Thermal Stability Enhancement of L-Asparaginase from Corynebacterium glutamicum Based on a Semi-Rational Design and Its Effect on Acrylamide Mitigation Capacity in Biscuits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Chemicals
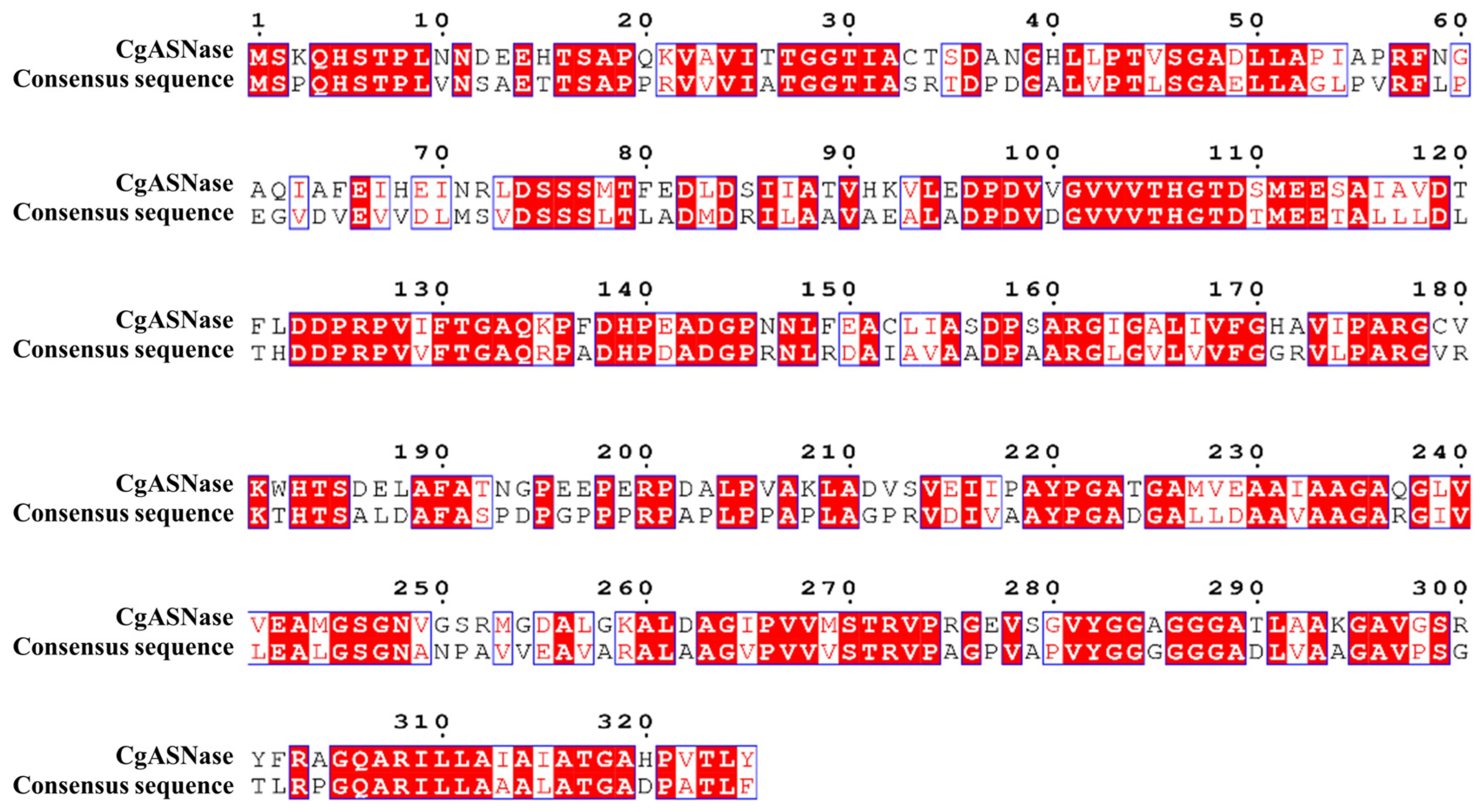
2.2. Identification of Critical Sites for Enhancing the Thermal Stability of CgASNase
2.3. Construction of Site-Point Saturation Mutations and Combinatorial Mutations
2.4. High-Throughput Screening for Mutants with Enhanced Thermal Stability
2.5. Expression, Purification, SDS-PAGE Analysis, and Protein Determination of L-Asparaginase
2.6. Measurement of L-Asparaginase Activities
2.7. Evaluation of Thermal Stability of CgASNase and Its Mutants
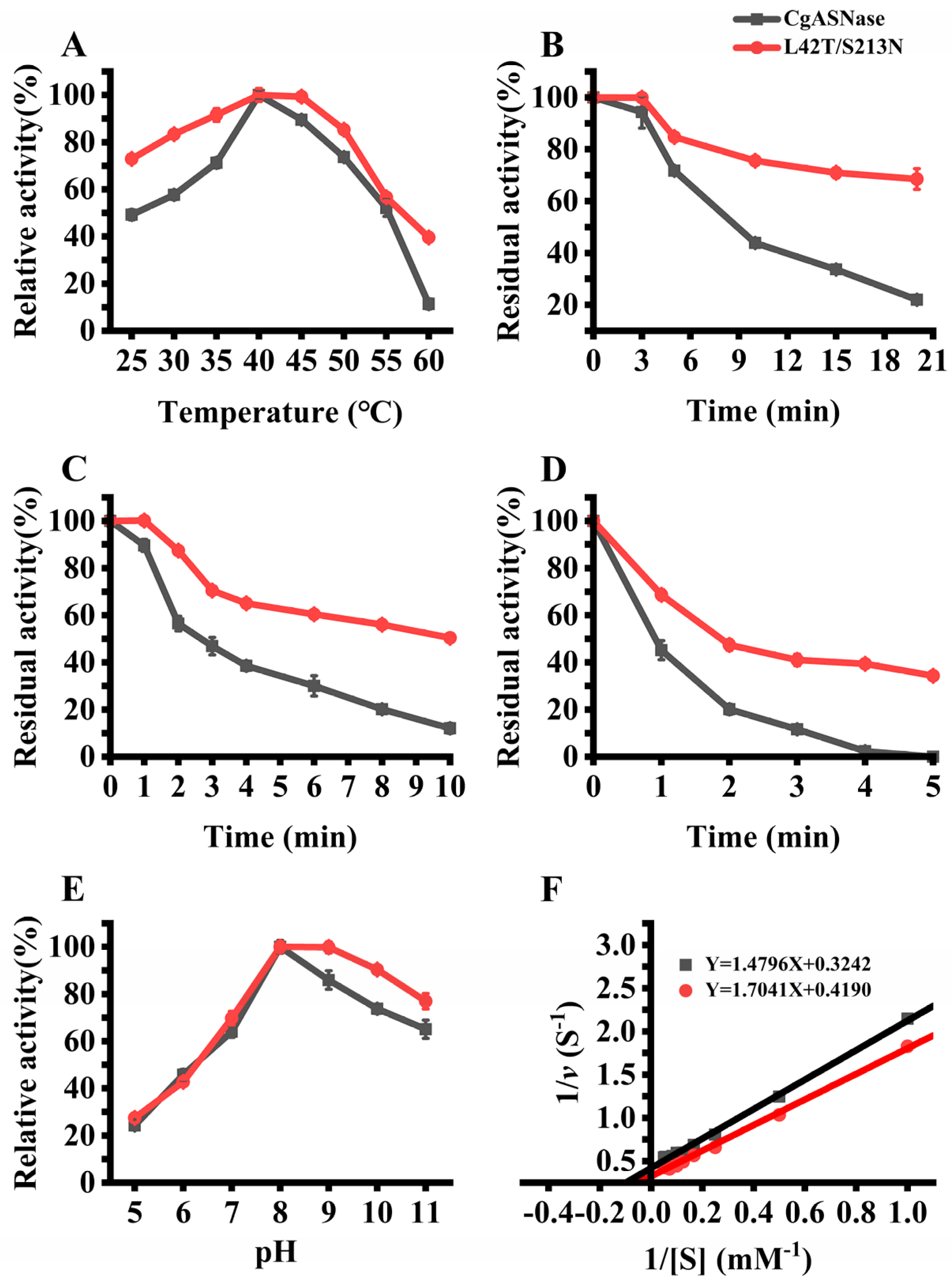
2.8. Enzymatic Characterization of CgASNase and Its Mutants
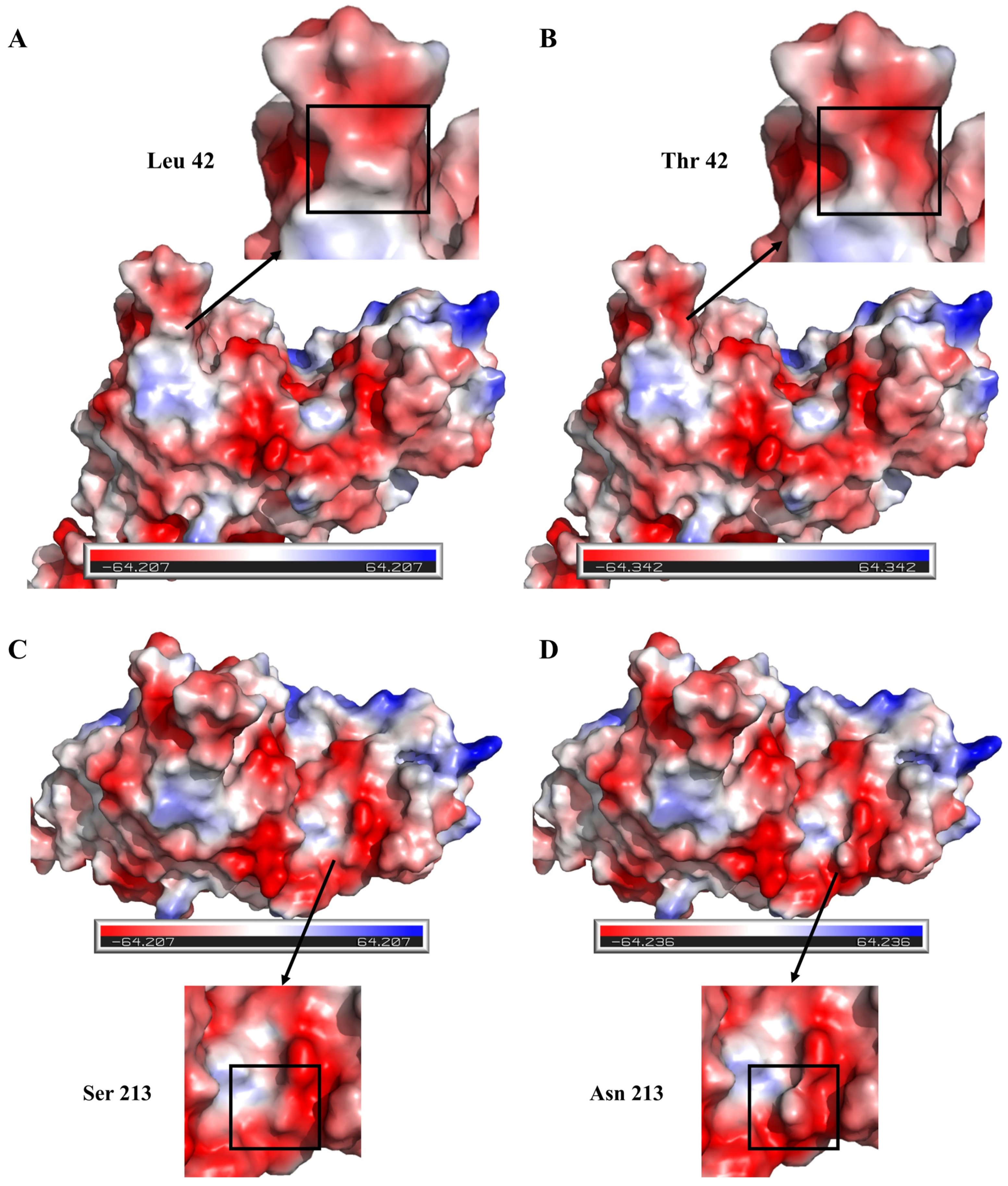
2.9. Molecular Modeling and Structural Analysis
2.10. Preparation of Biscuits
2.11. Application of CgASNase and Its Mutant in Biscuits
2.12. Extraction and Determination of Acrylamide
2.13. Statistical Analysis
3. Results and Discussion
3.1. Selection of Key Sites Affecting the Thermal Stabilization of CgASNase
3.2. The Effect of Key Amino Acid Residues on the Thermal Stability of CgASNase
3.3. Enzymatic Characteristics of the Wild-Type CgASNase and Its Mutant Enzymes
3.4. Investigating the Molecular Mechanism of the Enhanced Thermal Stability of the Double-Mutant Enzyme L42T/S213N
3.5. Application of Enzymes in Biscuits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Chen, Y.; Deng, P.; He, Z.; Qin, F.; Chen, Q.; Wang, Z.; Pan, H.; Chen, J.; Zeng, M. Research progress on generation, detection and inhibition of multiple hazards-acrylamide, 5-hydroxymethylfurfural, advanced glycation end products, methylimidazole-in baked goods. Food Chem. 2024, 431, 137152. [Google Scholar] [CrossRef]
- Gökmen, V. Acrylamide in thermally processed potato products. Potato Res. 2023. [Google Scholar] [CrossRef]
- IARC, Acrylamide. IARC monographs on the evaluation of the carcinogenic risks of chemicals to humans. Some Ind. Chem. 1994, 60, 389–433. [Google Scholar]
- Capuano, E.; Fogliano, V. Acrylamide and 5-hydroxymethylfurfural (HMF): A review on metabolism, toxicity, occurrence in food and mitigation strategies. LWT Food Sci. Technol. 2011, 44, 793–810. [Google Scholar] [CrossRef]
- Khalil, N.M.; Rodriguez-Couto, S.; El-Ghany, M.N.A. Characterization of Penicillium crustosum L-asparaginase and its acrylamide alleviation efficiency in roasted coffee beans at non-cytotoxic levels. Arch. Microbiol. 2021, 203, 2625–2637. [Google Scholar] [CrossRef]
- Kumari, A.; Bhattacharya, B.; Agarwal, T.; Paul, V.; Chakkaravarthi, S. Integrated approach towards acrylamide reduction in potato-based snacks: A critical review. Food Res. Int. 2022, 156, 111172. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Qin, R.; Li, D.; Ji, K.; Wang, T.; Cui, Z.; Huang, Y. A novel bacterial type II L-asparaginase and evaluation of its enzymatic acrylamide reduction in French fries. Int. J. Biol. Macromol. 2016, 92, 232–239. [Google Scholar] [CrossRef]
- Xu, F.; Oruna-Concha, M.-J.; Elmore, J.S. The use of asparaginase to reduce acrylamide levels in cooked food. Food Chem. 2016, 210, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, P.; Chen, F.; Yuan, Y.; Zhu, Y.; Yan, H.; Hu, X. Role of plant polyphenols in acrylamide formation and elimination. Food Chem. 2015, 186, 46–53. [Google Scholar] [CrossRef]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-asparaginase for application in acrylamide mitigation from food: Current research status and future perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Shahana Kabeer, S.; Francis, B.; Vishnupriya, S.; Kattatheyil, H.; Joseph, K.J.; Krishnan, K.P.; Mohamed Hatha, A.A. Characterization of L-asparaginase from Streptomyces koyangensis SK4 with acrylamide-minimizing potential in potato chips. Braz. J. Microbiol. 2023, 54, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Wu, H.; Zhang, W.; Guang, C.; Mu, W. Microbial production, molecular modification, and practical application of L-asparaginase: A review. Int. J. Biol. Macromol. 2021, 186, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Lincoln, L.; Niyonzima, F.N.; More, S.S. Isolation, purification and characterization of fungal extracellular L-asparaginase from mucor hiemalis. J. Biocatal. Biotransformation 2013, 2, 1000108. [Google Scholar]
- Ebrahiminezhad, A.; Rasoul-Amini, S.; Ghasemi, Y. L-asparaginase production by moderate halophilic bacteria isolated from maharloo salt lake. Indian J. Microbiol. 2011, 51, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Keramat, J.; LeBail, A.; Prost, C.; Jafari, M. Acrylamide in baking products: A review article. Food Bioprocess Technol. 2010, 4, 530–543. [Google Scholar] [CrossRef]
- Morales, F.; Capuano, E.; Fogliano, V. Mitigation strategies to reduce acrylamide formation in fried potato products. In Maillard Reaction: Recent Advances in Food and Biomedical Sciences; Wiley-Blackwell: Hoboken, NJ, USA, 2008; Volume 1126, pp. 89–100. [Google Scholar]
- Nunes, J.C.F.; Cristovao, R.O.; Freire, M.G.; Santos-Ebinuma, V.C.; Faria, J.L.; Silva, C.G.; Tavares, A.P.M. Recent strategies and applications for L-asparaginase confinement. Molecules 2020, 25, 5827. [Google Scholar] [CrossRef]
- Chi, H.; Chen, M.; Jiao, L.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F. Characterization of a novel L-asparaginase from Mycobacterium gordonae with acrylamide mitigation potential. Foods 2021, 10, 2819. [Google Scholar] [CrossRef]
- Bi, J.; Jing, X.; Wu, L.; Zhou, X.; Gu, J.; Nie, Y.; Xu, Y. Computational design of noncanonical amino acid-based thioether staples at N/C-terminal domains of multi-modular pullulanase for thermostabilization in enzyme catalysis. Comput. Struct. Biotechnol. J. 2021, 19, 577–585. [Google Scholar] [CrossRef]
- Bommarius, A.S.; Blum, J.K.; Abrahamson, M.J. Status of protein engineering for biocatalysts: How to design an industrially useful biocatalyst. Curr. Opin. Chem. Biol. 2011, 15, 194–200. [Google Scholar] [CrossRef]
- Bloom, J.D.; Labthavikul, S.T.; Otey, C.R.; Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 2006, 103, 5869–5874. [Google Scholar] [CrossRef]
- Che Hussian, C.H.A.; Leong, W.Y. Thermostable enzyme research advances: A bibliometric analysis. J. Genet. Eng. Biotechnol. 2023, 21, 37. [Google Scholar] [CrossRef] [PubMed]
- Arbab, S.; Ullah, H.; Khan, M.I.U.; Khattak, M.N.K.; Zhang, J.; Li, K.; Hassan, I.U. Diversity and distribution of thermophilic microorganisms and their applications in biotechnology. J. Basic Microbiol. 2022, 62, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, N.; Ma, J.; Zhou, Y.; Wei, Q.; Tian, C.; Fang, Y.; Zhong, R.; Chen, G.; Zhang, S. Advances in cold-adapted enzymes derived from microorganisms. Front. Microbiol. 2023, 14, 1152847. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, K.; Chen, H.; Wang, Z.; Zhao, W.; Zhu, L. Cold-adapted enzymes: Mechanisms, engineering and biotechnological application. Bioprocess Biosyst. Eng. 2023, 46, 1399–1410. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Wang, P.; Chen, W. Biochemical properties of a cold-active chitinase from marine Trichoderma gamsii R1 and its application to preparation of chitin oligosaccharides. Mar. Drugs 2010, 21, 332. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Wang, Y.; Li, X.; Zhu, M.; Zhang, H.; Xu, M.; Yang, T.; Rao, Z. Heterologous expression and rational design of L-asparaginase from Rhizomucor miehei to improve thermostability. Biology 2021, 10, 1346. [Google Scholar] [CrossRef]
- Long, S.; Zhang, X.; Rao, Z.; Chen, K.; Xu, M.; Yang, T.; Yang, S. Amino acid residues adjacent to the catalytic cavity of tetramer L-asparaginase II contribute significantly to its catalytic efficiency and thermostability. Enzym. Microb. Technol. 2016, 82, 15–22. [Google Scholar] [CrossRef]
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M. A novel l-asparaginase from hyperthermophilic archaeon Thermococcus sibiricus: Heterologous expression and characterization for biotechnology application. Int. J. Mol. Sci. 2021, 22, 9894. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Xu, S.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Rao, Z. Insight into the thermostability of thermophilic L-asparaginase and non-thermophilic l-asparaginase II through bioinformatics and structural analysis. Appl. Microbiol. Biotechnol. 2019, 103, 7055–7070. [Google Scholar] [CrossRef]
- Kamble, A.; Srinivasan, S.; Singh, H. In-silico bioprospecting: Finding better enzymes. Mol. Biotechnol. 2019, 61, 53–59. [Google Scholar] [CrossRef]
- Chi, H.; Zhu, X.; Shen, J.; Lu, Z.; Lu, F.; Lyu, Y.; Zhu, P. Thermostability enhancement and insight of L-asparaginase from Mycobacterium sp. via consensus-guided engineering. Appl. Microbiol. Biotechnol. 2023, 107, 2321–2333. [Google Scholar] [CrossRef]
- Qian, H.; Zhang, C.; Lu, Z.; Xia, B.; Bie, X.; Zhao, H.; Lu, F.; Yang, G.-Y. Consensus design for improved thermostability of lipoxygenase from Anabaena sp. PCC 7120. BMC Biotechnol. 2018, 18, 57. [Google Scholar] [CrossRef] [PubMed]
- Sternke, M.; Tripp, K.W.; Barrick, D. The use of consensus sequence information to engineer stability and activity in proteins. Enzym. Eng. Evol. Gen. Methods 2020, 643, 149–179. [Google Scholar]
- Xiong, N.; Lv, P.-J.; Song, J.-W.; Shen, Q.; Xue, Y.-P.; Zheng, Y.-G. Engineering of a nitrilase through consensus sequence analysis and conserved site substitution to improve its thermostability and activity. Biochem. Eng. J. 2022, 184, 108475. [Google Scholar] [CrossRef]
- Gomez-Fernandez, B.J.; Risso, V.A.; Sanchez-Ruiz, J.M.; Alcalde, M. Consensus design of an evolved high-redox potential laccase. Front. Bioeng. Biotechnol. 2020, 8, 354. [Google Scholar] [CrossRef]
- Sternke, M.; Tripp, K.W.; Barrick, D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 11275–11284. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Chi, H.; Xia, B.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F.; Chen, M. Thermostability improvement of L-asparaginase from Acinetobacter soli via consensus-designed cysteine residue substitution. Molecules 2022, 27, 6670. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Xia, B.; Shen, J.; Zhu, X.; Lu, Z.; Lu, F.; Zhu, P. Characterization of a novel and glutaminase-free type II L-asparaginase from Corynebacterium glutamicum and its acrylamide alleviation efficiency in potato chips. Int. J. Biol. Macromol. 2022, 221, 1384–1393. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, Y.; Zhang, C.; Bie, X.; Zhao, H.; Lu, F.; Lu, Z. Biochemical characterization of a novel L-asparaginase from Bacillus megaterium H-1 and its application in French fries. Food Res. Int. 2016, 77, 527–533. [Google Scholar] [CrossRef]
- Pedreschi, F.; Mariotti, S.; Granby, K.; Risum, J. Acrylamide reduction in potato chips by using commercial asparaginase in combination with conventional blanching. LWT Food Sci. Technol. 2011, 44, 1473–1476. [Google Scholar] [CrossRef]
- Ruslan, U.S.; Ahmad Raston, N.H.; Mohd Sharif, N.A.; Neoh, H.M.; Nathan, S.; Ramzi, A.B. Development of Corynebacterium glutamicum as staphylococcal-targeting chassis via the construction of autoinducing peptide (aip)-responsive expression system. Sains Malays. 2023, 52, 431–439. [Google Scholar] [CrossRef]
- Jeon, E.J.; Lee, Y.-M.; Choi, E.J.; Kim, S.-B.; Jeong, K.J. Production of tagatose by whole-cell bioconversion from fructose using Corynebacterium glutamicum. Biotechnol. Bioprocess Eng. 2023, 28, 419–427. [Google Scholar] [CrossRef]
- Ali, U.; Naveed, M.; Ullah, A.; Ali, K.; Shah, S.A.; Fahad, S.; Mumtaz, A.S. l-asparaginase as a critical component to combat Acute Lymphoblastic Leukaemia (ALL): A novel approach to target ALL. Eur. J. Pharmacol. 2016, 771, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Meena, B.; Anburajan, L.; Vinithkumar, N.V.; Shridhar, D.; Raghavan, R.V.; Dharani, G. Molecular expression of L-asparaginase gene from Nocardiopsis alba NIOT-VKMA08 in Escherichia coli: A prospective recombinant enzyme for leukaemia chemotherapy. Gene 2016, 590, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.J.; Kan, C.N.E.; Luo, C.; Kazlauskas, R.J. Consensus Finder web tool to predict stabilizing substitutions in proteins. Enzym. Eng. Evol. Gen. Methods 2020, 643, 129–148. [Google Scholar]
- Xavier, R.; Patrice, G. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar]
- Chi, H.; Wang, Y.; Xia, B.; Zhou, Y.; Lu, Z.; Lu, F.; Zhu, P. Enhanced thermostability and molecular insights for L-asparaginase from Bacillus licheniformis via structure- and computation-based rational design. J. Agric. Food Chem. 2022, 70, 14499–14509. [Google Scholar] [CrossRef] [PubMed]
- Anese, M.; Quarta, B.; Peloux, L.; Calligaris, S. Effect of formulation on the capacity of L-asparaginase to minimize acrylamide formation in short dough biscuits. Food Res. Int. 2011, 44, 2837–2842. [Google Scholar] [CrossRef]
- Anese, M.; Quarta, B.; Frias, J. Modelling the effect of asparaginase in reducing acrylamide formation in biscuits. Food Chem. 2011, 126, 435–440. [Google Scholar] [CrossRef]
- Pedreschi, F.; Kaack, K.; Granby, K. The effect of asparaginase on acrylamide formation in French fries. Food Chem. 2008, 109, 386–392. [Google Scholar] [CrossRef]
- Hendriksen, H.V.; Kornbrust, B.A.; Ostergaard, P.R.; Stringer, M.A. Evaluating the potential for enzymatic acrylamide mitigation in a range of food products using an asparaginase from Aspergillus oryzae. J. Agric. Food Chem. 2009, 57, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Chi, H.; Lu, Z.; Zhang, C.; Chia, S.R.; Show, P.L.; Tao, Y.; Lu, F. Characterization of a novel type I L-asparaginase from Acinetobacter soli and its ability to inhibit acrylamide formation in potato chips. J. Biosci. Bioeng. 2020, 129, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Yan, R.; Shen, J.; Zhu, X.; Meng, F.; Lu, Z.; Lu, F. Cis-element engineering promotes the expression of Bacillus subtilis type I l-asparaginase and its application in food. Int. J. Mol. Sci. 2022, 23, 6588. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Sim, B.R.; Li, J.; Yu, Y.; Qin, L.; Wu, L.; Shen, Y.; Nie, Y.; Zhao, Y.-L.; Xu, Y. Evolutionary coupling-inspired engineering of alcohol dehydrogenase reveals the influence of distant sites on its catalytic efficiency for stereospecific synthesis of chiral alcohols. Comput. Struct. Biotechnol. J. 2021, 19, 5864–5873. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, Y.; Wang, Y.; Huang, H.; Bai, Y.; Su, X.; Zhang, J.; Yao, B.; Tu, T.; Luo, H. Exploiting the activity-stability trade-off of glucose oxidase from Aspergillus niger using a simple approach to calculate thermostability of mutants. Food Chem. 2021, 342, 128270. [Google Scholar] [CrossRef]
- Gazi, S.; Tas, N.G.; Gorgulu, A.; Gokmen, V. Effectiveness of asparaginase on reducing acrylamide formation in bakery products according to their dough type and properties. Food Chem. 2023, 402, 134224. [Google Scholar] [CrossRef]



| Enzymes | Specific Activities (IU/mg) | Half-Lives (40 °C, min) |
|---|---|---|
| Wild type | 1970.15 ± 23.54 a | 3.24 ± 0.23 f |
| I70L | 1661.67 ± 25.31 c | 4.47 ± 0.42 e |
| C152I | 1354.95 ± 13.59 d | 7.71 ± 0.41 b |
| E187N | 1211.36 ± 12.75 e | 8.42 ± 0.19 b |
| N59S | 1206.90 ± 21.65 e | 9.36 ± 0.53 a |
| N59Q | 1124.77 ± 32.76 f | 9.75 ± 0.38 a |
| S213H | 966.94 ± 13.92 g | 5.77 ± 0.45 c |
| S213V | 1740.39 ± 23.82 b | 6.54 ± 0.25 d |
| L42T | 1922.50 ± 21.53 a | 6.71 ± 0.61 d |
| H171S | 1613.30 ± 31.06 c | 8.10 ± 0.54 b |
| Enzymes | Specific Activities (IU/mg) | Half-Lives (40 °C, min) |
|---|---|---|
| Wild type | 1970.15 ± 23.54 a | 3.24 ± 0.23 d |
| L42T | 1922.50 ± 21.53 a | 6.71 ± 0.61 c |
| L42T/S213H | 1627.86 ± 32.67 c | 12.05 ± 0.56 a |
| L42T/S213N | 1931.01 ± 25.84 a | 13.29 ± 0.91 a |
| L42T/S213C | 1763.49 ± 64.24 b | 8.84 ± 0.73 b |
| L42T/N59S | 1491.18 ± 36.57 d | 8.70 ± 0.82 b |
| Groups | Acrylamide Content (μg/kg) | Acrylamide Reduction (%) |
|---|---|---|
| Control group | 464.74 ± 6.68 a | - |
| CgASNase | 145.11 ± 4.19 b | 68.78 ± 2.73 a |
| L42T/S213N | 68.30 ± 1.67 c | 85.31 ± 1.01 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, H.; Jiang, Q.; Feng, Y.; Zhang, G.; Wang, Y.; Zhu, P.; Lu, Z.; Lu, F. Thermal Stability Enhancement of L-Asparaginase from Corynebacterium glutamicum Based on a Semi-Rational Design and Its Effect on Acrylamide Mitigation Capacity in Biscuits. Foods 2023, 12, 4364. https://doi.org/10.3390/foods12234364
Chi H, Jiang Q, Feng Y, Zhang G, Wang Y, Zhu P, Lu Z, Lu F. Thermal Stability Enhancement of L-Asparaginase from Corynebacterium glutamicum Based on a Semi-Rational Design and Its Effect on Acrylamide Mitigation Capacity in Biscuits. Foods. 2023; 12(23):4364. https://doi.org/10.3390/foods12234364
Chicago/Turabian StyleChi, Huibing, Qingwei Jiang, Yiqian Feng, Guizheng Zhang, Yilian Wang, Ping Zhu, Zhaoxin Lu, and Fengxia Lu. 2023. "Thermal Stability Enhancement of L-Asparaginase from Corynebacterium glutamicum Based on a Semi-Rational Design and Its Effect on Acrylamide Mitigation Capacity in Biscuits" Foods 12, no. 23: 4364. https://doi.org/10.3390/foods12234364
APA StyleChi, H., Jiang, Q., Feng, Y., Zhang, G., Wang, Y., Zhu, P., Lu, Z., & Lu, F. (2023). Thermal Stability Enhancement of L-Asparaginase from Corynebacterium glutamicum Based on a Semi-Rational Design and Its Effect on Acrylamide Mitigation Capacity in Biscuits. Foods, 12(23), 4364. https://doi.org/10.3390/foods12234364







