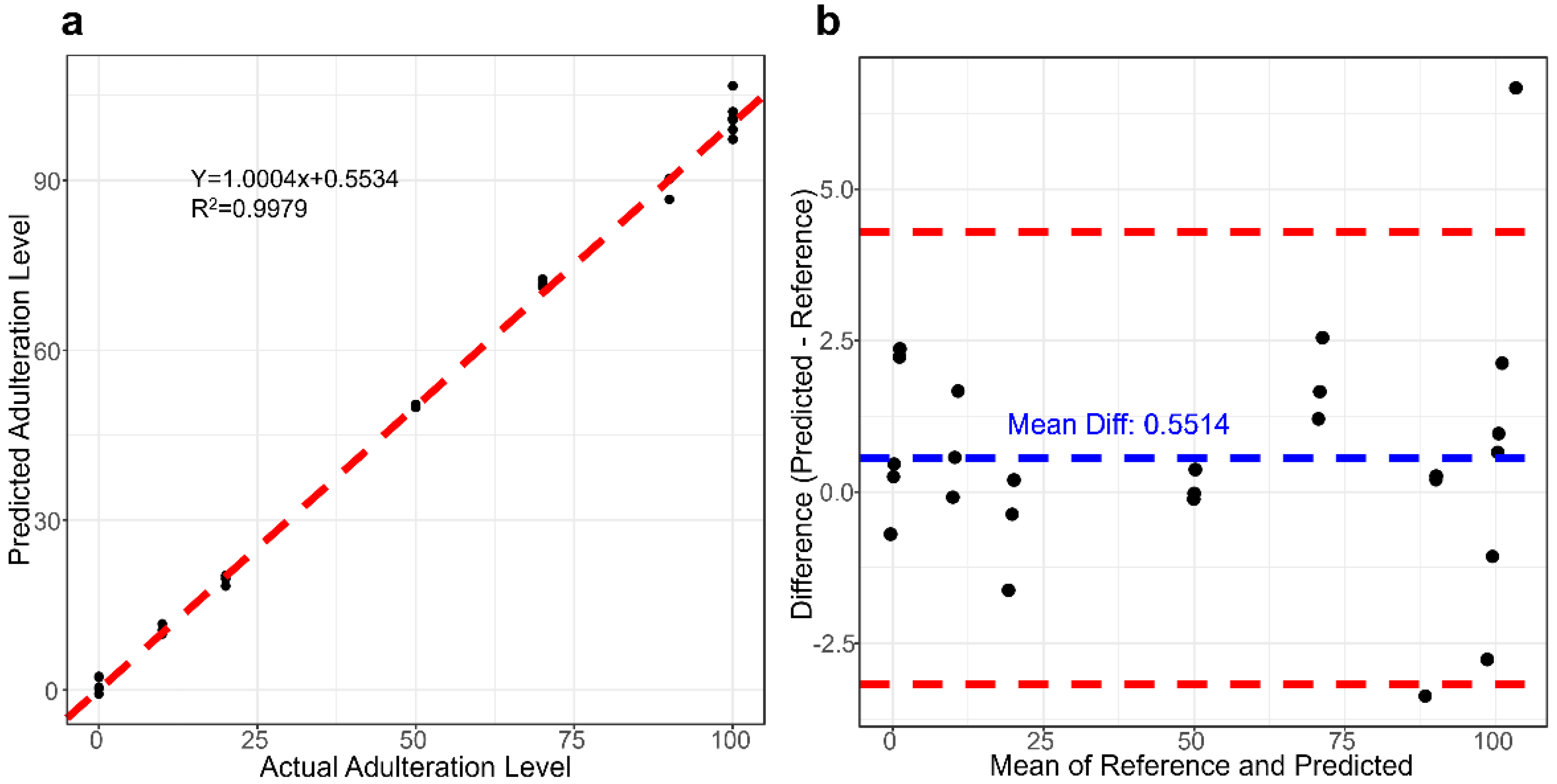
3.3. Determination of Adulteration in Camel Milk with Cow Milk
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mati, A.; Senoussi-Ghezali, C.; Zennia, S.S.A.; Almi-Sebbane, D.; El-Hatmi, H.; Girardet, J.-M. Dromedary camel milk proteins, a source of peptides having biological activities—A review. Int. Dairy J. 2017, 73, 25–37. [Google Scholar] [CrossRef]
- Konuspayeva, G.; Faye, B.; Bengoumi, M. Mineral status in camel milk: A critical review. Anim. Front. 2022, 12, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Benmeziane–Derradji, F. Evaluation of camel milk: Gross composition—A scientific overview. Trop. Anim. Health Prod. 2021, 53, 308. [Google Scholar] [CrossRef] [PubMed]
- Muthukumaran, M.S.; Mudgil, P.; Baba, W.N.; Ayoub, M.A.; Maqsood, S. A comprehensive review on health benefits, nutritional composition and processed products of camel milk. Food Rev. Int. 2023, 39, 3080–3116. [Google Scholar] [CrossRef]
- Shakeel, K.; Rabail, R.; Sehar, S.; Nawaz, A.; Manzoor, M.F.; Walayat, N.; Socol, C.T.; Maerescu, C.M.; Aadil, R.M. Camel milk protectiveness toward multiple liver disorders: A review. Front. Nutr. 2022, 9, 944842. [Google Scholar] [CrossRef]
- Al-wabel, N.A.; Hassan, A.; Abbas, H.; Muosa, H. Antiulcerogenic effect of camel milk against ethanol induced gastric ulcers in rats. WebmedCent. Vet. Med. 2012, 3, WMC002804. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Wang, J.; Li, M.; Yang, Y.; Wang, Z.; Miao, J.; Zhao, Z.; Yang, J. Detection of the adulteration of camel milk powder with cow milk by ultra-high performance liquid chromatography (UPLC). Int. Dairy J. 2021, 121, 105117. [Google Scholar] [CrossRef]
- Boukria, O.; Boudalia, S.; Bhat, Z.F.; Hassoun, A.; Aït-Kaddour, A. Evaluation of the adulteration of camel milk by non-camel milk using multispectral image, fluorescence and infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 300, 122932. [Google Scholar] [CrossRef]
- Zavoi, S.; Fetea, F.; Ranga, F.; Baciu, A.; Socaciu, C. Comparative fingerprint and extraction yield of medicinal herb phenolics with hepatoprotective potential, as determined by UV-Vis and FT-MIR spectroscopy. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 82–89. [Google Scholar] [CrossRef]
- Temizkan, R.; Can, A.; Dogan, M.A.; Mortas, M.; Ayvaz, H. Rapid detection of milk fat adulteration in yoghurts using near and mid-infrared spectroscopy. Int. Dairy J. 2020, 110, 104795. [Google Scholar] [CrossRef]
- Pereira, C.G.; Leite, A.I.N.; Andrade, J.; Bell, M.J.V.; Anjos, V. Evaluation of butter oil adulteration with soybean oil by FT-MIR and FT-NIR spectroscopies and multivariate analyses. LWT 2019, 107, 1–8. [Google Scholar] [CrossRef]
- Balan, B.; Dhaulaniya, A.S.; Jamwal, R.; Sodhi, K.K.; Kelly, S.; Cannavan, A.; Singh, D.K. Application of Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) spectroscopy coupled with chemometrics for detection and quantification of formalin in cow milk. Vib. Spectrosc. 2020, 107, 103033. [Google Scholar] [CrossRef]
- Friggens, N.; Ridder, C.; Løvendahl, P. On the use of milk composition measures to predict the energy balance of dairy cows. J. Dairy Sci. 2007, 90, 5453–5467. [Google Scholar] [CrossRef] [PubMed]
- Ferragina, A.; de Los Campos, G.; Vazquez, A.; Cecchinato, A.; Bittante, G. Bayesian regression models outperform partial least squares methods for predicting milk components and technological properties using infrared spectral data. J. Dairy Sci. 2015, 98, 8133–8151. [Google Scholar] [CrossRef] [PubMed]
- Mabood, F.; Jabeen, F.; Ahmed, M.; Hussain, J.; Al Mashaykhi, S.A.; Al Rubaiey, Z.M.; Farooq, S.; Boqué, R.; Ali, L.; Hussain, Z. Development of new NIR-spectroscopy method combined with multivariate analysis for detection of adulteration in camel milk with goat milk. Food Chem. 2017, 221, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, K.L.; Barbano, D.M. Prediction of fatty acid chain length and unsaturation of milk fat by mid-infrared milk analysis. J. Dairy Sci. 2016, 99, 8561–8570. [Google Scholar] [CrossRef] [PubMed]
- Lanjewar, M.G.; Morajkar, P.P.; Parab, J.S. Portable system to detect starch adulteration in turmeric using NIR spectroscopy. Food Control 2024, 155, 110095. [Google Scholar] [CrossRef]
- El Orche, A.; Mamad, A.; Elhamdaoui, O.; Cheikh, A.; El Karbane, M.; Bouatia, M. Comparison of Machine Learning Classification Methods for Determining the Geographical Origin of Raw Milk Using Vibrational Spectroscopy. J. Spectrosc. 2021, 2021, 5845422. [Google Scholar] [CrossRef]
- Amsaraj, R.; Ambade, N.D.; Mutturi, S. Variable selection coupled to PLS2, ANN and SVM for simultaneous detection of multiple adulterants in milk using spectral data. Int. Dairy J. 2021, 123, 105172. [Google Scholar] [CrossRef]
- Pérez-Calabuig, A.M.; Pradana-López, S.; Lopez-Ortega, S.; Sáenz, K.d.J.B.; Cancilla, J.C.; Torrecilla, J.S. Application of residual neural networks to detect and quantify milk adulterations. J. Food Compos. Anal. 2023, 122, 105427. [Google Scholar] [CrossRef]
- Aghili, N.S.; Rasekh, M.; Karami, H.; Edriss, O.; Wilson, A.D.; Ramos, J. Aromatic fingerprints: VOC analysis with E-nose and GC-MS for rapid detection of adulteration in sesame oil. Sensors 2023, 23, 6294. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Deng, T.; Huang, A.; Liu, N.; Jiang, X. Energy consumption prediction using the GRU-MMattention-LightGBM model with features of Prophet decomposition. PLoS ONE 2023, 18, e0277085. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F.; Chang, C.; Lin, C. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), R package version 1.7–3; TU Wien: Vienna, Austria, 2019. [Google Scholar]
- Wang, N.; Er, M.J.; Han, M. Generalized single-hidden layer feedforward networks for regression problems. IEEE Trans. Neural Netw. Learn. Syst. 2014, 26, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Soyeurt, H.; Grelet, C.; McParland, S.; Calmels, M.; Coffey, M.; Tedde, A.; Delhez, P.; Dehareng, F.; Gengler, N. A comparison of 4 different machine learning algorithms to predict lactoferrin content in bovine milk from mid-infrared spectra. J. Dairy Sci. 2020, 103, 11585–11596. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wen, Y.; Dong, N.; Lai, C.L.; Zhao, G.H. Authentication of lotus root powder adulterated with potato starch and/or sweet potato starch using Fourier transform mid-infrared spectroscopy. Food Chem. 2013, 141, 3103–3109. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Nie, P.; Zhang, X.; Chen, C.; An, Z.; Wei, K.; Zhao, J.; Lv, H.; Niu, K.; Yang, Y. Establishment and Validation of Fourier Transform Infrared Spectroscopy (FT–MIR) Methodology for the Detection of Linoleic Acid in Buffalo Milk. Foods 2023, 12, 1199. [Google Scholar] [CrossRef] [PubMed]
- Socrates, G. Infrared Characteristic Group Frequencies: Tables and Charts; Wiley: New York, NY, USA, 1995. [Google Scholar]
- Grelet, C.; Pierna, J.F.; Dardenne, P.; Baeten, V.; Dehareng, F. Standardization of milk mid-infrared spectra from a European dairy network. J. Dairy Sci. 2015, 98, 2150–2160. [Google Scholar] [CrossRef]
- Konuspayeva, G.; Faye, B.; Loiseau, G. The composition of camel milk: A meta-analysis of the literature data. J. Food Compos. Anal. 2009, 22, 95–101. [Google Scholar] [CrossRef]
- Khaskheli, M.; Arain, M.; Chaudhry, S.; Soomro, A.; Qureshi, T. Physico-chemical quality of camel milk. J. Agric. Soc. Sci. 2005, 2, 164–166. [Google Scholar]
- Musaad, A.M.; Faye, B.; Al-Mutairi, S.E. Seasonal and physiological variation of gross composition of camel milk in Saudi Arabia. Emir. J. Food Agric. 2013, 25, 618–624. [Google Scholar] [CrossRef]
- Mehaia, M.A.; Hablas, M.A.; Abdel-Rahman, K.M.; El-Mougy, S.A. Milk composition of majaheim, wadah and hamra camels in Saudi Arabia. Food Chem. 1995, 52, 115–122. [Google Scholar] [CrossRef]
- Alhaj, O.A.; Altooq, N.J.; Alenezi, A.F.; Janahi, A.I.; Janahi, M.I.; Humood, A.M.; AlRasheed, M.M.; Bragazzi, N.L.; Jahrami, H.A.; Faye, B. Camel milk composition by breed, season, publication year, and country: A global systematic review, meta-analysis, and meta-regression. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2520–2559. [Google Scholar] [CrossRef]
- Haddadin, M.S.; Gammoh, S.I.; Robinson, R.K. Seasonal variations in the chemical composition of camel milk in Jordan. J. Dairy Res. 2008, 75, 8–12. [Google Scholar] [CrossRef]
- Farag, S.; Kebary, K. Chemical composition and physical properties of camel’s milk and milk fat. In Proceedings of the 5th Egyptian Conference for Dairy Science and Technology, Cairo, Egypt, 19–21 October 1992; pp. 57–67. [Google Scholar]
- Yagil, R.; Etzion, Z. Effect of drought condition on the quality of camel milk. J. Dairy Res. 1980, 47, 159–166. [Google Scholar] [CrossRef]
- Konuspayeva, G.; Lemarie, É.; Faye, B.; Loiseau, G.; Montet, D. Fatty acid and cholesterol composition of camel’s (Camelus bactrianus, Camelus dromedarius and hybrids) milk in Kazakhstan. Dairy Sci. Technol. 2008, 88, 327–340. [Google Scholar] [CrossRef]
- Choubey, D.K.; Kumar, M.; Shukla, V.; Tripathi, S.; Dhandhania, V.K. Comparative analysis of classification methods with PCA and LDA for diabetes. Curr. Diabetes Rev. 2020, 16, 833–850. [Google Scholar]
- Tominaga, Y. Comparative study of class data analysis with PCA-LDA, SIMCA, PLS, ANNs, and k-NN. Chemom. Intell. Lab. 1999, 49, 105–115. [Google Scholar] [CrossRef]
- Sorokina, D.; Caruana, R.; Riedewald, M.; Fink, D. Detecting statistical interactions with additive groves of trees. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008; pp. 1000–1007. [Google Scholar]
- Tsang, M.; Cheng, D.; Liu, Y. Detecting statistical interactions from neural network weights. arXiv 2017, arXiv:1705.04977. [Google Scholar]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, S49. [Google Scholar]
- Wu, X.; Na, Q.; Hao, S.; Ji, R.; Ming, L. Detection of ovine or bovine milk components in commercial camel milk powder using a pcr-based method. Molecules 2022, 27, 3017. [Google Scholar] [CrossRef]
- Souhassou, S.; Bassbasi, M.; Hirri, A.; Kzaiber, F.; Oussama, A. Detection of camel milk adulteration using Fourier transformed infrared spectroscopy FT-IR coupled with chemometrics methods. Int. Food Res. J. 2018, 25, 1213–1218. [Google Scholar]
- Meng, F.; Duan, M.; Wu, W.; Shao, S.; Qin, Y.; Zhang, M. Enzymatic construction Au NPs-rGO based MIP electrochemical sensor for adulteration detection of bovine-derived allergen in camel milk. Food Chem. 2024, 436, 137638. [Google Scholar] [CrossRef]


| Item | Camel Milk (Mean ± SD) | Coefficient of Variation % | Cow Milk (Mean ± SD) | Coefficient of Variation % | Adulterated Milk (Mean ± SD) | Coefficient of Variation % |
|---|---|---|---|---|---|---|
| Fat (%) | 3.91 ± 0.46 | 11.89 | 3.40 ± 0.33 | 9.90 | 3.58 ± 0.31 | 8.85 |
| Protein (%) | 3.69 ± 0.20 | 5.43 | 3.19 ± 0.23 | 7.34 | 3.41 ± 0.21 | 6.02 |
| Lactose (%) | 5.63 ± 0.27 | 4.79 | 5.16 ± 0.16 | 3.13 | 5.37 ± 0.20 | 3.79 |
| TS (%) | 14.15 ± 0.53 | 3.75 | 12.46 ± 0.53 | 4.26 | 13.19 ± 4.68 | 4.68 |
| SNF (%) | 10.07 ± 0.36 | 3.61 | 8.92 ± 0.34 | 3.91 | 9.43 ± 0.41 | 4.38 |
| Urea (mg/100 mL) | 36.58 ± 8.62 | 23.57 | 13.08 ± 1.26 | 9.67 | 24.57 ± 8.56 | 34.84 |
| β-Casein (%) | 2.89 ± 0.14 | 4.83 | 2.54 ± 0.19 | 7.79 | 2.66 ± 0.13 | 5.08 |
| SFA (g/100 g) | 2.23 ± 0.43 | 17.78 | 2.25 ± 0.26 | 11.51 | 2.29 ± 0.25 | 11.24 |
| MUFA (g/100 g) | 1.21 ± 0.39 | 32.72 | 0.85 ± 0.13 | 15.8 | 1.02 ± 0.22 | 22.33 |
| PUFA (g/100 g) | 0.09 ± 0.03 | 33.32 | 0.08 ± 0.01 | 20.58 | 0.07 ± 0.03 | 73.57 |
| Re-Processing | Model Type | LV 1 | RMSECV 2 | R2cv 3 | RMSEP 4 | R2P 5 | PRD 6 |
|---|---|---|---|---|---|---|---|
| Base | PLS | 19 | 4.456 | 0.991 | 3.404 | 0.993 | 12.380 |
| PCA | 25 | 5.366 | 0.987 | 3.528 | 0.992 | 11.947 | |
| SVM | 0.5 | 6.188 | 0.983 | 3.289 | 0.993 | 12.814 | |
| ANN | 3; 0.01 | 4.318 | 0.992 | 3.298 | 0.994 | 12.778 | |
| SG | PLS | 14 | 4.765 | 0.989 | 3.181 | 0.994 | 13.243 |
| PCA | 25 | 5.320 | 0.986 | 3.433 | 0.993 | 12.276 | |
| SVM | 0.5 | 6.162 | 0.983 | 3.406 | 0.993 | 12.374 | |
| ANN | 3; 0.01 | 4.302 | 0.993 | 2.902 | 0.995 | 14.526 | |
| SG1 | PLS | 14 | 5.525 | 0.986 | 3.818 | 0.991 | 11.039 |
| PCA | 25 | 5.998 | 0.984 | 3.926 | 0.991 | 10.374 | |
| SVM | 0.05 | 7.426 | 0.982 | 4.687 | 0.981 | 8.992 | |
| ANN | 6; 0.001 | 4.695 | 0.992 | 3.634 | 0.992 | 11.413 | |
| SG2 | PLS | 8 | 6.997 | 0.980 | 4.417 | 0.980 | 9.543 |
| PCA | 16 | 6.879 | 0.982 | 4.587 | 0.988 | 9.188 | |
| SVM | 0.001 | 6.747 | 0.986 | 6.493 | 0.976 | 6.493 | |
| ANN | 6; 0.001 | 5.597 | 0.988 | 3.747 | 0.992 | 11.249 | |
| SNV | PLS | 14 | 7.793 | 0.959 | 5.459 | 0.983 | 7.721 |
| PCA | 25 | 7.979 | 0.966 | 5.570 | 0.982 | 7.567 | |
| SVM | 2.5 | 7.841 | 0.966 | 6.632 | 0.975 | 6.354 | |
| ANN | 3; 0.3 | 6.872 | 0.976 | 3.410 | 0.993 | 12.359 | |
| SSG | PLS | 13 | 7.840 | 0.958 | 5.553 | 0.982 | 7.591 |
| PCA | 25 | 8.076 | 0.963 | 5.553 | 0.982 | 7.591 | |
| SVM | 2.5 | 7.820 | 0.967 | 6.795 | 0.974 | 6.203 | |
| ANN | 5; 0.4 | 5.479 | 0.987 | 3.083 | 0.994 | 13.672 | |
| SSG1 | PLS | 25 | 31.281 | 0.655 | 33.286 | 0.376 | 1.266 |
| PCA | 18 | 35.301 | 0.389 | 36.297 | 0.259 | 1.611 | |
| SVM | 0.005 | 11.774 | 0.949 | 9.594 | 0.948 | 4.393 | |
| ANN | 3; 0.5 | 27.937 | 0.576 | 36.101 | 0.266 | 1.167 | |
| SSG2 | PLS | 2 | 33.484 | 0.433 | 39.983 | 0.100 | 1.054 |
| PCA | 8 | 33.689 | 0.422 | 38.610 | 0.161 | 1.091 | |
| SVM | 0.005 | 11.190 | 0.962 | 9.954 | 0.944 | 4.234 | |
| ANN | 3; 0.5 | 31.159 | 0.511 | 34.193 | 0.342 | 1.122 |
| Theoretical Value | Predicted Value | Trueness | Precision | |
|---|---|---|---|---|
| Level (g/100 g) | Mean ± SD (%) | Bias (%) | Recovery (%) | Repeatability (RSD%) |
| 10 | 10.71 ± 0.72 | 7.19 | 107.19 | 6.75 |
| 20 | 19.40 ± 0.76 | 2.97 | 97.02 | 3.91 |
| 50 | 50.07 ± 0.21 | 0.15 | 100.15 | 0.42 |
| 70 | 71.80 ± 0.55 | 2.57 | 102.57 | 0.77 |
| 90 | 89.03 ± 1.69 | 1.07 | 98.92 | 1.90 |
| Technique | Advantages | Disadvantages | Detection Effect | References |
|---|---|---|---|---|
| PCR | Very selective and sensitive | The sample DNA extraction stage requires contamination prevention, and specific primers need to be designed and synthesized. | Recoveries ranging from 80% to 110% with a coefficient of variation of less than 7% | Wu et al. [44] |
| Ultra-high performance liquid chromatography | High Resolution High Sensitivity | Expensive Equipment Complex Sample Pre-treatment | Recoveries ranging from 94% to 105% with a coefficient of variation of less than 5% | Li et al. [7] |
| NIR spectroscopy | Convenient, rapid, automated and simplify sample handling | Limited sensitivity Expensive instrumentation. | The detection limit is 0.5%, and the quantification limit is 2%. The R-squared value is 0.94. | Mabood et al. [15] |
| FTIR spectroscopy | Convenient, rapid, automated and simplify sample handling | Limited sensitivity Expensive instrumentation. | The relative error is 3.8%, and the detection limit is 2.59%. The R-squared value is 0.994. | Souhassou et al. [45] |
| Electrochemical sensor | Good speed, sensitivity and stability | Expensive instrumentation. | Identification of β-lactoglobulin within the range of 4–100 ng/mL, with a detection limit of 3.58 ng/mL. | Meng et al. [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Z.; Zhang, X.; Nie, P.; Lv, H.; Yang, Y.; Zou, W.; Yang, L. Identification of Milk Adulteration in Camel Milk Using FT-Mid-Infrared Spectroscopy and Machine Learning Models. Foods 2023, 12, 4517. https://doi.org/10.3390/foods12244517
Yao Z, Zhang X, Nie P, Lv H, Yang Y, Zou W, Yang L. Identification of Milk Adulteration in Camel Milk Using FT-Mid-Infrared Spectroscopy and Machine Learning Models. Foods. 2023; 12(24):4517. https://doi.org/10.3390/foods12244517
Chicago/Turabian StyleYao, Zhiqiu, Xinxin Zhang, Pei Nie, Haimiao Lv, Ying Yang, Wenna Zou, and Liguo Yang. 2023. "Identification of Milk Adulteration in Camel Milk Using FT-Mid-Infrared Spectroscopy and Machine Learning Models" Foods 12, no. 24: 4517. https://doi.org/10.3390/foods12244517
APA StyleYao, Z., Zhang, X., Nie, P., Lv, H., Yang, Y., Zou, W., & Yang, L. (2023). Identification of Milk Adulteration in Camel Milk Using FT-Mid-Infrared Spectroscopy and Machine Learning Models. Foods, 12(24), 4517. https://doi.org/10.3390/foods12244517









