The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Culture Conditions
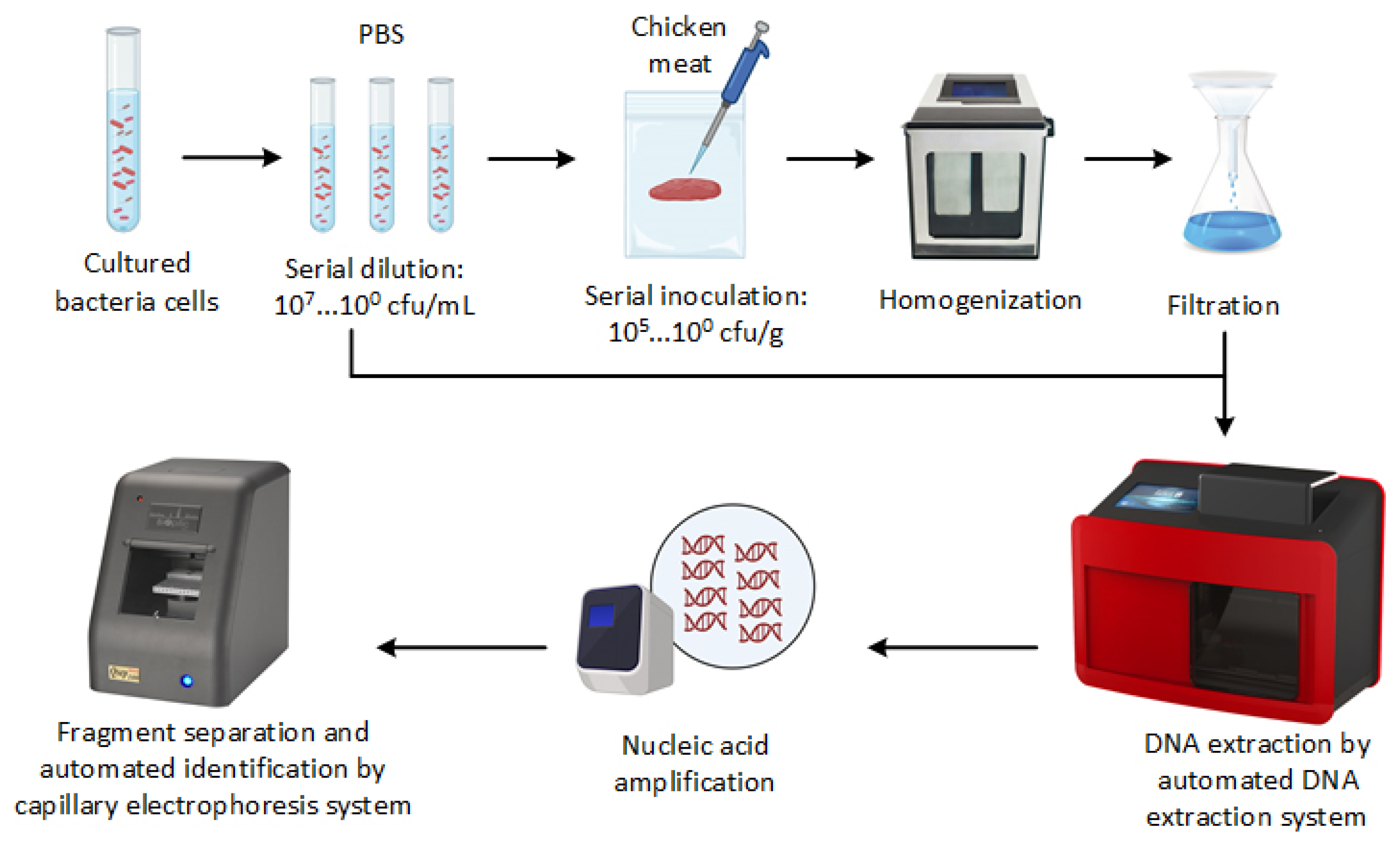
2.2. Inoculation of Chicken Meat Samples
2.3. Genomic DNA Extraction
2.4. Optimization of the PCR-CE
3. Results
3.1. Identification of Pathogens via the PCR-CE
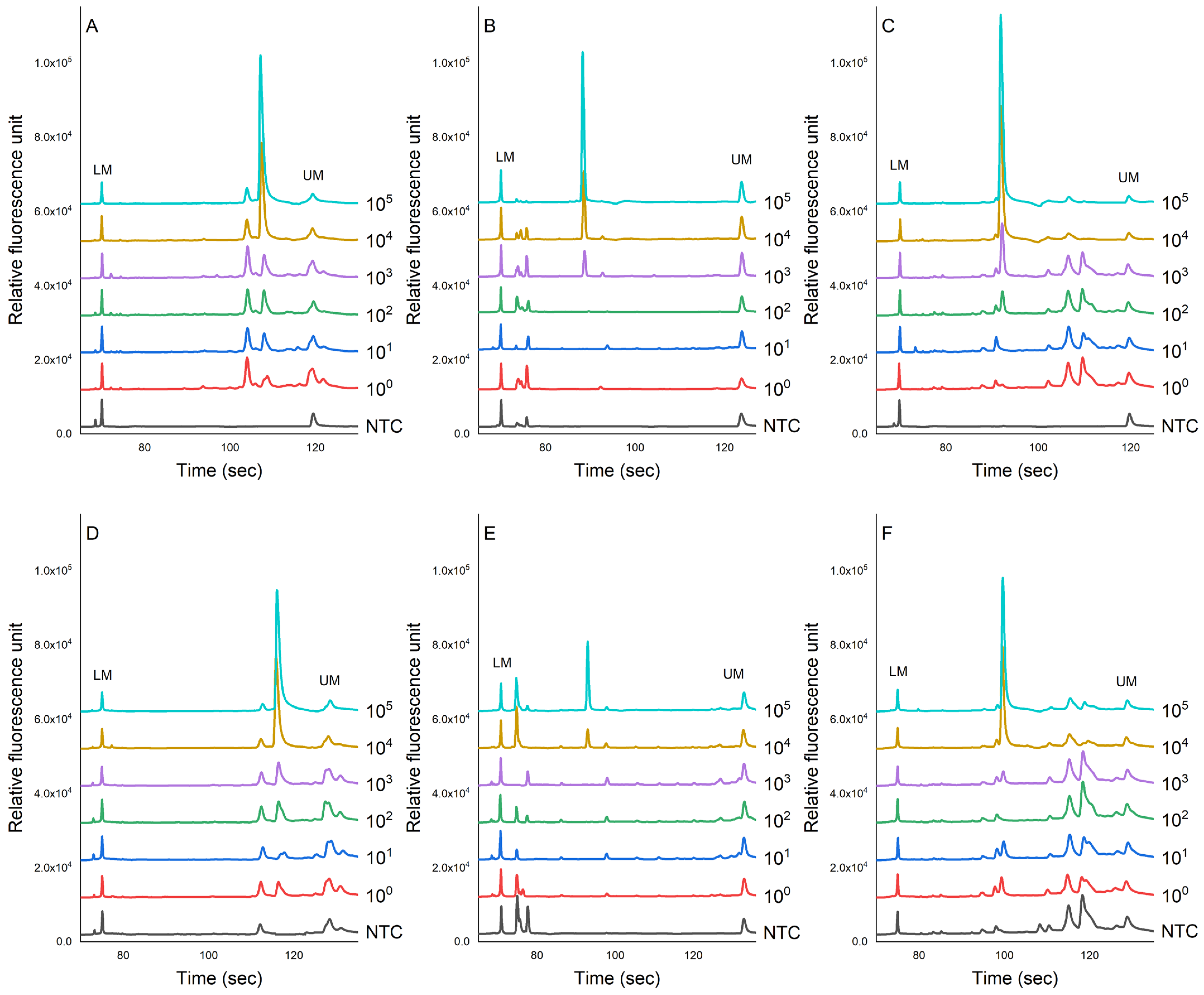
3.2. Sensitivity Evaluation of PCR-CE System in Pure Culture of Bacteria
3.3. Evaluation of the Sensitivity of the PCR-CE System in Artificially Contaminated Samples
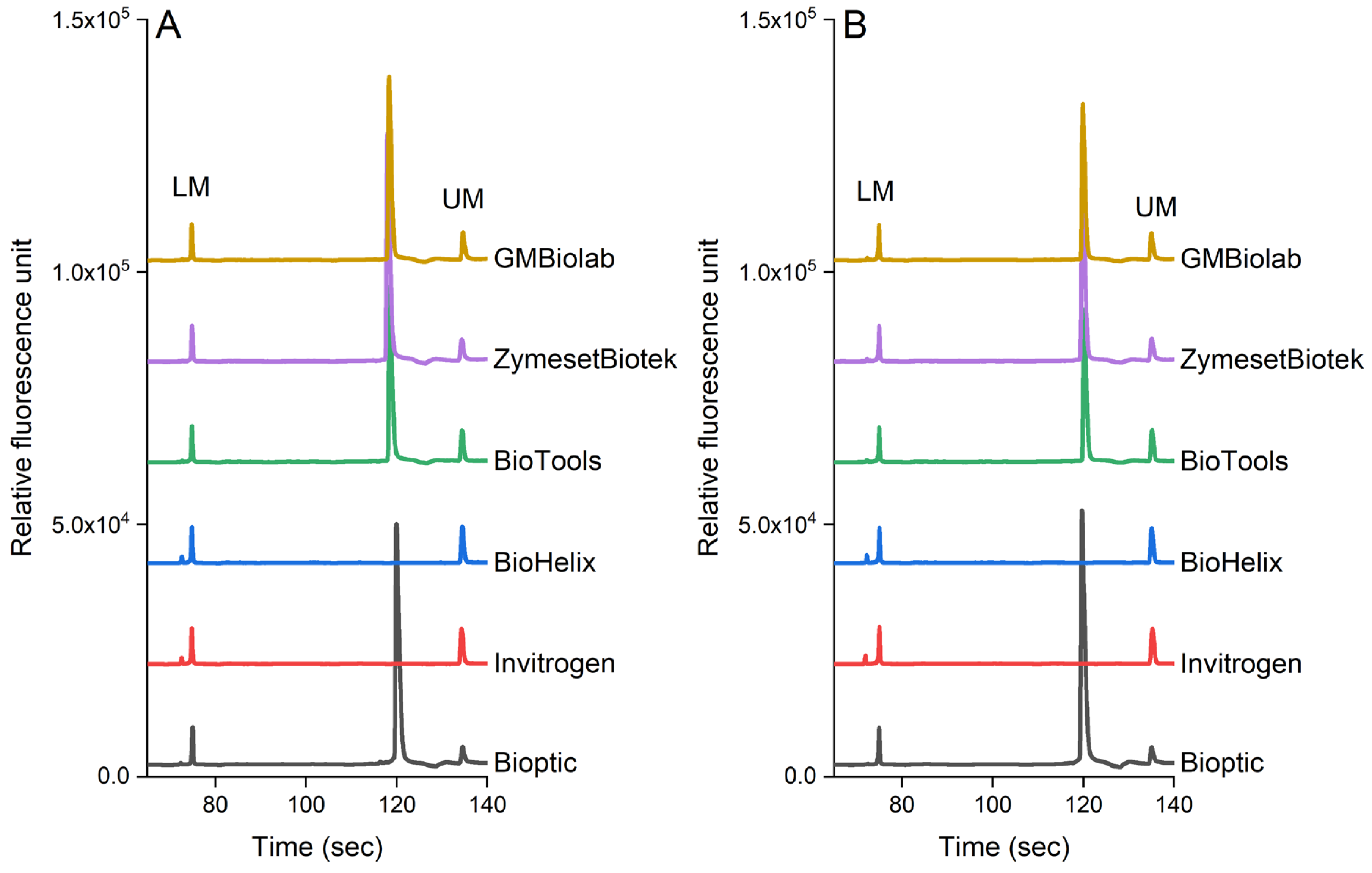
3.4. Evaluation of PCR-CE Using Multiple Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO’s First Ever Global Estimates of Foodborne Diseases Find Children under 5 Account for almost One Third of Deaths. Available online: https://www.who.int/news/item/03-12-2015-who-s-first-ever-global-estimates-of-foodborne-diseases-find-children-under-5-account-for-almost-one-third-of-deaths (accessed on 6 September 2023).
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. European Food Safety Authority Foodborne Outbreaks Report; The European Food Safety Authority: Parma, Italy, 2020.
- Chen, L.; Sun, L.; Zhang, R.; Liao, N.; Qi, X.; Chen, J. Surveillance for Foodborne Disease Outbreaks in Zhejiang Province, China, 2015–2020. BMC Public Health 2022, 22, 135. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Tenório, A.; Silva, B.; Rodrigues, V.; Cadavez, V.; Gonzales-Barron, U. Prevalence of Pathogens in Poultry Meat: A Meta-Analysis of European Published Surveys. Foods 2018, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Siriken, B.; Türk, H.; Yildirim, T.; Durupinar, B.; Erol, I. Prevalence and Characterization of Salmonella Isolated from Chicken Meat in Turkey. J. Food Sci. 2015, 80, M1044–M1050. [Google Scholar] [CrossRef] [PubMed]
- Thung, T.Y.; Mahyudin, N.A.; Basri, D.F.; Wan Mohamed Radzi, C.W.J.; Nakaguchi, Y.; Nishibuchi, M.; Radu, S. Prevalence and Antibiotic Resistance of Salmonella Enteritidis and Salmonella Typhimurium in Raw Chicken Meat at Retail Markets in Malaysia. Poult. Sci. 2016, 95, 1888–1893. [Google Scholar] [CrossRef]
- Rortana, C.; Nguyen-Viet, H.; Tum, S.; Unger, F.; Boqvist, S.; Dang-Xuan, S.; Koam, S.; Grace, D.; Osbjer, K.; Heng, T.; et al. Prevalence of Salmonella spp. and Staphylococcus aureus in Chicken Meat and Pork from Cambodian Markets. Pathogens 2021, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Farhoumand, P.; Hassanzadazar, H.; Soltanpour, M.S.; Aminzare, M.; Abbasi, Z. Prevalence, Genotyping and Antibiotic Resistance of Listeria monocytogenes and Escherichia coli in Fresh Beef and Chicken Meats Marketed in Zanjan, Iran. Iran. J. Microbiol. 2020, 12, 537. [Google Scholar] [CrossRef]
- Elmali, M.; Can, H.Y.; Yaman, H. Prevalence of Listeria monocytogenes in Poultry Meat. Food Sci. Technol. 2015, 35, 672–675. [Google Scholar] [CrossRef]
- Panera-Martínez, S.; Rodríguez-Melcón, C.; Serrano-Galán, V.; Alonso-Calleja, C.; Capita, R. Prevalence, Quantification and Antibiotic Resistance of Listeria monocytogenes in Poultry Preparations. Food Control 2022, 135, 108608. [Google Scholar] [CrossRef]
- Mangal, M.; Bansal, S.; Sharma, S.K.; Gupta, R.K. Molecular Detection of Foodborne Pathogens: A Rapid and Accurate Answer to Food Safety. Crit. Rev. Food Sci. Nutr. 2016, 56, 1568–1584. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, J.; Shi, Z.; Cao, A.; Fang, W.; Yan, D.; Wang, Q.; Li, Y. Molecular Methods for Identification and Quantification of Foodborne Pathogens. Molecules 2022, 27, 8262. [Google Scholar] [CrossRef] [PubMed]
- Ndraha, N.; Lin, H.-Y.; Wang, C.-Y.; Hsiao, H.-I.; Lin, H.-J. Rapid Detection Methods for Foodborne Pathogens Based on Nucleic Acid Amplification: Recent Advances, Remaining Challenges, and Possible Opportunities. Food Chem. Mol. Sci. 2023, 7, 100183. [Google Scholar] [CrossRef]
- Panwar, S.; Duggirala, K.S.; Yadav, P.; Debnath, N.; Yadav, A.K.; Kumar, A. Advanced Diagnostic Methods for Identification of Bacterial Foodborne Pathogens: Contemporary and Upcoming Challenges. Crit. Rev. Biotechnol. 2022, 43, 1–19. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR Inhibitors—Occurrence, Properties and Removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in QPCR, DPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Maurer, J.J. Rapid Detection and Limitations of Molecular Techniques. Annu. Rev. Food Sci. Technol. 2011, 2, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, Y.; Huang, L.; Guo, J.; Wang, A.; Ma, C.; Shi, C. Direct Capture and Amplification of Nucleic Acids Using a Universal, Elution-Free Magnetic Bead-Based Method for Rapid Pathogen Detection in Multiple Types of Biological Samples. Anal. Bioanal. Chem. 2023, 415, 427–438. [Google Scholar] [CrossRef]
- He, Z.; Tang, C.; Chen, X.; Liu, H.; Yang, G.; Xiao, Z.; Li, W.; Deng, Y.; Jin, L.; Chen, H.; et al. Based on Magnetic Beads to Develop the Kit for Extraction of High-Quality Cell-Free DNA from Blood of Breast Cancer Patients. Mater. Express 2019, 9, 956–961. [Google Scholar] [CrossRef]
- Jin, B.; Ma, B.; Li, J.; Hong, Y.; Zhang, M. Simultaneous Detection of Five Foodborne Pathogens Using a Mini Automatic Nucleic Acid Extractor Combined with Recombinase Polymerase Amplification and Lateral Flow Immunoassay. Microorganisms 2022, 10, 1352. [Google Scholar] [CrossRef]
- Hedman, J.; Rådström, P. Overcoming Inhibition in Real-Time Diagnostic PCR. In PCR Detection of Microbial Pathogens; Wilks, M., Ed.; Humana: Totowa, NJ, USA, 2013; pp. 17–48. [Google Scholar]
- Butler, J.M. Separation of DNA Restriction Fragments and PCR Products. In Analysis of Nucleic Acids by Capillary Electrophoresis; Heller, C., Ed.; Chromatographia CE Series; Vieweg + Teubner Verlag: Wiesbaden, Germany, 1997; Volume 1, pp. 195–212. ISBN 978-3-322-91017-2. [Google Scholar]
- Lian, D.-S.; Zeng, H.-S. Capillary Electrophoresis Based on Nucleic Acid Detection as Used in Food Analysis. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1281–1295. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Zhang, Y.; He, P.; Wang, Q. Simultaneous Detection of Three Foodborne Pathogenic Bacteria in Food Samples by Microchip Capillary Electrophoresis in Combination with Polymerase Chain Reaction. J. Chromatogr. A 2018, 1555, 100–105. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Zheng, B.; Qu, L.; Li, C. Determination of Foodborne Pathogenic Bacteria by Multiplex PCR-Microchip Capillary Electrophoresis with Genetic Algorithm-Support Vector Regression Optimization. Anal. Chim. Acta 2009, 643, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, B.; García-Cañas, V.; Cifuentes, A.; González, R.; Aznar, R.; Alarcon, B.; Garcia-Canas, V.; Cifuentes, A.; Gonzalez, R.; Aznar, R. Simultaneous and Sensitive Detection of Three Foodborne Pathogens by Multiplex PCR, Capillary Gel Electrophoresis, and Laser-Induced Fluorescence. J. Agric. Food Chem. 2004, 52, 7180–7186. [Google Scholar] [CrossRef]
- Denis, E.; Bielińska, K.; Wieczorek, K.; Osek, J. Multiplex Real-Time PCRs for Detection of Salmonella, Listeria monocytogenes, and Verotoxigenic Escherichia coli in Carcasses of Slaughtered Animals. J. Vet. Res. 2016, 60, 287–292. [Google Scholar] [CrossRef]
- Leader, B.T.; Frye, J.G.; Hu, J.; Fedorka-Cray, P.J.; Boyle, D.S. High-Throughput Molecular Determination of Salmonella enterica Serovars by Use of Multiplex PCR and Capillary Electrophoresis Analysis. J. Clin. Microbiol. 2009, 47, 1290–1299. [Google Scholar] [CrossRef]
- Chen, J.; Sun, Y.; Peng, X.; Ni, Y.; Wang, F.; Dou, X. Rapid Analysis for Staphylococcus aureus via Microchip Capillary Electrophoresis. Sensors 2021, 21, 1334. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Huang, Y.; Ma, Y.; Yu, H.; Pang, B.; Liu, X.; Yin, C.; Wang, X.; Wei, Y.; Tian, Y.; et al. Detection of Four Foodborne Pathogens Based on Magnetic Separation Multiplex PCR and Capillary Electrophoresis. Biotechnol. J. 2022, 17, 2100335. [Google Scholar] [CrossRef]
- Research and Markets Global Chicken Market, Size, Forecast 2023–2028, Industry Trends, Growth, Share, Outlook, Impact of Inflation, Opportunity Company Analysis. Available online: https://www.researchandmarkets.com/report/chicken (accessed on 7 September 2023).
- Miller, M.; Gerval, A.; Hansen, J.; Grossen, G. Poultry Expected to Continue Leading Global Meat Imports as Demand Rises. Available online: https://www.ers.usda.gov/amber-waves/2022/august/poultry-expected-to-continue-leading-global-meat-imports-as-demand-rises/ (accessed on 7 September 2023).
- Joint FAO/WHO Codex Alimentarius Commission. General Standard for Irradiated Foods—CODEX STAN 106–1983, REV. 1–2003; Joint FAO/WHO Codex Alimentarius Commission: Rome, Italy, 2003; pp. 1–3. [Google Scholar]
- Wang, K.; Pang, X.; Zeng, Z.; Xiong, H.; Du, J.; Li, G.; Baidoo, I.K. Research on Irradiated Food Status and Consumer Acceptance: A Chinese Perspective. Food Sci. Nutr. 2023, 11, 4964–4974. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. Irradiation in The Production, Processing and Handling of Food; 21 C.F.R. § 179; The Code of Federal Regulations: New York, NY, USA, 2023; pp. 460–466.
- Suklim, K.; Flick, G.J.; Vichitphan, K. Effects of Gamma Irradiation on the Physical and Sensory Quality and Inactivation of Listeria monocytogenes in Blue Swimming Crab Meat (Portunas pelagicus). Radiat. Phys. Chem. 2014, 103, 22–26. [Google Scholar] [CrossRef]
- Spoto, M.H.F.; Gallo, C.R.; Alcarde, A.R.; Gurgel, M.S.d.A.; Blumer, L.; Walder, J.M.M.; Domarco, R.E. Gamma Irradiation in the Control of Pathogenic Bacteria in Refrigerated Ground Chicken Meat. Sci. Agric. 2000, 57, 389–394. [Google Scholar] [CrossRef]
- Gezgin, Z.; Gunes, G. Influence of Gamma Irradiation on Growth and Survival of Escherichia coli O157:H7 and Quality of Cig Kofte, a Traditional Raw Meat Product. Int. J. Food Sci. Technol. 2007, 42, 1067–1072. [Google Scholar] [CrossRef]
- Lee, N.; Kwon, K.Y.; Oh, S.K.; Chang, H.-J.; Chun, H.S.; Choi, S.-W. A Multiplex PCR Assay for Simultaneous Detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus in Korean Ready-to-Eat Foo. Foodborne Pathog. Dis. 2014, 11, 574–580. [Google Scholar] [CrossRef]
- Molina, F.; Lopez-Acedo, E.; Tabla, R.; Roa, I.; Gomez, A.; Rebollo, J. Improved Detection of Escherichia coli and Coliform Bacteria by Multiplex PCR. BMC Biotechnol. 2015, 15, 48. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q. Sensitive and Specific Detection of E. coli, Listeria monocytogenes, and Salmonella enterica Serovar Typhimurium in Milk by Microchip Electrophoresis Combined with Multiplex PCR Amplification. Microchem. J. 2020, 157, 104876. [Google Scholar] [CrossRef]
- Chin, W.H.; Sun, Y.; Høgberg, J.; Quyen, T.L.; Engelsmann, P.; Wolff, A.; Bang, D.D. Direct PCR—A Rapid Method for Multiplexed Detection of Different Serotypes of Salmonella in Enriched Pork Meat Samples. Mol. Cell Probes 2017, 32, 24–32. [Google Scholar] [CrossRef]
- Lai, Y.-H.; Chung, Y.-A.; Wu, Y.-C.; Fang, C.-T.; Chen, P.-J. Disease Burden from Foodborne Illnesses in Taiwan, 2012–2015. J. Formos. Med. Assoc. 2020, 119, 1372–1381. [Google Scholar] [CrossRef]
- Yu, C.-P.; Chou, Y.-C.; Wu, D.-C.; Cheng, C.-G.; Cheng, C.-A. Surveillance of Foodborne Diseases in Taiwan. Medicine 2021, 100, e24424. [Google Scholar] [CrossRef]
- Salazar, J.K.; Wang, Y.; Yu, S.; Wang, H.; Zhang, W. Polymerase Chain Reaction-Based Serotyping of Pathogenic Bacteria in Food. J. Microbiol. Methods 2015, 110, 18–26. [Google Scholar] [CrossRef]
- Lukinmaa, S.; Nakari, U.-M.; Eklund, M.; Siitonen, A. Application of Molecular Genetic Methods in Diagnostics and Epidemiology of Food-Borne Bacterial Pathogens. APMIS 2004, 112, 908–929. [Google Scholar] [CrossRef]
- Priyanka, B.; Patil, R.; Dwarakanath, S. A Review on Detection Methods Used for Foodborne Pathogens. Indian J. Med. Res. 2016, 144, 327. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Yen, S.-C.; Tsai, S.-K.; Shen, F.; Lin, J.H.-Y.; Lin, H.-J. Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp. Life 2022, 12, 276. [Google Scholar] [CrossRef]
- Emaus, M.N.; Varona, M.; Eitzmann, D.R.; Hsieh, S.-A.; Zeger, V.R.; Anderson, J.L. Nucleic Acid Extraction: Fundamentals of Sample Preparation Methodologies, Current Advancements, and Future Endeavors. TrAC Trends Anal. Chem. 2020, 130, 115985. [Google Scholar] [CrossRef]
- Barbosa, C.; Nogueira, S.; Gadanho, M.; Chaves, S. DNA Extraction: Finding the Most Suitable Method. In Molecular Microbial Diagnostic Methods: Pathways to Implementation for the Food and Water Industries; Cook, N., D’Agostino, M., Thompson, K.C., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 135–154. ISBN 9780124171701. [Google Scholar]
- Gheorghiu, E. Detection of Pathogenic Bacteria by Magneto-Immunoassays: A Review. J. Biomed. Res. 2021, 35, 277. [Google Scholar] [CrossRef]
- Herzig, G.P.D.; Aydin, M.; Dunigan, S.; Shah, P.; Jeong, K.C.; Park, S.H.; Ricke, S.C.; Ahn, S. Magnetic Bead-based Immunoassay Coupled with Tyramide Signal Amplification for Detection of Salmonella in Foods. J. Food Saf. 2016, 36, 383–391. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Chen, J.; Hua, D.; Li, Y.; Deng, H.; Li, Y.; Liang, Z.; Huang, J. Rapid Detection of Food-Borne Salmonella Contamination Using IMBs-QPCR Method Based on PagC Gene. Braz. J. Microbiol. 2018, 49, 320–328. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.; Liu, S.; Yang, J.; Wang, Y.; Nie, F.; Zhou, Q.; Li, Y.; Zhao, G. Simultaneous Detection of Six Food-Borne Pathogens by Multiplex PCR with a GeXP Analyzer. Food Control 2013, 32, 198–204. [Google Scholar] [CrossRef]
- Ruan, J.; Li, M.; Liu, Y.-P.; Li, Y.-Q.; Li, Y.-X. Rapid and Sensitive Detection of Cronobacter spp. (Previously Enterobacter sakazakii) in Food by Duplex PCR Combined with Capillary Electrophoresis-Laser-Induced Fluorescence Detector. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 921, 15–20. [Google Scholar] [CrossRef]
- Rossen, L.; Nørskov, P.; Holmstrøm, K.; Rasmussen, O.F. Inhibition of PCR by Components of Food Samples, Microbial Diagnostic Assays and DNA-Extraction Solutions. Int. J. Food Microbiol. 1992, 17, 37–45. [Google Scholar] [CrossRef]
- Bicart-See, A.; Rottman, M.; Cartwright, M.; Seiler, B.; Gamini, N.; Rodas, M.; Penary, M.; Giordano, G.; Oswald, E.; Super, M.; et al. Rapid Isolation of Staphylococcus aureus Pathogens from Infected Clinical Samples Using Magnetic Beads Coated with Fc-Mannose Binding Lectin. PLoS ONE 2016, 11, e0156287. [Google Scholar] [CrossRef]
- Fukushima, H.; Katsube, K.; Hata, Y.; Kishi, R.; Fujiwara, S. Rapid Separation and Concentration of Food-Borne Pathogens in Food Samples Prior to Quantification by Viable-Cell Counting and Real-Time PCR. Appl. Environ. Microbiol. 2007, 73, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Matsuda, Y.; Yamazaki, Y. Separation of Viable Lactic Acid Bacteria from Fermented Milk. Heliyon 2018, 4, e00597. [Google Scholar] [CrossRef]
- Fukushima, H.; Shimizu, S.; Inatsu, Y. Yersinia enterocolitica and Yersinia pseudotuberculosis Detection in Foods. J. Pathog. 2011, 2011, 735308. [Google Scholar] [CrossRef]
- Wang, M.; Gong, Q.; Liu, W.; Tan, S.; Xiao, J.; Chen, C. Applications of Capillary Electrophoresis in the Fields of Environmental, Pharmaceutical, Clinical, and Food Analysis (2019–2021). J. Sep. Sci. 2022, 45, 1918–1941. [Google Scholar] [CrossRef]
- Buszewski, B.; Maślak, E.; Złoch, M.; Railean-Plugaru, V.; Kłodzińska, E.; Pomastowski, P. A New Approach to Identifying Pathogens, with Particular Regard to Viruses, Based on Capillary Electrophoresis and Other Analytical Techniques. TrAC Trends Anal. Chem. 2021, 139, 116250. [Google Scholar] [CrossRef]
- Musso, M.; Bocciardi, R.; Parodi, S.; Ravazzolo, R.; Ceccherini, I. Betaine, Dimethyl Sulfoxide, and 7-Deaza-DGTP, a Powerful Mixture for Amplification of GC-Rich DNA Sequences. J. Mol. Diagn. 2006, 8, 544–550. [Google Scholar] [CrossRef]
- Grunenwald, H. Optimization of Polymerase Chain Reactions. In PCR Protocols; Bartlett, J.M.S., Stirling, D., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 89–100. [Google Scholar]
- Jensen, M.A.; Fukushima, M.; Davis, R.W. DMSO and Betaine Greatly Improve Amplification of GC-Rich Constructs in de Novo Synthesis. PLoS ONE 2010, 5, e11024. [Google Scholar] [CrossRef]
- Strien, J.; Sanft, J.; Mall, G. Enhancement of PCR Amplification of Moderate GC-Containing and Highly GC-Rich DNA Sequences. Mol. Biotechnol. 2013, 54, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Karunanathie, H.; Kee, P.S.; Ng, S.F.; Kennedy, M.A.; Chua, E.W. PCR Enhancers: Types, Mechanisms, and Applications in Long-Range PCR. Biochimie 2022, 197, 130–143. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Methods for Detection of Viable Foodborne Pathogens: Current State-of-Art and Future Prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef]
- Becskei, A.; Rahaman, S. The Life and Death of RNA across Temperatures. Comput. Struct. Biotechnol. J. 2022, 20, 4325–4336. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, H.; Li, J.; Hu, J.; Lin, G.; Tan, B.K.; Lin, S. Current Perspectives on Viable but Non-Culturable Foodborne Pathogenic Bacteria: A Review. Foods 2023, 12, 1179. [Google Scholar] [CrossRef]
- Wang, A.; Liu, L.; Zhang, S.; Ye, W.; Zheng, T.; Xie, J.; Wu, S.; Wu, Z.; Feng, Q.; Dong, H.; et al. Development of a Duplex Real-Time Reverse Transcription-Polymerase Chain Reaction Assay for the Simultaneous Detection of Goose Astrovirus Genotypes 1 and 2. J. Virol. Methods 2022, 310, 114612. [Google Scholar] [CrossRef]
- Chen, M.; Lan, X.; Zhu, L.; Ru, P.; Xu, W.; Liu, H. PCR Mediated Nucleic Acid Molecular Recognition Technology for Detection of Viable and Dead Foodborne Pathogens. Foods 2022, 11, 2675. [Google Scholar] [CrossRef]


| Target Pathogens | Nucleotide Sequence | Target Gene | Size (bp) | Ref |
|---|---|---|---|---|
| S. aureus | F: AAA TTA CAT AAA GAA CCT GCG AC | nuc | 164 | [40] |
| R: GCA CTT GCT TCA GGA CCA TA | ||||
| L. monocytogenes | F: CGC AAC AAA CTG AAG CAA AG | hly | 210 | [40] |
| R: TTG GCG GCA CAT TTG TCA C | ||||
| S. enterica | F: GCC GCC AAA CCT AA | invA | 392 | [42] |
| R: CTG CTA CCT TGC TGA TG |
| Enzyme | Trade Name | Source | Volume (µL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 10× Taq Buffer | 10 mM dNTP | Hot Start Taq | F-Primer | R-Primer | DNA Template | Water | |||
| GMbiolab | GenHot Taq DNA Polymerase | Gmbiolab Co., Ltd., Taipei, Taiwan | 2.5 | 0.5 | 0.50 | 0.5 | 0.5 | 2.0 | 18.50 |
| ZymesetBiotek | Hot start Taq DNA polymerase | ZymesetBiotek, Taipei, Taiwan | 2.5 | 0.5 | 0.25 | 0.5 | 0.5 | 2.0 | 18.75 |
| BioTools | TOOLS Hotstart Polymerase | Biotools, New Taipei City, Taiwan | 2.5 | 0.5 | 0.50 | 0.5 | 0.5 | 2.0 | 18.50 |
| BioHelix | nanoTaq Hot-Start DNA Polymerase | Bio-Helix Co., Ltd., New Taipei City, Taiwan | 2.5 | 0.5 | 1.25 | 0.5 | 0.5 | 2.0 | 17.75 |
| ThermoFisher | Invitrogen™ Platinum™ Taq DNA Polymerase 1 | ThermoFisher, Waltham, MA, USA | 2.5 | 0.5 | 0.10 | 0.5 | 0.5 | 2.0 | 18.15 |
| BiOptic | DirectGOTM PreMix-CE 2 | BiOptic Inc., New Taipei City, Taiwan | Premix: 12.5 | 0.5 | 0.5 | 2.0 | 9.50 | ||
| Species Detected | Sample | Detection Limit, Direct | Detection Method | Reference |
|---|---|---|---|---|
| L. monocytogenes, Salmonella and S. aureus | Stomached raw beef filtrate | Lm: 5.7 × 102, Sal: 7.9 × 102, Sa: 2.6 × 103 cfu/mL | Multiplex PCR | [27] |
| Salmonella, E. coli O157:H7, L. monocytogenes, S. aureus, Shigella spp. and C. jejuni | Broth | Sal: 4.2 × 102, Ec: 9.3 × 101, Lm: 3.1 × 102, Sa: 2.7 × 102, Sh: 8.5 × 101, and Cj: 6.6 × 101 cfu/mL | Multiplex PCR | [55] |
| Cronobacter spp. | Broth | Cr: 1.6 × 101 cfu/mL | Duplex PCR | [56] |
| S. enterica, L. monocytogenes, and S. aureus | Broth | Sal: 7.3 × 101, Lm: 6.7 × 102, Sa: 6.9 × 102 cfu/mL | Simplex PCR | This study |
| S. enterica, L. monocytogenes, and S. aureus | Stomached raw chicken filtrate | Sal: 7.3 × 104, Lm: 6.7 × 103, Sa: 6.9 × 102 cfu/g | Simplex PCR | This study |
| Steps | Culture-Based Method 1 | General PCR | PCR-CE Method |
|---|---|---|---|
| Sample preparation and dilution | ~10 min | ~30 min | ~30 min |
| Pre-enrichment | ~18 ± 2 h | − | − |
| Selective enrichment | ~24 ± 3 h | − | − |
| Plating out | ~24 ± 3 h | − | − |
| Confirmation 2 | ~24 ± 2 h | − | − |
| Serotyping | 4–6 h | − | − |
| DNA isolation by commercial kit | − | ~1 h (manual) | ~1 h (automatic) |
| Nucleic acid amplification | − | ~2 h | ~2 h |
| Agarose preparation | − | ~10 min | − |
| Amplicon analysis | − | ~30 min | ~4 min |
| Total spent time | ~94 h (3.9 days) | ~4 h and 10 min | ~3 h and 34 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndraha, N.; Lin, H.-Y.; Tsai, S.-K.; Hsiao, H.-I.; Lin, H.-J. The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis. Foods 2023, 12, 3895. https://doi.org/10.3390/foods12213895
Ndraha N, Lin H-Y, Tsai S-K, Hsiao H-I, Lin H-J. The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis. Foods. 2023; 12(21):3895. https://doi.org/10.3390/foods12213895
Chicago/Turabian StyleNdraha, Nodali, Hung-Yun Lin, Shou-Kuan Tsai, Hsin-I Hsiao, and Han-Jia Lin. 2023. "The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis" Foods 12, no. 21: 3895. https://doi.org/10.3390/foods12213895
APA StyleNdraha, N., Lin, H.-Y., Tsai, S.-K., Hsiao, H.-I., & Lin, H.-J. (2023). The Rapid Detection of Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus via Polymerase Chain Reaction Combined with Magnetic Beads and Capillary Electrophoresis. Foods, 12(21), 3895. https://doi.org/10.3390/foods12213895








