High Prevalence of Multidrug-Resistant, Biofilm-Forming Virulent Clostridium perfringens in Broiler Chicken Retail Points in Northeast India
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation
2.2. Toxinotyping of Confirmed Isolates
2.3. Antimicrobial Susceptibility Testing and MAR Indexing
2.4. Evaluation of Biofilm-Forming Ability
2.5. Hierarchical Clustering, Heatmap, and Correlation Plot Analysis
2.6. Statistical Analysis
3. Results
3.1. Occurrence of C. perfringens
3.2. Molecular Toxinotyping
3.3. Antimicrobial Resistance Profiling and MAR Index
3.4. Evaluation of Biofilm-Forming Ability
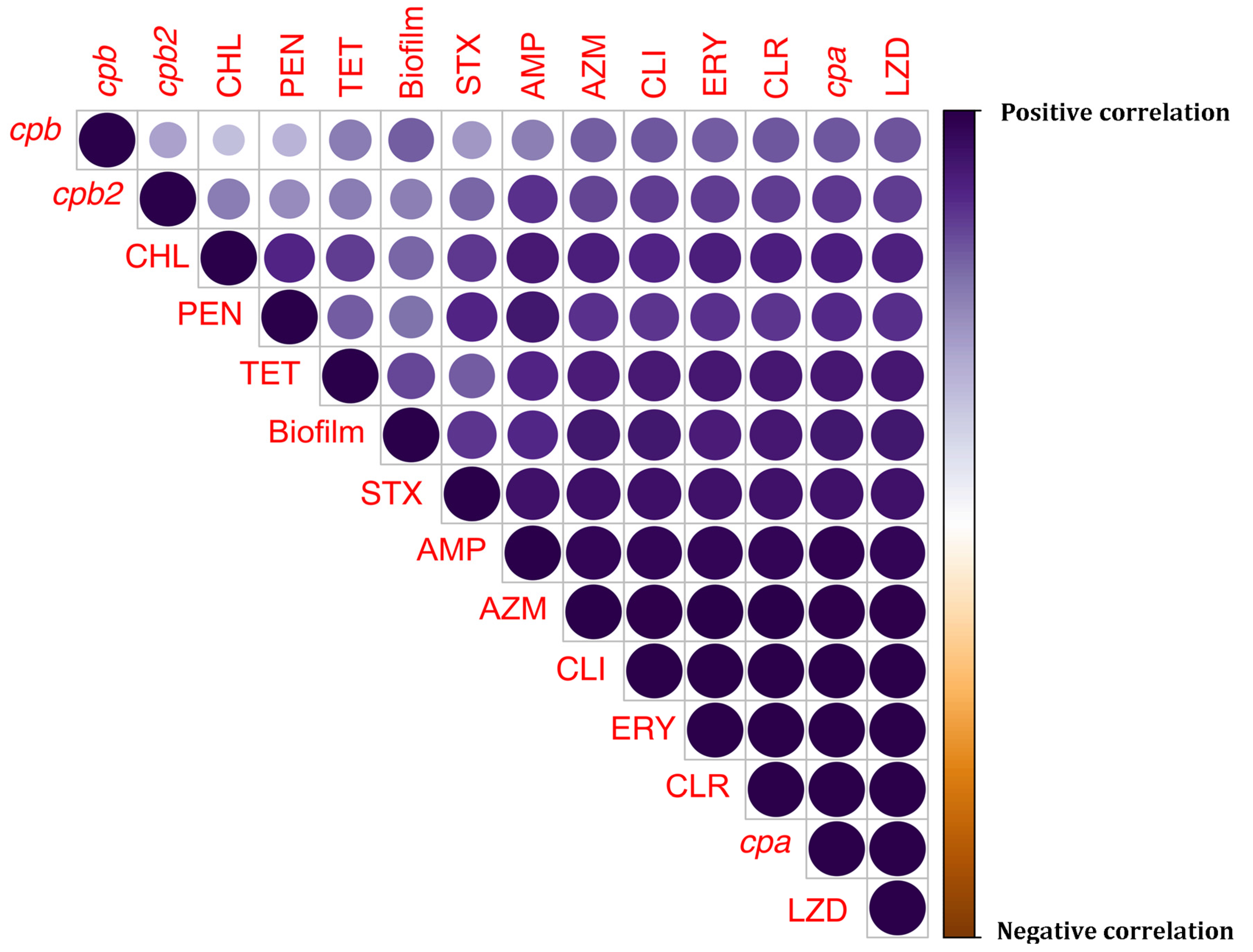
3.5. Heatmap-Based Hierarchical Clustering and Correlation Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milton, A.A.P.; Momin, A.G.; Gandhale, P.N.; Das, S.; Ghatak, S.; Priya, G.B.; Firake, D.M.; Srinivas, K.; Momin, K.M.; Hussain, Z.; et al. Prevalence, toxinotyping, antimicrobial susceptibility and biofilm-forming ability of Clostridium perfringens isolated from free-living rodents and shrews. Anaerobe 2022, 77, 102618. [Google Scholar] [CrossRef] [PubMed]
- Van Immerseel, F.; De Buck, J.; Pasmans, F.; Huyghebaert, G.; Haesebrouck, F.; Ducatelle, R. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004, 33, 537–549. [Google Scholar] [CrossRef]
- Thomas, M.; Murray, R. Estimating the burden of food-borne illness in Canada. Can. Commun. Dis. Rep. 2014, 40, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001940. [Google Scholar]
- Abdelrahim, A.M.; Radomski, N.; Delannoy, S.; Djellal, S.; Le Négrate, M.; Hadjab, K.; Fach, P.; Hennekinne, J.A.; Mistou, M.Y.; Firmesse, O. Large-Scale genomic analyses and toxinotyping of Clostridium perfringens implicated in foodborne outbreaks in France. Front. Microbiol. 2019, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Petit, L.; Gibert, M.; Popoff, M.R. Clostridium perfringens: Toxinotype and genotype. Trends Microbiol. 1999, 7, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.C.; Shrestha, A.; McClane, B.A. Clostridium perfringens enterotoxin: Action, genetics, and translational applications. Toxins 2016, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yang, Q.; Huang, X.; Yan, Z.; Zhang, S.; Luo, R.; Wang, P.; Wang, W.; Xie, K.; Jiang, T.; et al. Effects of Clostridium perfringens beta2 toxin on apoptosis, inflammation, and barrier function of intestinal porcine epithelial cells. Microb. Pathog. 2020, 147, 104379. [Google Scholar] [CrossRef]
- Coursodon, C.F.; Glock, R.D.; Moore, K.L.; Cooper, K.K.; Songer, J.G. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 2012, 18, 117–121. [Google Scholar] [CrossRef]
- Adams, V.; Han, X.; Lyras, D.; Rood, J.I. Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens. Plasmid 2018, 99, 32–39. [Google Scholar] [CrossRef]
- Hassani, S.; Pakbin, B.; Brück, W.M.; Mahmoudi, R.; Mousavi, S. Prevalence, Antibiotic Resistance, Toxin-Typing and Genotyping of Clostridium perfringens in Raw Beef Meats Obtained from Qazvin City, Iran. Antibiotics 2022, 11, 340. [Google Scholar] [CrossRef]
- Pantaléon, V.; Bouttier, S.; Soavelomandroso, A.P.; Janoir, C.; Candela, T. Biofilms of Clostridium species. Anaerobe 2014, 30, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, A.; Jacques, M.; Archambault, M. Comparative transcriptomic analysis of Clostridium perfringens biofilms and planktonic cells. Avian Pathol. 2016, 45, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Milton, A.A.P.; Sanjukta, R.; Gogoi, A.P.; Momin, K.M.; Priya, G.B.; Das, S.; Ghatak, S.; Sen, A.; Kandpal, B.K. Prevalence, molecular typing and antibiotic resistance of Clostridium perfringens in free range ducks in Northeast India. Anaerobe 2020, 64, 102242. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, H.; Hazarika, R.; Deka, P.; Sharma, R. Isolation and Characterization of Clostridium perfringens from Suspected Cases of Necrotic Enteritis (NE) in Broiler Chicken. Int. J. Livest. Res. 2019, 9, 221–232. [Google Scholar] [CrossRef]
- Das, A.; Mazumder, Y.; Dutta, B.K.; Shome, B.R.; Bujarbaruah, K.M.; Kumar, A. Clostridium perfringens Type A from Broiler Chicken with Necrotic Enteritis. Int. J. Poult. Sci. 2008, 7, 601–609. [Google Scholar] [CrossRef][Green Version]
- Van Asten, A.J.A.M.; van der Wiel, C.W.; Nikolaou, G.; Houwers, D.J.; Gröne, A. A multiplex PCR for toxin typing of Clostridium perfringens isolates. Vet. Microbiol. 2009, 136, 411–412. [Google Scholar] [CrossRef]
- Tolooe, A.; Shojadoost, B.; Peighambari, S.M.; Tamaddon, Y. Prevalence of netB Gene among Clostridium perfringens Isolates Obtained from Healthy and Diseased Chickens. J. Anim. Vet. Adv. 2011, 10, 106–110. [Google Scholar] [CrossRef]
- M100 ED32; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2022.
- Guran, H.S.; Oksuztepe, G. Detection and typing of Clostridium perfringens from retail chicken meat parts. Lett. Appl. Microbiol. 2013, 57, 77–82. [Google Scholar] [CrossRef]
- Svobodová, I.; Steinhauserová, I.; Nebola, M. Incidence of Clostridium perfringens in broiler chickens in the Czech Republic. Acta Vet. Brno 2007, 76, S25–S30. [Google Scholar] [CrossRef]
- Fan, Y.C.; Wang, C.L.; Wang, C.; Chen, T.C.; Chou, C.H.; Tsai, H.J. Incidence and Antimicrobial Susceptibility to Clostridium perfringens in Premarket Broilers in Taiwan. Avian Dis. 2016, 60, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, H.; Chen, S.; Chen, Y.; Liu, L.; Wu, W. Tracing Clostridium perfringens strains along the chicken production chain from farm to slaughter by multilocus sequence typing. Zoonoses Public Health 2021, 68, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.S.; Kim, D.H.; Bae, D.; Kim, S.H.; Kim, H.; Moon, J.S.; Song, K.Y.; Chon, J.W.; Seo, K.H. Prevalence, toxin-typing, and antimicrobial susceptibility of Clostridium perfringens from retail meats in Seoul, Korea. Anaerobe 2020, 64, 102235. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.S.; Hwang, E.H.; Kim, G.; Park, J.Y.; Oh, H.; Lim, K.S.; Kang, P.; Lee, H.Y.; Jeong, K.J.; Mo, I.; et al. Prevalence and characterization of Clostridium perfringens isolated from feces of captive cynomolgus monkeys (Macaca fascicularis). Anaerobe 2020, 64, 102236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, P.; Wang, Y.; Shen, Z.; Wang, S. Multiresistance gene cfr(C) in Clostridium perfringens of cattle origin from China. J. Antimicrob. Chemother. 2021, 76, 3310–3312. [Google Scholar] [CrossRef]
- Osman, K.M.; Elhariri, M. Antibiotic resistance of Clostridium perfringens isolates from broiler chickens in Egypt. OIE Rev. Sci. Tech. 2013, 32, 841–850. [Google Scholar] [CrossRef]
- Doyle, M.E. Multidrug-resistant pathogens in the food supply. Foodborne Pathog. Dis. 2015, 12, 261–279. [Google Scholar] [CrossRef]
- Hu, W.S.; Woo, D.U.; Kang, Y.J.; Koo, O.K. Biofilm and spore formation of Clostridium perfringens and its resistance to disinfectant and oxidative stress. Antibiotics 2021, 10, 396. [Google Scholar] [CrossRef]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef]

| Isolate ID | Source | Toxin Genes | Designated Toxinotype | Antimicrobial Susceptibility # | MAR Index | Biofilm-Forming Ability ## | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cpa | cpb | cpb2 | ERY | CLR | AMP | CHL | CLI | LZD | OFX | PEN | STX | TET | AZM | |||||
| SI_F | Intestine | + | − | − | Type A | I | I | R | I | R | R | S | R | S | I | S | 0.36 | ++ |
| SI_A | Intestine | + | − | − | Type A | R | R | R | I | R | R | S | S | R | S | R | 0.64 | ++ |
| SI_B | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | R | R | I | R | 0.73 | − |
| S4_C | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | R | I | R | 0.55 | ++ |
| S6 _D | Intestine | + | − | − | Type A | R | R | R | R | R | R | S | R | R | S | R | 0.82 | ++ |
| S9_H | Intestine | + | − | − | Type A | R | R | R | R | R | R | I | R | R | R | R | 0.91 | + |
| S4_A | Intestine | + | − | − | Type A | I | R | S | S | R | R | S | S | S | S | R | 0.36 | ++ |
| S3_G | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | S | S | I | R | 0.55 | ++ |
| S1_C | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | R | I | R | 0.55 | + |
| S10_E | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | S | R | I | 0.45 | − |
| S7_F | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | R | S | I | R | 0.64 | − |
| BR6_E | Intestine | + | − | − | Type A | R | S | S | S | R | R | S | S | S | R | R | 0.45 | − |
| BR6_F | Intestine | + | − | − | Type A | R | R | S | S | I | R | S | S | S | I | I | 0.27 | + |
| BR14_C | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | S | R | I | 0.45 | + |
| CPN15 | Intestine | + | − | + | Type A with cpb2 | R | R | R | R | R | R | S | S | R | S | R | 0.72 | ++ |
| CP3 | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | S | I | R | 0.45 | ++ |
| BR3_H | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | S | R | R | 0.55 | − |
| S1 | Intestine | + | − | − | Type A | R | R | R | R | R | R | S | S | S | R | R | 0.72 | ++ |
| CP6 | Intestine | + | − | + | Type A with cpb2 | R | R | R | R | R | R | S | R | R | I | R | 0.82 | − |
| CS1 | Intestine | + | − | + | Type A with cpb2 | I | I | R | I | I | R | S | R | S | S | I | 0.27 | ++ |
| CPS1 | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | R | R | I | R | 0.73 | ++ |
| CS4 | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | R | R | R | 0.64 | +++ |
| N10_A | Intestine | + | − | + | Type A with cpb2 | R | R | S | S | R | R | S | S | R | I | R | 0.55 | − |
| C6_E | Intestine | + | − | − | Type A | R | R | S | S | R | R | S | S | S | R | R | 0.55 | ++ |
| CS7 | Intestine | + | − | − | Type A | R | R | S | R | S | R | S | S | S | I | R | 0.45 | ++ |
| CP14 C | Intestine | + | − | − | Type A | R | R | S | R | R | R | S | S | S | I | R | 0.55 | +++ |
| CPS6 | Intestine | + | − | + | Type A with cpb2 | R | R | R | S | R | R | S | S | R | I | R | 0.64 | + |
| CPS1B | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | S | R | I | R | 0.64 | + |
| CP8 | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | S | R | I | R | 0.64 | + |
| CP6_1 | Intestine | + | − | − | Type A | R | R | R | S | R | R | S | S | R | R | R | 0.73 | + |
| CP3_2 | Intestine | + | − | − | Type A | R | R | R | R | R | R | S | R | S | R | R | 0.82 | + |
| RUC9_2 | Meat | + | − | − | Type A | R | R | R | I | R | R | S | S | R | S | R | 0.64 | + |
| ENC2N2_1 | Meat | + | − | + | Type A with cpb2 | R | R | R | S | R | R | S | R | R | R | R | 0.82 | +++ |
| EIC4_1 | Meat | + | − | − | Type A | R | R | R | R | R | R | S | S | S | I | R | 0.64 | − |
| RUC4_1 | Meat | + | − | + | Type A with cpb2 | R | R | R | S | R | R | S | S | S | S | R | 0.55 | + |
| RUC9 _1 | Meat | + | − | − | Type A | R | R | R | I | R | R | S | S | R | S | R | 0.64 | − |
| L3 _E | Meat | + | − | − | Type A | S | S | R | S | S | S | S | R | S | S | S | 0.18 | + |
| L1_I | Meat | + | + | − | Type C | R | R | S | S | R | R | S | S | S | S | R | 0.45 | ++ |
| MC2 | Meat | + | + | + | Type C with cpb2 | R | R | R | R | S | R | S | R | S | R | R | 0.73 | − |
| N10_D | Meat | + | + | − | Type C | R | R | S | S | R | R | S | S | S | S | R | 0.45 | + |
| EIC4_2 | Meat | + | − | − | Type A | I | R | S | S | R | R | S | S | S | I | I | 0.27 | ++ |
| ELC3_1 | Meat | + | + | − | Type C | R | R | R | I | R | R | S | R | R | I | R | 0.73 | ++ |
| RUC4_2 | Meat | + | − | − | Type A | R | R | R | I | R | R | S | S | S | R | R | 0.64 | +++ |
| ENCINI_1 | Meat | + | − | + | Type A with cpb2 | I | R | R | S | R | R | S | S | S | R | S | 0.45 | − |
| EIC3_1 | Meat | + | − | − | Type A | R | R | R | S | R | R | S | R | R | I | R | 0.72 | + |
| ENC2N2_2 | Meat | + | + | + | Type C with cpb2 | R | R | R | S | R | R | S | S | S | S | R | 0.55 | ++ |
| EIC3_2 | Meat | + | − | − | Type A | R | R | R | R | R | R | S | R | R | I | S | 0.72 | + |
| ENC3 | Meat | + | − | − | Type A | R | R | R | R | R | R | S | R | R | I | R | 0.82 | − |
| M3 | Meat | + | + | − | Type C | R | R | S | S | R | R | S | S | S | R | S | 0.45 | − |
| N15 | Meat | + | − | − | Type A | R | R | R | S | R | R | S | S | S | S | R | 0.55 | − |
| ENC3_2 | Meat | + | − | − | Type A | R | R | R | R | R | R | S | S | R | I | R | 0.73 | ++ |
| RUC8_2 | Meat | + | − | − | Type A | R | S | R | S | I | R | S | S | S | I | S | 0.27 | − |
| RUC10_1 | Meat | + | − | + | Type A with cpb2 | R | R | R | I | R | R | S | S | S | I | S | 0.45 | − |
| RUC8_1 | Meat | + | − | − | Type A | R | R | R | R | I | R | S | R | I | R | R | 0.73 | − |
| CPL4 | Water | + | − | − | Type A | R | R | R | R | R | R | S | S | S | R | R | 0.73 | − |
| U2_G | Water | + | − | − | Type A | R | R | R | R | R | R | S | R | S | R | R | 0.82 | + |
| CP1_A | Water | + | − | − | Type A | R | R | R | R | R | R | S | R | R | R | R | 0.91 | − |
| CP1_B | Water | + | − | − | Type A | R | R | R | R | R | R | S | R | R | R | R | 0.91 | ++ |
| U3 | Water | + | + | − | Type C | I | R | R | S | R | R | S | R | R | R | R | 0.73 | ++ |
| CP3H | Water | + | − | − | Type A | R | R | R | S | R | R | S | S | S | R | R | 0.64 | ++ |
| CP14D | Water | + | − | + | Type A with cpb2 | R | R | R | R | R | S | S | S | S | I | R | 0.55 | + |
| CPL1 | Water | + | − | + | Type A with cpb2 | R | R | R | S | R | R | S | S | S | R | R | 0.64 | − |
| CPL3 | Water | + | − | − | Type A | R | R | R | S | I | R | S | S | S | R | R | 0.55 | ++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Priya, G.B.; Srinivas, K.; Shilla, H.; Milton, A.A.P. High Prevalence of Multidrug-Resistant, Biofilm-Forming Virulent Clostridium perfringens in Broiler Chicken Retail Points in Northeast India. Foods 2023, 12, 4185. https://doi.org/10.3390/foods12224185
Priya GB, Srinivas K, Shilla H, Milton AAP. High Prevalence of Multidrug-Resistant, Biofilm-Forming Virulent Clostridium perfringens in Broiler Chicken Retail Points in Northeast India. Foods. 2023; 12(22):4185. https://doi.org/10.3390/foods12224185
Chicago/Turabian StylePriya, Govindarajan Bhuvana, Kandhan Srinivas, Heiborkie Shilla, and Arockiasamy Arun Prince Milton. 2023. "High Prevalence of Multidrug-Resistant, Biofilm-Forming Virulent Clostridium perfringens in Broiler Chicken Retail Points in Northeast India" Foods 12, no. 22: 4185. https://doi.org/10.3390/foods12224185
APA StylePriya, G. B., Srinivas, K., Shilla, H., & Milton, A. A. P. (2023). High Prevalence of Multidrug-Resistant, Biofilm-Forming Virulent Clostridium perfringens in Broiler Chicken Retail Points in Northeast India. Foods, 12(22), 4185. https://doi.org/10.3390/foods12224185






