Epidemiological Data Mining for Assisting with Foodborne Outbreak Investigation
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Data Source
3.2. Experiment Design
3.3. Data Preprocessing
3.4. Statistical Analysis and Regression Models
3.5. Data Mining with Association Rules
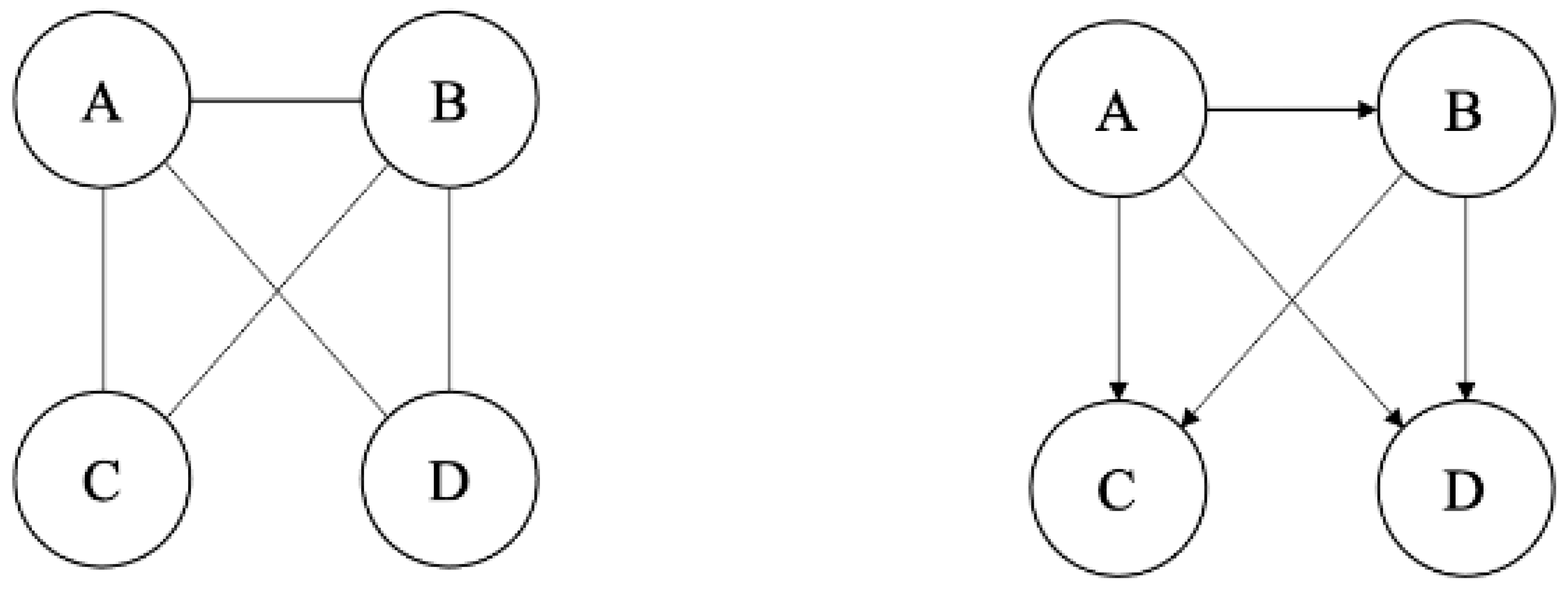
3.6. Network Construction and Centrality Measures
- Degree centrality: Degree centrality is the simplest centrality measure, defined as the number of other nodes connected with a node (i.e., the number of links that a node has). The degree can be interpreted in terms of the immediate risk of a node for catching whatever is flowing through the network.
- Betweenness centrality: Betweenness centrality quantifies the number of times a node acts as a bridge along the shortest path between two other nodes.
- Closeness centrality: Closeness centrality is the average length of the shortest paths between the node and all other nodes in the graph. Thus, the more central a node is, the closer it is to all other nodes.
- Eigenvector centrality: Eigenvector centrality assigns relative scores to all nodes in the network based on the concept that connections to high-scoring nodes contribute more to the score of the node in question than equal connections to low-scoring nodes.
3.7. Monte Carlo Simulation Modeling
4. Results and Discussions
4.1. Univariant Analysis between Attributes
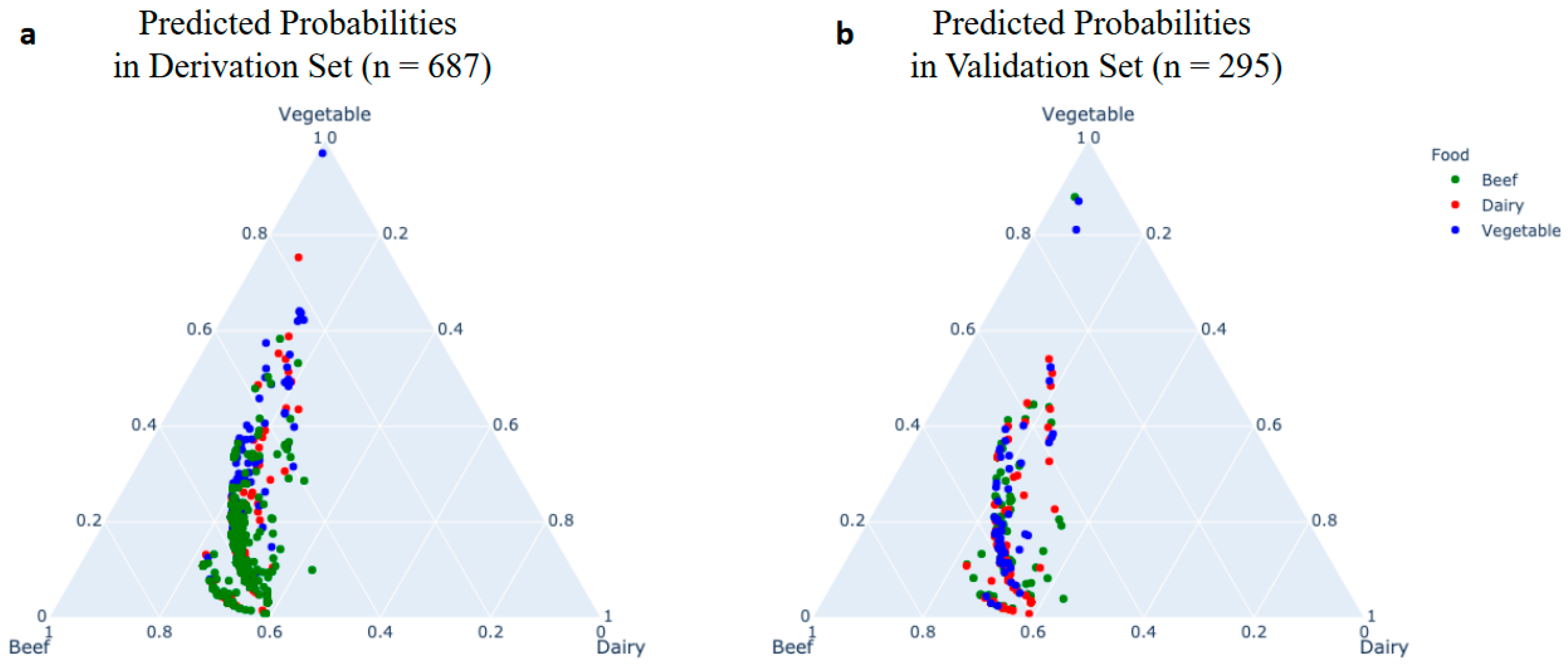
4.2. Multinomial Regression for Predicting Food Sources
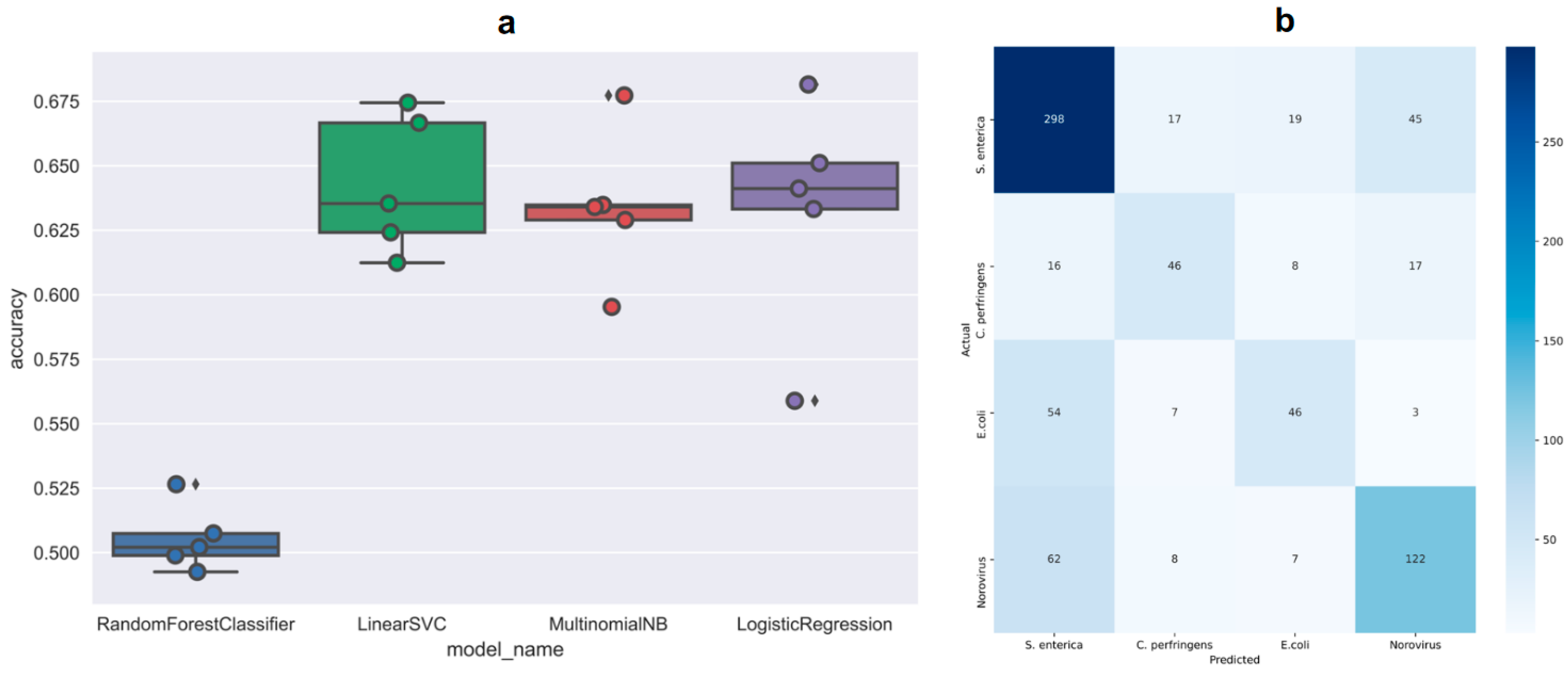
4.3. Classification Models for Predicting Etiology Based on Food Vehicles
4.4. Association Rule Mining between Food Vehicles and Etiologies
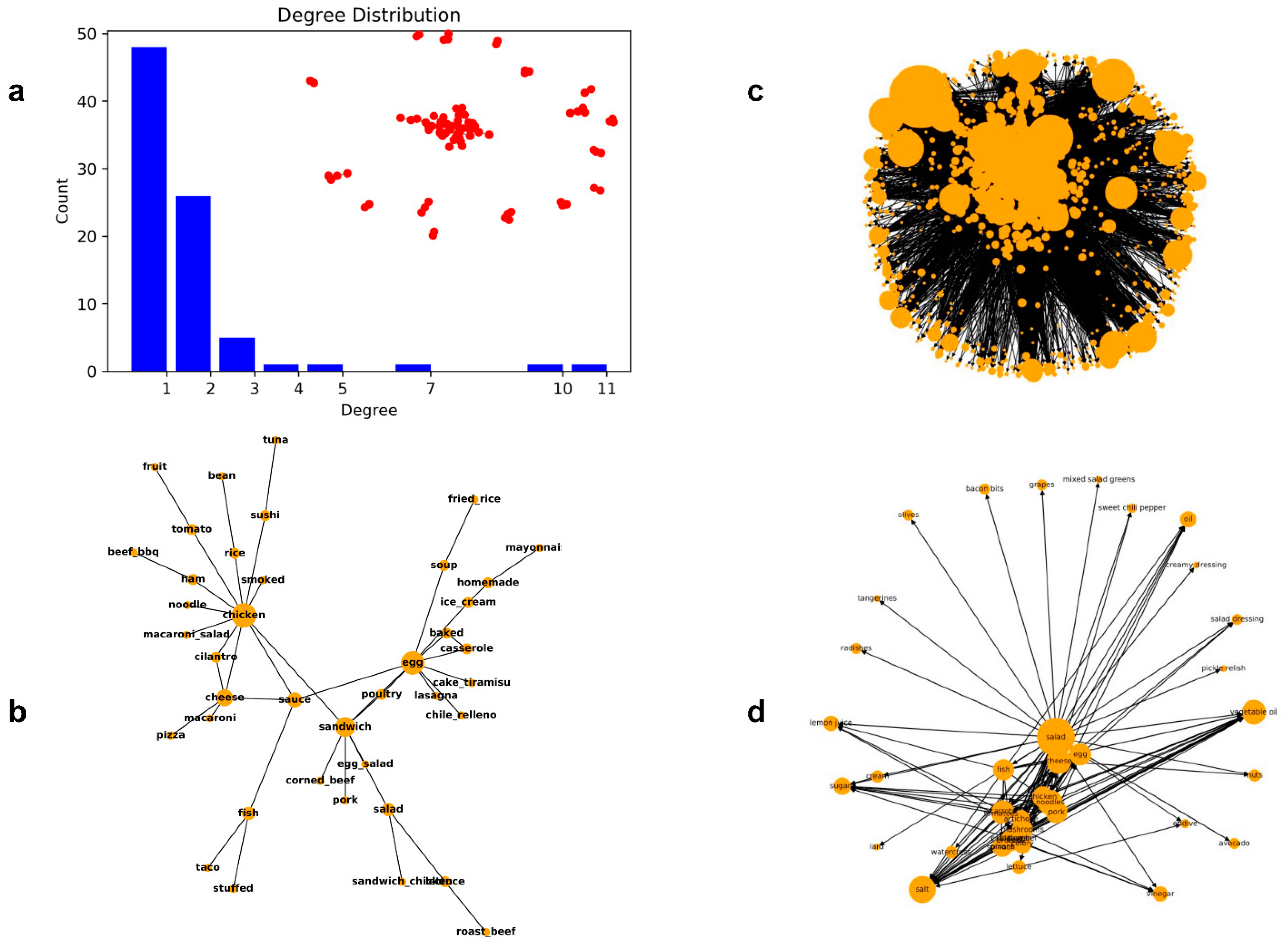
4.5. Characteristics of the Food Co-Occurrence Network
4.6. Characteristics of the Food Ingredient Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.F.; Rosenberg, L.; Kubota, K.; Ingram, L.A. Variability among states in investigating foodborne disease outbreaks. Foodborne Pathog. Dis. 2013, 10, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Greig, J.D.; Ravel, A. Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol. 2009, 130, 77–87. [Google Scholar] [CrossRef]
- Pires, S.M.; Vieira, A.R.; Hald, T.; Cole, D. Source attribution of human salmonellosis: An overview of methods and estimates. Foodborne Pathog. Dis. 2014, 11, 667–676. [Google Scholar] [CrossRef]
- Morgan, O. How decision makers can use quantitative approaches to guide outbreak responses. Philos. Trans. R. Soc. B 2019, 374, 20180365. [Google Scholar] [CrossRef]
- White, A.; Cronquist, A.; Bedrick, E.J.; Scallan, E. Food source prediction of Shiga toxin–producing Escherichia coli outbreaks using demographic and outbreak characteristics, United States, 1998–2014. Foodborne Pathog. Dis. 2016, 13, 527–534. [Google Scholar] [CrossRef]
- Adak, G.K.; Meakins, S.M.; Yip, H.; Lopman, B.A.; O’Brien, S.J. Disease risks from foods, England and Wales, 1996–2000. Emerg. Infect. Dis. 2005, 11, 365. [Google Scholar] [CrossRef]
- Hoffmann, S.; Devleesschauwer, B.; Aspinall, W.; Cooke, R.; Corrigan, T.; Havelaar, A.; Angulo, F.; Gibb, H.; Kirk, M.; Lake, R.; et al. Attribution of global foodborne disease to specific foods: Findings from a World Health Organization structured expert elicitation. PLoS ONE 2017, 12, e0183641. [Google Scholar] [CrossRef]
- Batz, M.B.; Hoffmann, S.; Morris, J.G. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J. Food Prot. 2012, 75, 1278–1291. [Google Scholar] [CrossRef]
- Thakur, M.; Olafsson, S.; Lee, J.S.; Hurburgh, C.R. Data mining for recognizing patterns in foodborne disease outbreaks. J. Food Eng. 2010, 97, 213–227. [Google Scholar] [CrossRef]
- Nsoesie, E.O.; Kluberg, S.A.; Brownstein, J.S. Online reports of foodborne illness capture foods implicated in official foodborne outbreak reports. Prev. Med. 2014, 67, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Sadilek, A.; Caty, S.; DiPrete, L.; Mansour, R.; Schenk, T.; Bergtholdt, M.; Jha, A.; Ramaswami, P.; Gabrilovich, E. Machine-learned epidemiology: Real-time detection of foodborne illness at scale. NPJ Digit. Med. 2018, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Pires, S.M.; Vieira, A.R.; Perez, E.; Wong, D.L.F.; Hald, T. Attributing human foodborne illness to food sources and water in Latin America and the Caribbean using data from outbreak investigations. Int. J. Food Microbiol. 2012, 152, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407. [Google Scholar] [CrossRef]
- Han, J.; Pei, J.; Tong, H. Data Mining: Concepts and Techniques; Morgan Kaufmann: San Francisco, CA, USA, 2022. [Google Scholar]
- Ahn, Y.Y.; Ahnert, S.E.; Bagrow, J.P.; Barabási, A.L. Flavor network and the principles of food pairing. Sci. Rep. 2011, 1, 196. [Google Scholar] [CrossRef]
- Simas, T.; Ficek, M.; Diaz-Guilera, A.; Obrador, P.; Rodriguez, P.R. Food-bridging: A new network construction to unveil the principles of cooking. Front. ICT 2017, 4, 14. [Google Scholar] [CrossRef]
- Jensen, K.; Panagiotou, G.; Kouskoumvekaki, I. NutriChem: A systems chemical biology resource to explore the medicinal value of plant-based foods. Nucleic Acids Res. 2015, 43, D940–D945. [Google Scholar] [CrossRef]
- Kim, S.; Sung, J.; Foo, M.; Jin, Y.S.; Kim, P.J. Uncovering the nutritional landscape of food. PLoS ONE 2015, 10, e0118697. [Google Scholar] [CrossRef]
- Yang, H.; Swaminathan, R.; Sharma, A.; Ketkar, V.; Jason, D.S. Mining biomedical text towards building a quantitative food-disease-gene network. In Learning Structure and Schemas from Documents; Springer: Berlin/Heidelberg, Germany, 2011; pp. 205–225. [Google Scholar]
- Cooper, K.M. The ingredient co-occurrence network of packaged foods distributed in the United States. J. Food Compos. Anal. 2020, 86, 103391. [Google Scholar] [CrossRef]
- Smid, J.H.; Verloo, D.; Barker, G.C.; Havelaar, A.H. Strengths and weaknesses of Monte Carlo simulation models and Bayesian belief networks in microbial risk assessment. Int. J. Food Microbiol. 2010, 139, S57–S63. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, A.M.; Fazil, A. Hazard identification and exposure assessment for microbial food safety risk assessment. Int. J. Food Microbiol. 2000, 58, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Chavez, R.A.; Stasiewicz, M.J. When to use one-dimensional, two-dimensional, and Shifted Transversal Design pooling in mycotoxin screening. PLoS ONE 2020, 15, e0236668. [Google Scholar] [CrossRef]
- Hu, D.; Sun, T.; Yao, L.; Yang, Z.; Wang, A.; Ying, Y. Monte carlo: A flexible and accurate technique for modeling light transport in food and agricultural products. Trends Food Sci. Technol. 2020, 102, 280–290. [Google Scholar] [CrossRef]
- Djekic, I.; Bozickovic, I.; Djordjevic, V.; Smetana, S.; Terjung, N.; Ilic, J.; Doroski, A.; Tomasevic, I. Can we associate environmental footprints with production and consumption using Monte Carlo simulation? Case study with pork meat. J. Sci. Food Agric. 2021, 101, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Coyle, D.H.; Wu, J.H.; Di Tanna, G.L.; Shahid, M.; Taylor, F.; Neal, B.; Trevena, H. The effects of a supermarket-based intervention on the nutritional quality of private-label foods: A prospective study. Nutrients 2020, 12, 1692. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.J.; Wikswo, M.E.; Pringle, K.; Gould, L.H.; Parashar, U.D. Vital signs: Foodborne norovirus outbreaks—United States, 2009–2012. Morb. Mortal. Wkly. Rep. 2014, 63, 491. [Google Scholar]
- Tao, D.; Yang, P.; Feng, H. Utilization of text mining as a big data analysis tool for food science and nutrition. Compr. Rev. Food Sci. Food Saf. 2020, 19, 875–894. [Google Scholar] [CrossRef]
- Agrawal, R.; Mannila, H.; Srikant, R.; Toivonen, H.; Verkamo, A.I. Fast discovery of association rules. Adv. Knowl. Discov. Data Min. 1996, 12, 307–328. [Google Scholar]
- Hagberg, A.; Conway, D. Networkx: Network Analysis with Python. 2020. Available online: https://networkx.github.io (accessed on 15 January 2019).
- Foreman-Mackey, D.; Hogg, D.W.; Lang, D.; Goodman, J. emcee: The MCMC hammer. Publ. Astron. Soc. Pac. 2013, 125, 306. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.; Valero, A.; Carrasco, E.; García, R.M.; Zurera, G. Understanding and modelling bacterial transfer to foods: A review. Trends Food Sci. Technol. 2008, 19, 131–144. [Google Scholar] [CrossRef]
- Carrasco, E.; Morales-Rueda, A.; García-Gimeno, R.M. Cross-contamination and recontamination by Salmonella in foods: A review. Food Res. Int. 2012, 45, 545–556. [Google Scholar] [CrossRef]
- Herrera, J.C. The Contribution of Network Science to the Study of Food Recipes. A Review Paper. Appetite 2020, 159, 105048. [Google Scholar] [CrossRef] [PubMed]
| Attribute | Type | Description |
|---|---|---|
| Etiology | Nominal | Cause of outbreak, e.g., Salmonella enterica |
| State | Nominal | State where the outbreak occurred |
| Month | Nominal | Month when the outbreak occurred |
| Illnesses | Numeric | Number of illnesses reported |
| Hospitalizations | Numeric | Number of hospitalizations reported |
| Deaths | Numeric | Number of deaths reported |
| Food Vehicle | Text | Food item/s that caused the outbreak |
| Setting | Text | Location where food was consumed, e.g., restaurant |
| IFSAC Category | Number of Outbreaks |
|---|---|
| Multiple | 4045 |
| Undetermined | 1126 |
| Fish | 892 |
| Beef | 678 |
| Chicken | 501 |
| Unclassifiable | 433 |
| Mollusks | 339 |
| Dairy | 324 |
| Pork | 318 |
| Fruits | 245 |
| Vegetable Row Crops | 215 |
| Turkey | 190 |
| Eggs | 186 |
| Other | 182 |
| Grains-Beans | 178 |
| Seeded Vegetables | 136 |
| Crustaceans | 109 |
| Root/Underground | 79 |
| Sprouts | 53 |
| Fungi | 40 |
| Herbs | 33 |
| Game | 30 |
| Nuts-Seeds | 26 |
| Other Meat | 16 |
| Oils-Sugars | 12 |
| Other Poultry | 11 |
| Other Aquatic Animals | 11 |
| Identified Factors | Beef | Dairy | Vegetable | p-Value |
|---|---|---|---|---|
| Number of outbreaks | 678 | 324 | 215 | |
| Total cases | 11,649 | 6979 | 9910 | |
| Gender a | ||||
| Male | 50 (33, 60) | 50 (38, 61) | 37 (25, 50) | <0.01 |
| Female | 50 (40, 66) | 50 (36, 60) | 62 (50, 75) | <0.01 |
| Missing b | 121 (18) | 26 (8) | 27 (13) | <0.01 |
| Age (years) a | ||||
| % < 5 | 0 (0, 0) | 0 (0, 11) | 0 (0, 0) | <0.01 |
| % 5–19 | 0 (0, 14) | 0 (0, 22) | 0 (0, 11) | <0.01 |
| % 20–49 | 33 (12, 57) | 34 (10, 50) | 43 (21, 50) | 0.37 |
| % >= 50 | 0 (0, 24) | 0 (0, 12) | 8 (0, 29) | <0.01 |
| % Unknown | 0 (0, 5) | 0 (0, 0) | 0 (0, 4) | 0.42 |
| Missing b | 556 (82) | 158 (49) | 126 (59) | <0.01 |
| Number of cases a | 6 (3, 20) | 7 (3, 15) | 16 (9, 30) | <0.01 |
| Exposure setting b | <0.01 | |||
| Private | 125 (18) | 53 (16) | 18 (8) | |
| Non-private | 533 (79) | 252 (78) | 185 (86) | |
| Missing | 20 (3) | 19 (6) | 12 (6) | |
| Multistate Outbreak b | 50 (7) | 24 (7) | 45 (21) | <0.01 |
| Season b | 0.1 | |||
| Spring | 154 (23) | 62 (19) | 43 (20) | |
| Summer | 182 (27) | 103 (32) | 73 (34) | |
| Fall | 204 (30) | 88 (27) | 44 (20) | |
| Winter | 204 (30) | 71 (22) | 55 (26) | |
| Duration (days) a | 3 (2, 14) | 11 (4, 36) | 6 (3, 16) | <0.01 |
| Missing b | 437 (64) | 91 (28) | 84 (39) | <0.01 |
| Etiology | Food Vehicle | Confidence | Lift |
|---|---|---|---|
| C. perfringens | Roast beef | 0.69 | 5.35 |
| Gravy | 0.56 | 4.37 | |
| Beef | 0.33 | 2.55 | |
| Turkey | 0.36 | 2.76 | |
| Chicken | 0.21 | 1.63 | |
| STEC | Ground beef | 0.60 | 4.80 |
| Beef | 0.27 | 2.15 | |
| Lettuce | 0.26 | 2.05 | |
| Ice | 0.94 | 3.55 | |
| Raw oyster | 0.88 | 3.34 | |
| Sandwich | 0.86 | 3.25 | |
| Green salad | 0.75 | 2.85 | |
| Norovirus | Salad | 0.74 | 2.81 |
| Fruit | 0.70 | 2.64 | |
| Lettuce | 0.62 | 2.38 | |
| Coleslaw | 0.59 | 2.24 | |
| Cake | 0.56 | 2.11 | |
| Cheese | 0.32 | 1.22 | |
| S. enterica | Egg | 0.98 | 2.02 |
| Cantaloupe | 0.81 | 1.68 | |
| Fish | 0.80 | 1.66 | |
| Ice cream | 0.79 | 1.64 | |
| Alfalfa sprout | 0.73 | 1.51 | |
| Tomato | 0.71 | 1.47 | |
| Sushi | 0.71 | 1.46 | |
| Chicken | 0.69 | 1.43 | |
| Pork | 0.63 | 1.30 | |
| Cheese | 0.61 | 1.26 | |
| Salsa | 0.60 | 1.24 | |
| Turkey | 0.53 | 1.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, D.; Zhang, D.; Hu, R.; Rundensteiner, E.; Feng, H. Epidemiological Data Mining for Assisting with Foodborne Outbreak Investigation. Foods 2023, 12, 3825. https://doi.org/10.3390/foods12203825
Tao D, Zhang D, Hu R, Rundensteiner E, Feng H. Epidemiological Data Mining for Assisting with Foodborne Outbreak Investigation. Foods. 2023; 12(20):3825. https://doi.org/10.3390/foods12203825
Chicago/Turabian StyleTao, Dandan, Dongyu Zhang, Ruofan Hu, Elke Rundensteiner, and Hao Feng. 2023. "Epidemiological Data Mining for Assisting with Foodborne Outbreak Investigation" Foods 12, no. 20: 3825. https://doi.org/10.3390/foods12203825
APA StyleTao, D., Zhang, D., Hu, R., Rundensteiner, E., & Feng, H. (2023). Epidemiological Data Mining for Assisting with Foodborne Outbreak Investigation. Foods, 12(20), 3825. https://doi.org/10.3390/foods12203825





