Generation of an Ovomucoid-Immune scFv Library for the Development of Novel Immunoassays in Hen’s Egg Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Immunization
2.2. Materials and Bacterial Strains
2.3. Preparation of Antigen and Food Protein Extracts
2.4. DNA Isolation, Quantification, and Cloning
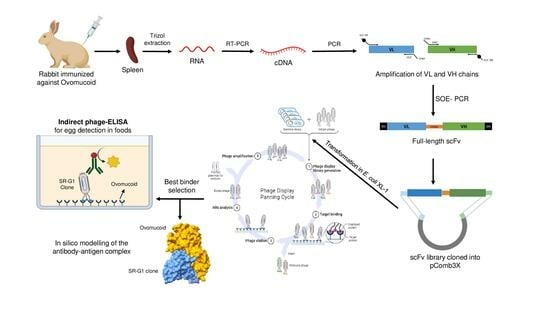
2.5. Construction of the Recombinant scFv Immune Library
2.6. Biopanning of the scFv Library against Ovomucoid and Egg White Extract
2.7. Characterization of the Ovomucoid-Binding Phage-scFvs
2.8. Indirect Phage-scFv ELISA
2.9. In Silico Modeling of SR-G1 scFv Structure and Its Interaction with Ovomucoid
3. Results and Discussion
3.1. Construction of the Immune scFv Library
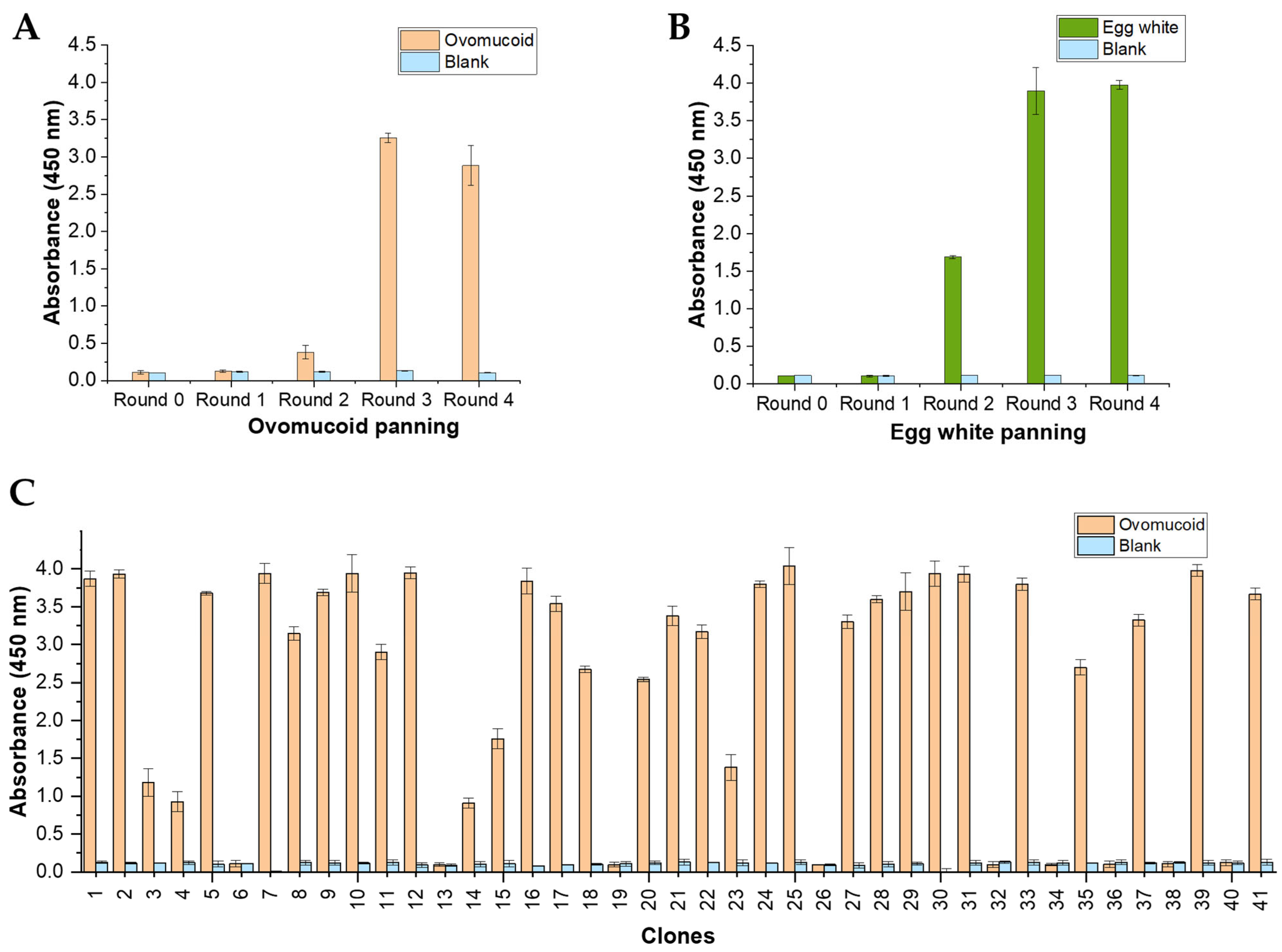
3.2. Biopanning of Anti-Ovomucoid Phage-scFv
3.3. Polyclonal Phage-ELISA
3.4. Screening of Anti-Ovomucoid Binders by Monoclonal Phage-ELISA
3.5. Sequence Analysis of the Positive Clones
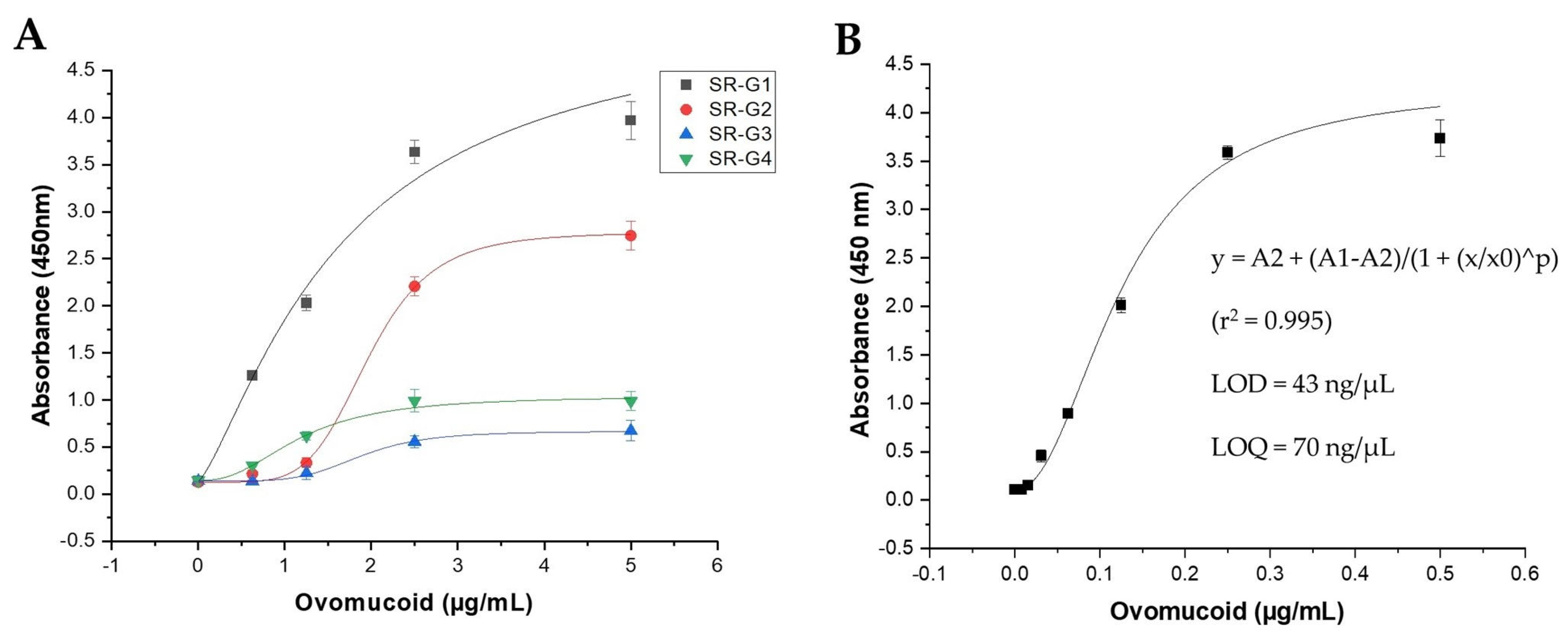
3.6. Characterization of the Anti-Ovomucoid Phage-scFvs Clones
3.7. Determination of Ovomucoid in Commercial Food Products
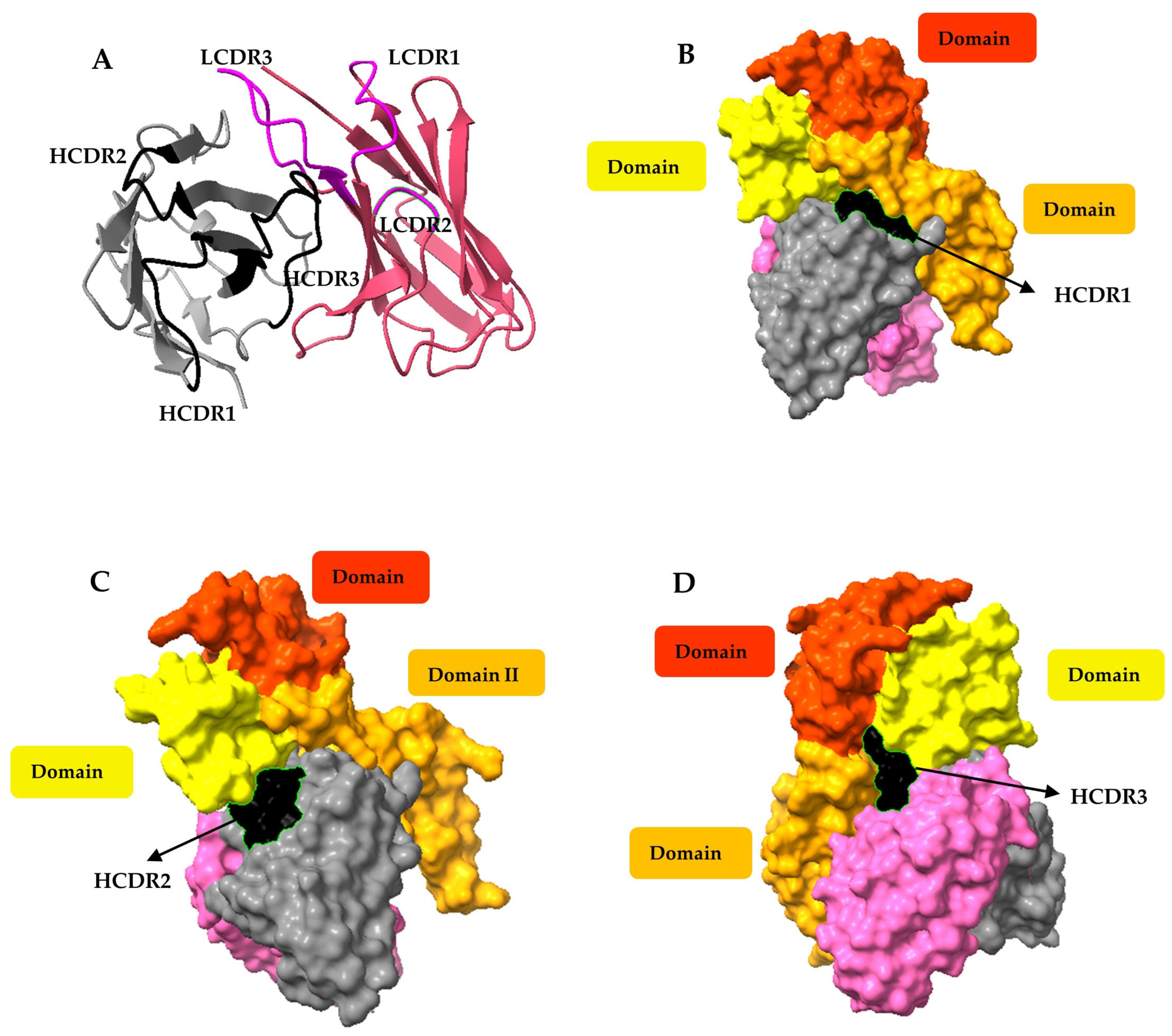
3.8. Molecular Docking of the Ovomucoid-Binding scFv SR-G1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Caubet, J.C.; Wang, J. Current understanding of egg allergy. Pediatr. Clin. North Am. 2011, 58, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Rona, R.J.; Keil, T.; Summers, C.; Gislason, D.; Zuidmeer, L.; Sodergren, E.; Sigurdardottir, S.T.; Lindner, T.; Goldhahn, K.; Dahlstrom, J.; et al. The prevalence of food allergy: A Meta-Analysis. J. Allergy Clin. Immunol. 2007, 120, 638–646. [Google Scholar] [CrossRef]
- Samady, W.; Warren, C.; Wang, J.; Das, R.; Gupta, R.S. Egg allergy in US children. J. Allergy Clin. Immunol. Pract. 2020, 8, 3066–3073.e6. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.H.; Matsui, E.C.; Skripak, J.M.; Wood, R.A. The natural history of egg allergy. J. Allergy Clin. Immunol. 2007, 120, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Caffarelli, C.; Giannetti, A.; Rossi, A.; Ricci, G. Egg allergy in children and weaning diet. Nutrients 2022, 14, 1540. [Google Scholar] [CrossRef] [PubMed]
- Kato, I.; Schrode, J.; Kohr, W.J.; Laskowski, M. Chicken Ovomucoid: Determination of its amino acid sequence, determination of the trypsin reactive site, and preparation of all three of its domains? Biochemistry 1987, 26, 193–201. [Google Scholar] [CrossRef]
- Matsuda, T.; Watanabe, K.; Nakamura, R. Immunochemical and Physical properties of peptic-digested ovomucoid. J. Agric. Food Chem. 1983, 31, 942–946. [Google Scholar] [CrossRef]
- Takagi, K.; Teshima, R.; Okunuki, H.; Itoh, S.; Kawasaki, N.; Kawanishi, T.; Hayakawa, T.; Kohno, Y.; Urisu, A.; Sawada, J.I. Kinetic analysis of pepsin digestion of chicken egg white ovomucoid and allergenic potential of pepsin fragments. Int. Arch. Allergy Immunol. 2005, 136, 23–32. [Google Scholar] [CrossRef]
- BernhiseI-Broadbent, J.; Dintzis, H.M.; Dintzis, R.Z.; Sampson, H.A.; Hopkins, J. Allergenicity and antigenicity of chicken egg ovomucoid (Gal d III) compared with ovalbumin (Gal d I) in children with egg allergy and in mice. J. Allergy Clin. Immunol. 1994, 93, 1047–1059. [Google Scholar] [CrossRef]
- Matsuda, T.; Nakamura, R.; Nakashima, I.; Hasegawa, Y.; Shimokata, K. Human IgE antibody to the carbohydrate-containing third domain of chicken ovomucoid. Biochem. Biophys. Res. Commun. 1985, 129, 505–510. [Google Scholar] [CrossRef]
- European Parliament and Council. Regulation (EU) No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the provision of food information to consumers, amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC and Commission Regulation (EC) No 608/2004. Off. J. Eur. Union 2011, 304, 18–63. [Google Scholar]
- Shon, D.-H.; Kim, H.-J.; Kim, S.-H.; Kwak, B.-Y. Enzyme-Linked Immunosorbent Assay for the detection of Hen’s egg proteins in processed foods. Korean. J. Food. Sci. Anim. Resour. 2010, 30, 36–42. [Google Scholar] [CrossRef][Green Version]
- Peltomaa, R.; Barderas, R.; Benito-Peña, E.; Moreno-Bondi, M.C. Recombinant antibodies and their use for food immunoanalysis. Anal. Bioanal. Chem. 2022, 414, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Phagen-Display: Einfache evolution in der petrischale (Nobel-Vortrag). Angew. Chem. 2019, 131, 14566–14576. [Google Scholar] [CrossRef]
- Vu, N.X.; Pruksametanan, N.; Srila, W.; Yuttavanichakul, W.; Teamtisong, K.; Teaumroong, N.; Boonkerd, N.; Tittabutr, P.; Yamabhai, M. Generation of a rabbit Single-Chain Fragment Variable (scFv) antibody for specific detection of Bradyrhizobium Sp. DOA9 in both free-living and bacteroid forms. PLoS ONE 2017, 12, e0179983. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv Antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef]
- de la Cruz, S.; Cubillos-Zapata, C.; López-Calleja, I.M.; Ghosh, S.; Alcocer, M.; González, I.; Martín, R.; García, T. Isolation of recombinant antibody fragments (scFv) by phage display technology for detection of almond allergens in food products. Food Control 2015, 54, 322–330. [Google Scholar] [CrossRef][Green Version]
- Madrid, R.; de la Cruz, S.; García-García, A.; Alcocer, M.J.C.; González, I.; García, T.; Martín, R. Multimeric recombinant antibody (scFv) for ELISA detection of allergenic walnut. An alternative to animal antibodies. J. Food Compos. Anal. 2018, 67, 201–210. [Google Scholar] [CrossRef]
- Pansri, P.; Jaruseranee, N.; Rangnoi, K.; Kristensen, P.; Yamabhai, M.A. Compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 2009, 9, 6. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Guan, Q.; Wang, L.; Yuan, H. Advances in the isolation of specific monoclonal rabbit antibodies. Front. Immunol. 2017, 8, 494. [Google Scholar] [CrossRef]
- Weber, J.; Peng, H.; Rader, C. From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 2017, 49, e305. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Nerreter, T.; Chang, J.; Qi, J.; Li, X.; Karunadharma, P.; Martinez, G.J.; Fallahi, M.; Soden, J.; Freeth, J.; et al. Mining naïve rabbit antibody repertoires by phage display for monoclonal antibodies of therapeutic utility. J. Mol. Biol. 2017, 429, 2954–2973. [Google Scholar] [CrossRef] [PubMed]
- Madrid, R.; García-García, A.; González, I.; Martín, R.; García, T. Phage displayed domain antibodies (dAb) for detection of allergenic pistachio proteins in foods. Foods 2020, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Calvo, E.; García-García, A.; Rodríguez, S.; Farrais, S.; Martín, R.; García, T. Construction of a Fab library merging chains from semisynthetic and immune origin, suitable for developing new tools for gluten immunodetection in food. Foods 2023, 12, 149. [Google Scholar] [CrossRef]
- Lim, B.N.; Chin, C.F.; Choong, Y.S.; Ismail, A.; Lim, T.S. Generation of a naïve human single chain variable fragment (scFv) library for the identification of monoclonal scFv against Salmonella Typhi Hemolysin antigen. Toxicon 2016, 117, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Calvo, E.; García-García, A.; Rodríguez-Gómez, S.; Farrais, S.; Martín, R.; García, T. Development of a new recombinant antibody, selected by phage-display technology from a celiac patient library, for detection of gluten in foods. Curr. Res. Food Sci. 2023, 7, 100578. [Google Scholar] [CrossRef]
- Barbas, C.F. Phage Display: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Brochet, X.; Lefranc, M.P.; Giudicelli, V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic. Acids Res. 2008, 36, 503–508. [Google Scholar] [CrossRef]
- Dunbar, J.; Krawczyk, K.; Leem, J.; Marks, C.; Nowak, J.; Regep, C.; Georges, G.; Kelm, S.; Popovic, B.; Deane, C.M. SAbPred: A structure-based antibody prediction server. Nucleic. Acids Res. 2016, 44, W474–W478. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; Van Dijk, M.; De Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural biology in the clouds: The WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Vangone, A.; Bonvin, A.M.J.J. PRODIGY: A contact-based predictor of binding affinity in protein-protein complexes. Bio. Protoc. 2017, 7, e2124. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Finlay, W.J.J.; Almagro, J.C. Natural and man-made V-Gene repertoires for antibody discovery. Front. Immunol. 2012, 3, 342. [Google Scholar] [CrossRef] [PubMed]
- Mage, R.G.; Lanning, D.; Knight, K.L. B cell and antibody repertoire development in rabbits: The requirement of gut-associated lymphoid tissues. Dev. Comp. Immunol. 2006, 30, 137–153. [Google Scholar] [CrossRef]
- Kodangattil, S.; Huard, C.; Ross, C.; Li, J.; Gao, H.; Mascioni, A.; Hodawadekar, S.; Naik, S.; Min-, J.; Visintin, A.; et al. Antibody discovery via next-generation sequencing the functional repertoire of rabbit antibodies and antibody discovery via next-generation sequencing. mAbs 2014, 6, 628–636. [Google Scholar] [CrossRef]
- Lee, Y.; Kyun, D.; Noh, J.; Ju, S.; Lee, E.; Lee, H.; Kwon, S.; Chung, J. Amplification of a minimally biased antibody repertoire for in vitro display using a universal primer-based amplification method. J. Immunol. Methods 2021, 496, 113089. [Google Scholar] [CrossRef]
- Zemlin, M.; Klinger, M.; Link, J.; Zemlin, C.; Bauer, K.; Engler, J.A.; Schroeder, H.W., Jr.; Kirkham, P.M. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003, 334, 733–749. [Google Scholar] [CrossRef]
- Lavinder, J.J.; Hoi, K.H.; Reddy, S.T.; Wine, Y.; Georgiou, G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS ONE 2014, 9, e101322. [Google Scholar] [CrossRef]
- Fæste, C.K.; Løvberg, K.E.; Lindvik, H.; Egaas, E. Extractability, stability, and allergenicity of egg white proteins in differently heat-processed foods. J. AOAC Int. 2007, 90, 427–436. [Google Scholar] [CrossRef]
- Morisset, M.; Moneret-Vautrin, D.A.; Kanny, G.; Guénard, L.; Beaudouin, E.; Flabbée, J.; Hatahet, R. Thresholds of clinical reactivity to milk, egg, peanut and sesame in immunoglobulin E-dependent allergies: Evaluation by double-blind or single-blind placebo-controlled oral challenges. Clin. Exp. Allergy 2003, 33, 1046–1051. [Google Scholar] [CrossRef]
- Hirose, J.; Kitabatake, N.; Kimura, A.; Narita, H. Recognition of native and/or thermally induced denatured forms of the major food allergen, ovomucoid, by human IgE and mouse monoclonal IgG antibodies. Biosci. Biotechnol. Biochem. 2004, 68, 2490–2497. [Google Scholar] [CrossRef] [PubMed]
- Hirose, J.; Murakami-Yamaguchi, Y.; Ikeda, M.; Kitabatake, N.; Narita, H. Oligoclonal enzyme-linked immunosorbent assay capable of determining the major food allergen, ovomucoid, irrespective of the degree of heat denaturation. Cytotechnology 2005, 47, 145–149. [Google Scholar] [CrossRef]
- Li, Y.; Song, C.; Zhang, K.; Wang, M.; Yang, K.; Yang, A.; Jin, B. Establishment of a highly sensitive sandwich enzyme-linked immunosorbent assay specific for ovomucoid from hen’s egg white. J. Agric. Food Chem. 2008, 56, 337–342. [Google Scholar] [CrossRef]
- Hwan, J.; Jung, T.; Seop, M.; Pil, J. A phage virus-based electrochemical biosensor for highly sensitive detection of ovomucoid. Food Chem. 2022, 378, 132061. [Google Scholar] [CrossRef]
- Abanades, B.; Wong, W.K.; Deane, C.M.; Boyles, F.; Georges, G.; Bujotzek, A. ImmuneBuilder: Deep-learning models for predicting the structures of immune proteins. Commun. Biol. 2023, 6, 575. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Yang, S.; Han, S.; Che, H. Food science and human wellness the amino acids differences in epitopes may promote the different allergenicity of ovomucoid derived from hen eggs and quail eggs. Food Sci. Hum. Wellness 2023, 12, 861–870. [Google Scholar] [CrossRef]
- Mine, Y.; Wei Zhang, J. Identification and fine mapping of IgG and IgE epitopes in ovomucoid. Biochem. Biophys. Res. Commun. 2002, 292, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- López-Camacho, E.; García-Godoy, M.J.; García-Nieto, J.; Nebro, A.J.; Aldana-Montes, J.F. A New Multi-Objective Approach for Molecular Docking Based on RMSD and Binding Energy; Springer International Publishing: Cham, Switzerland, 2016; pp. 65–77. [Google Scholar] [CrossRef]
- Selvam, R.; Lim, I.H.Y.; Lewis, J.C.; Lim, C.H.; Yap, M.K.K.; Tan, H.S. Selecting antibacterial aptamers against the BamA protein in Pseudomonas aeruginosa by incorporating genetic algorithm to optimise computational screening method. Sci. Rep. 2023, 13, 7582. [Google Scholar] [CrossRef]

| Vegetal Species | ||
| Almond (Prunus dulcis) | Kiwifruit (Actinidia deliciosa) | Rice (Oryza sativa) |
| Apple (Malus domestica) | Maize (Zea mays) | Sesame (Sesamum indicum) |
| Banana (Musa acuminate) | Mandarin orange (Citrus reticulate) | Spinach (Spinacea oleracea) |
| Carrot (Daucus carota) | Oats (Avena sativa) | Soy (Glycine max) |
| Cashew nut (Anacardium occidentale) | Onion (Allium cepa) | Tomato (Solanum lycopersicum) |
| Chia (Salvia hispanica) | Pear (Pyrus communis) | Walnut (Juglans regia) |
| Chickpea (Cicer arietinum) | Poppy seed (Papaver rhoeas) | Wheat (Triticum aestivum) |
| Flaxseed (Linum usitatissimum) | Quinoa (Chenopodium quinoa) | |
| Garlic (Allium sativum) | Red peppers (Capsicum annuum) | |
| Hazelnut (Corylus avellana) | ||
| Animal species | ||
| Beef (Bos taurus) | Pork (Sus scrofa domestica) | |
| Chicken (Gallus gallus domesticus) | Turkey (Meleagris gallopavo) | |
| Others | ||
| Milk (Bos taurus) | Ostrich’s egg (Struthio camelus) | Quail’s egg (Coturnix coturnix) |
| Hen’s egg (Gallus gallus domesticus) | Duck’s egg (Anas platyrhynchos domesticus) | Goose’s egg (Anas pedes sulfurate) |
| Clone | Light Chain | Heavy Chain |
|---|---|---|
| SR-G1 | IGKV1S36*01 | IGHV1S69*01 |
| IGKJ1-2*04 | IGHJ6*02 | |
| IGHD2-1*01 | ||
| SR-G2 | IGKV1S34*01 | IGHV1S69*01 |
| IGKJ1-2*01 | IGHJ6*02 | |
| IGHD2-1*01 | ||
| SR-G3 | IGKV1S44*01 | IGHV1S69*01 |
| IGKJ1-2*02 | IGHJ4*02 | |
| IGHD7-1*01 | ||
| SR-G4 | IGKV1S4*01 | IGHV1S69*01 |
| IGKJ1-2*02 | IGHJ4*02 | |
| IGHD7-1*01 |
| Label Statement | Product | Number of Samples Analysed | Phage-ELISA | Polyclonal ELISA |
|---|---|---|---|---|
| Egg declared as ingredie+nt | Biscuits | 1 | +(1) | +(1) |
| Cake | 1 | +(1) | +(1) | |
| Flan | 2 | +(2) | +(2) | |
| Meat product | 3 | +(3) | +(3) | |
| Omelet | 2 | +(2) | +(2) | |
| Sandwich | 1 | +(1) | +(1) | |
| Salad | 2 | +(2) | +(2) | |
| Egg yolk declared as ingredient | Bakery | 1 | −(1) | +(1) |
| Salad | 2 | +(1)/−(1) | +(2) | |
| May contain egg traces | Meat product | 3 | +(1)/(−2) | +(2)/(−1) |
| Not declaring to contain egg or traces | Breakfast cereal | 1 | −(1) | −(1) |
| Seeds | 1 | −(1) | −(1) | |
| Snack | 1 | −(1) | −(1) | |
| Meat | 2 | +(1)/−(1) | +(1)/−(1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, S.; García-García, A.; Garcia-Calvo, E.; Esteban, V.; Pastor-Vargas, C.; Díaz-Perales, A.; García, T.; Martín, R. Generation of an Ovomucoid-Immune scFv Library for the Development of Novel Immunoassays in Hen’s Egg Detection. Foods 2023, 12, 3831. https://doi.org/10.3390/foods12203831
Rodríguez S, García-García A, Garcia-Calvo E, Esteban V, Pastor-Vargas C, Díaz-Perales A, García T, Martín R. Generation of an Ovomucoid-Immune scFv Library for the Development of Novel Immunoassays in Hen’s Egg Detection. Foods. 2023; 12(20):3831. https://doi.org/10.3390/foods12203831
Chicago/Turabian StyleRodríguez, Santiago, Aina García-García, Eduardo Garcia-Calvo, Vanesa Esteban, Carlos Pastor-Vargas, Araceli Díaz-Perales, Teresa García, and Rosario Martín. 2023. "Generation of an Ovomucoid-Immune scFv Library for the Development of Novel Immunoassays in Hen’s Egg Detection" Foods 12, no. 20: 3831. https://doi.org/10.3390/foods12203831
APA StyleRodríguez, S., García-García, A., Garcia-Calvo, E., Esteban, V., Pastor-Vargas, C., Díaz-Perales, A., García, T., & Martín, R. (2023). Generation of an Ovomucoid-Immune scFv Library for the Development of Novel Immunoassays in Hen’s Egg Detection. Foods, 12(20), 3831. https://doi.org/10.3390/foods12203831