Screening of Lactiplantibacillus plantarum 67 with Strong Adhesion to Caco-2 Cells and the Effects of Protective Agents on Its Adhesion Ability during Vacuum Freeze Drying
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains of Bacteria
2.2. Oro-Gastrointestinal Transit Stress Tolerance of LAB
2.3. Adhesion of LAB to Caco-2 Cells
2.4. Key Adhesin of LAB
2.5. Vacuum Freeze-Drying of LAB
2.6. Scanning Electron Microscopy
2.7. Protein Extraction
2.8. Protein Digestion and Tandem Mass Tag Labeling
2.9. Mass Spectrometry Identification
2.10. Protein Identification and Quantification and Bioinformatics
2.11. Statistical Analysis
3. Results
3.1. Survival of LAB after Simulated OGT Stress
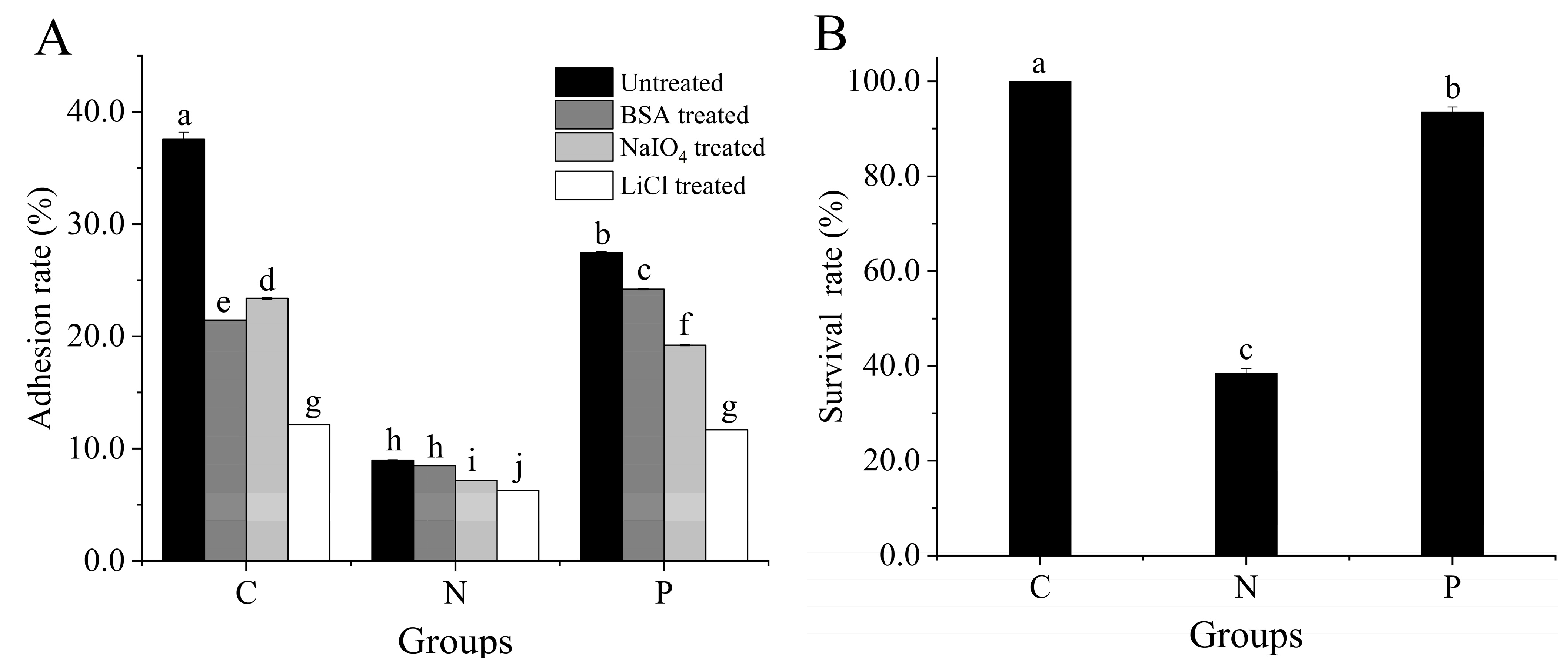
3.2. Adhesion to Caco-2 Cells
3.3. Key Adhesin and Survival Rate of L. plantarum 67
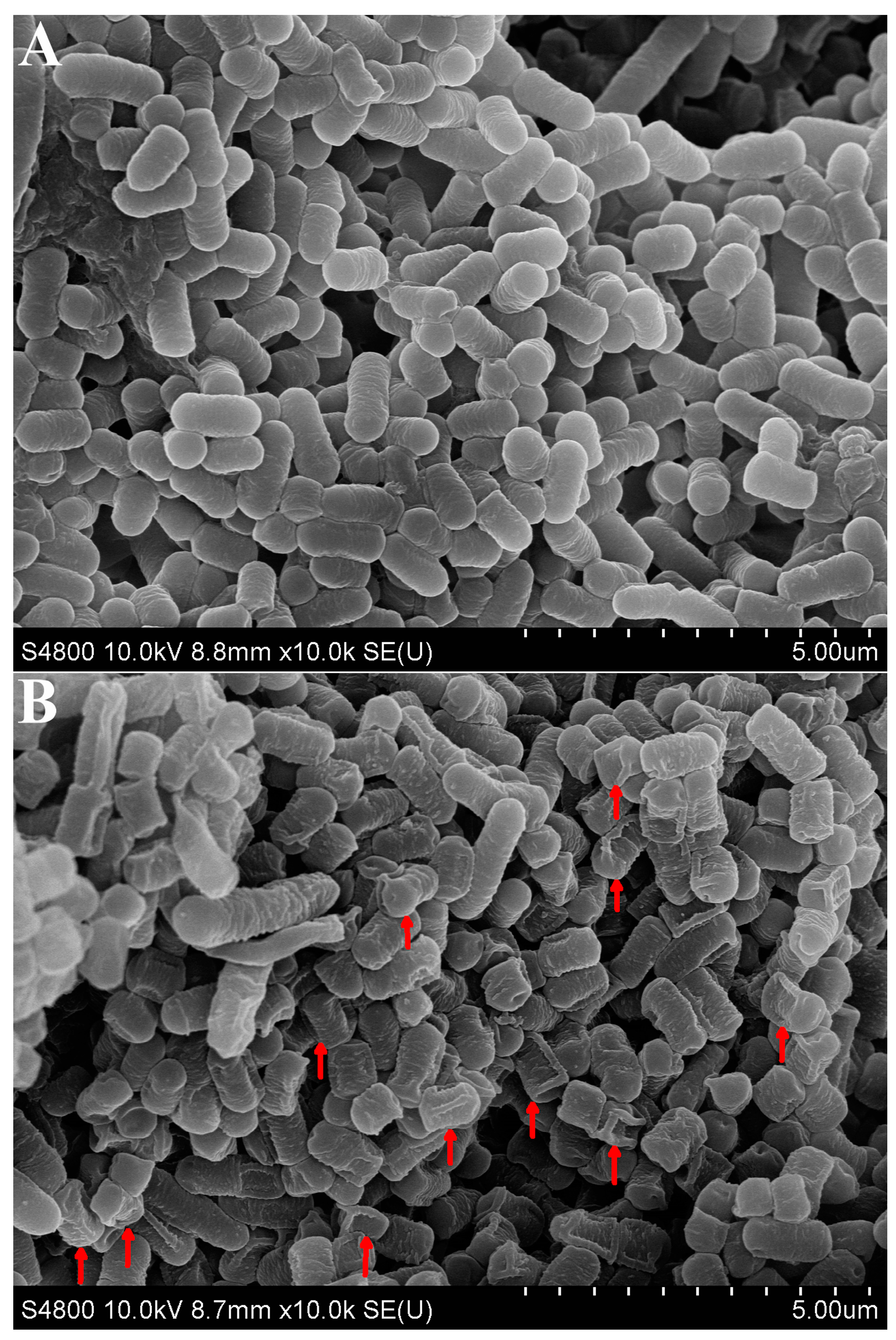
3.4. SEM Observation of L. plantarum 67 Adhering to Caco-2 Cells
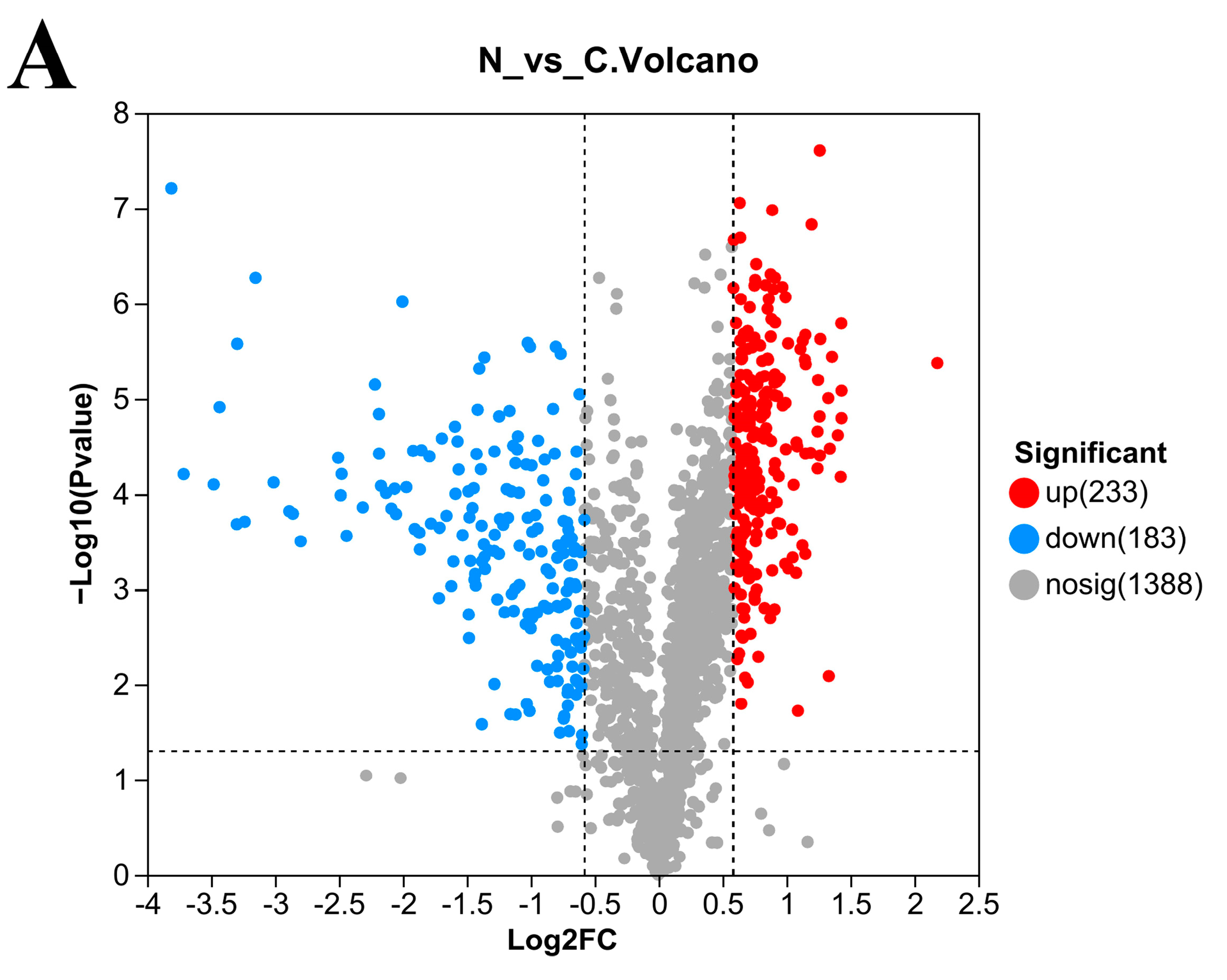
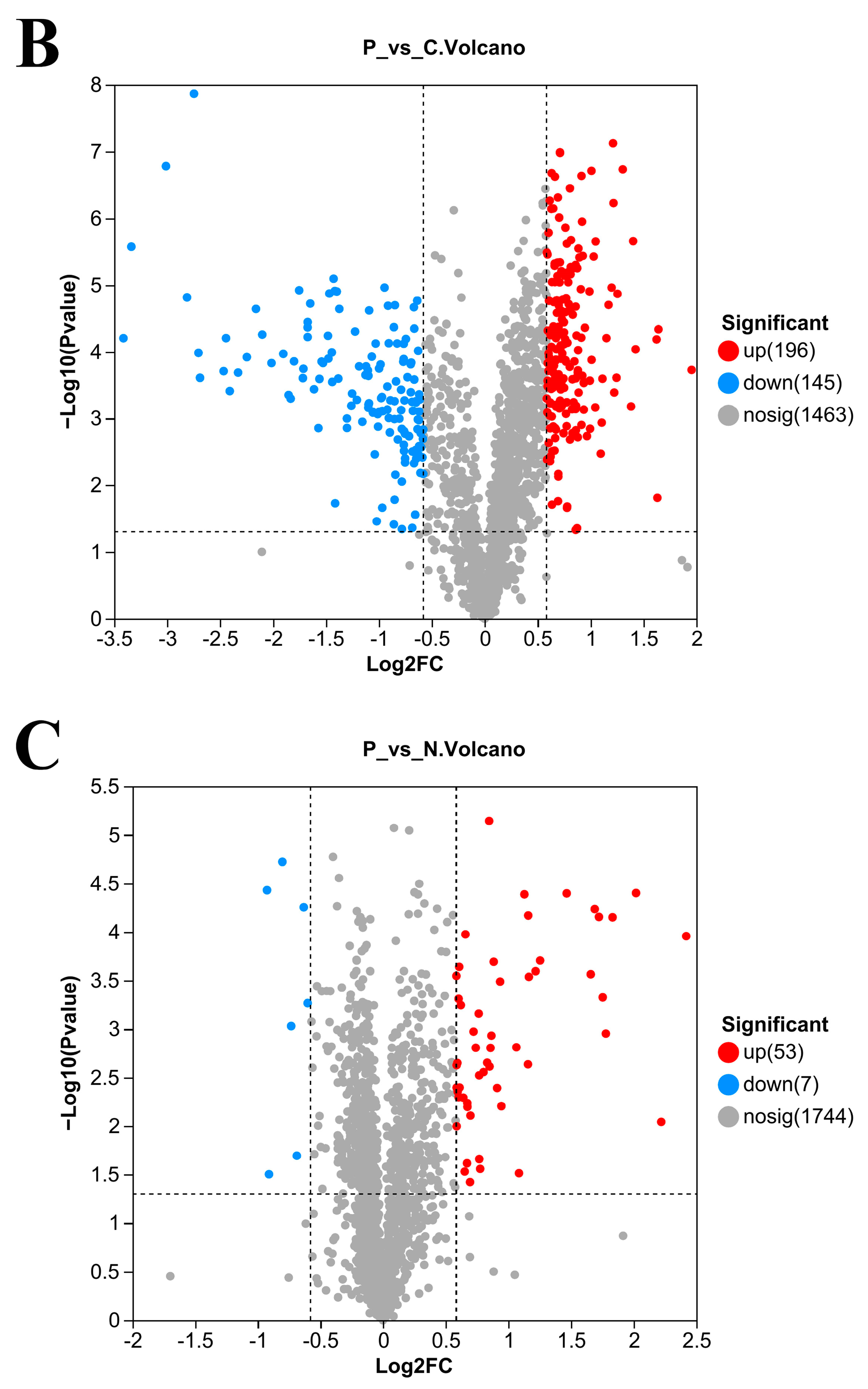
3.5. Number of DEPs in L. plantarum 67 after Vacuum Freeze-Drying
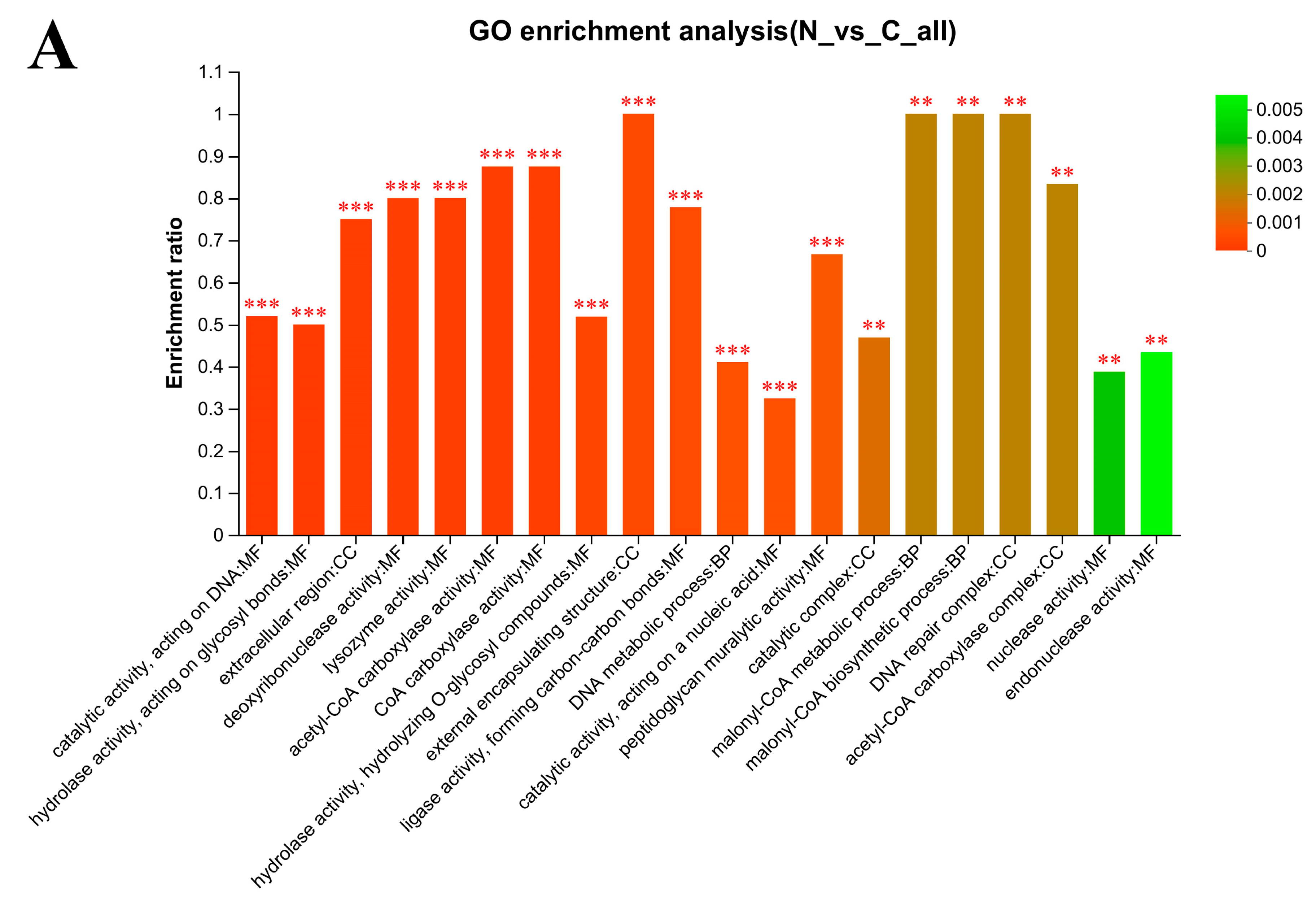
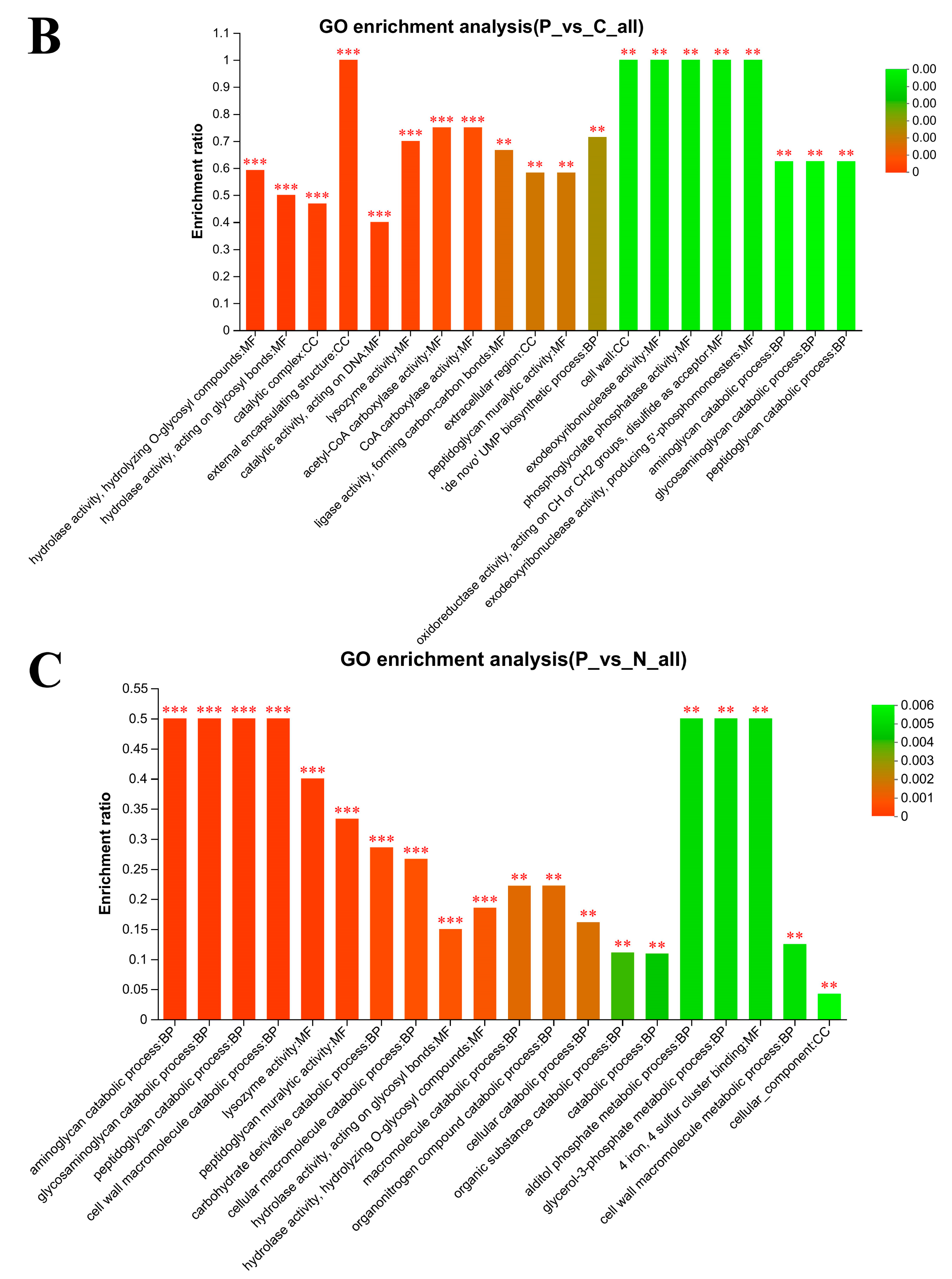
3.6. Gene Ontology (GO) Enrichment Analysis
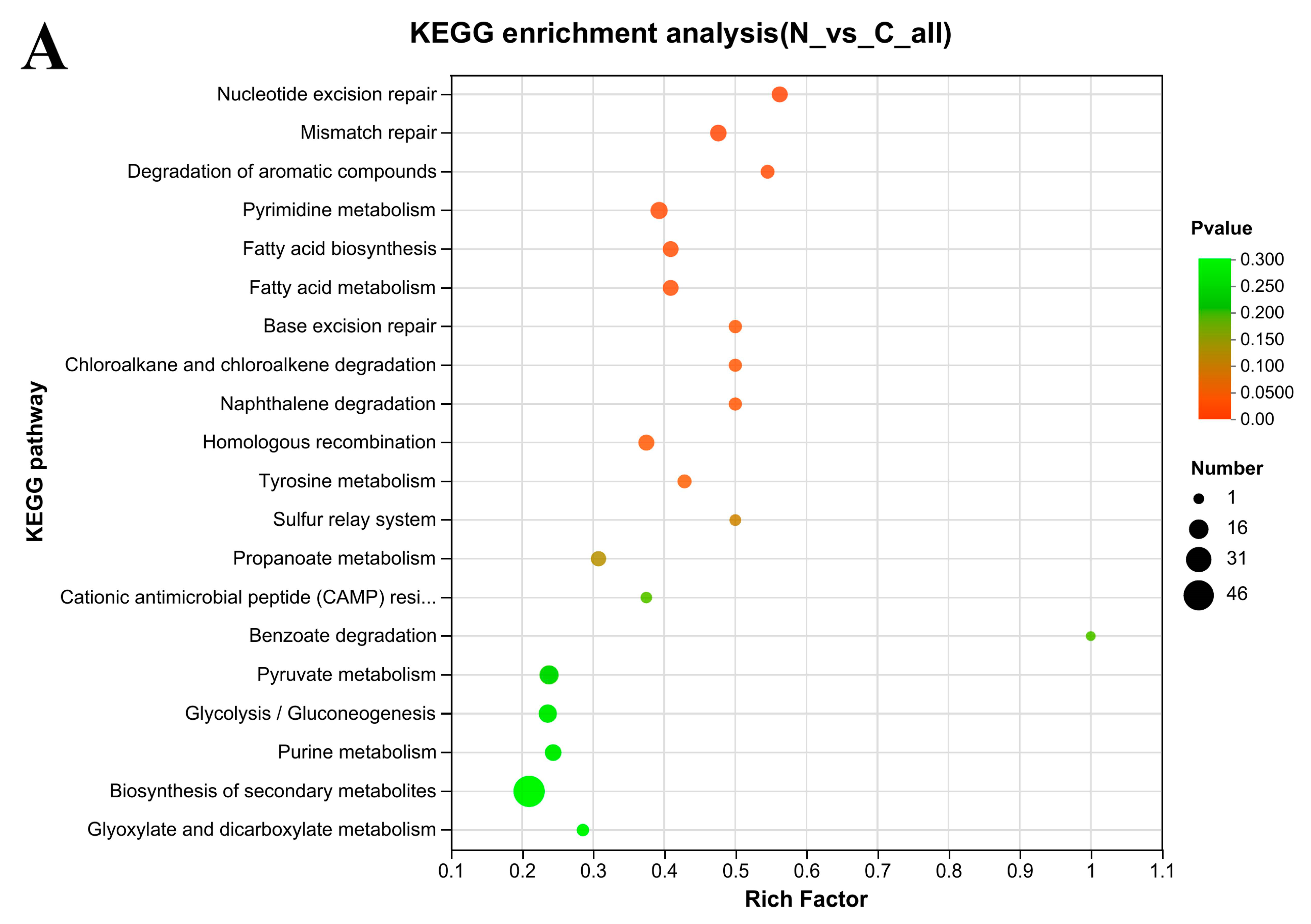
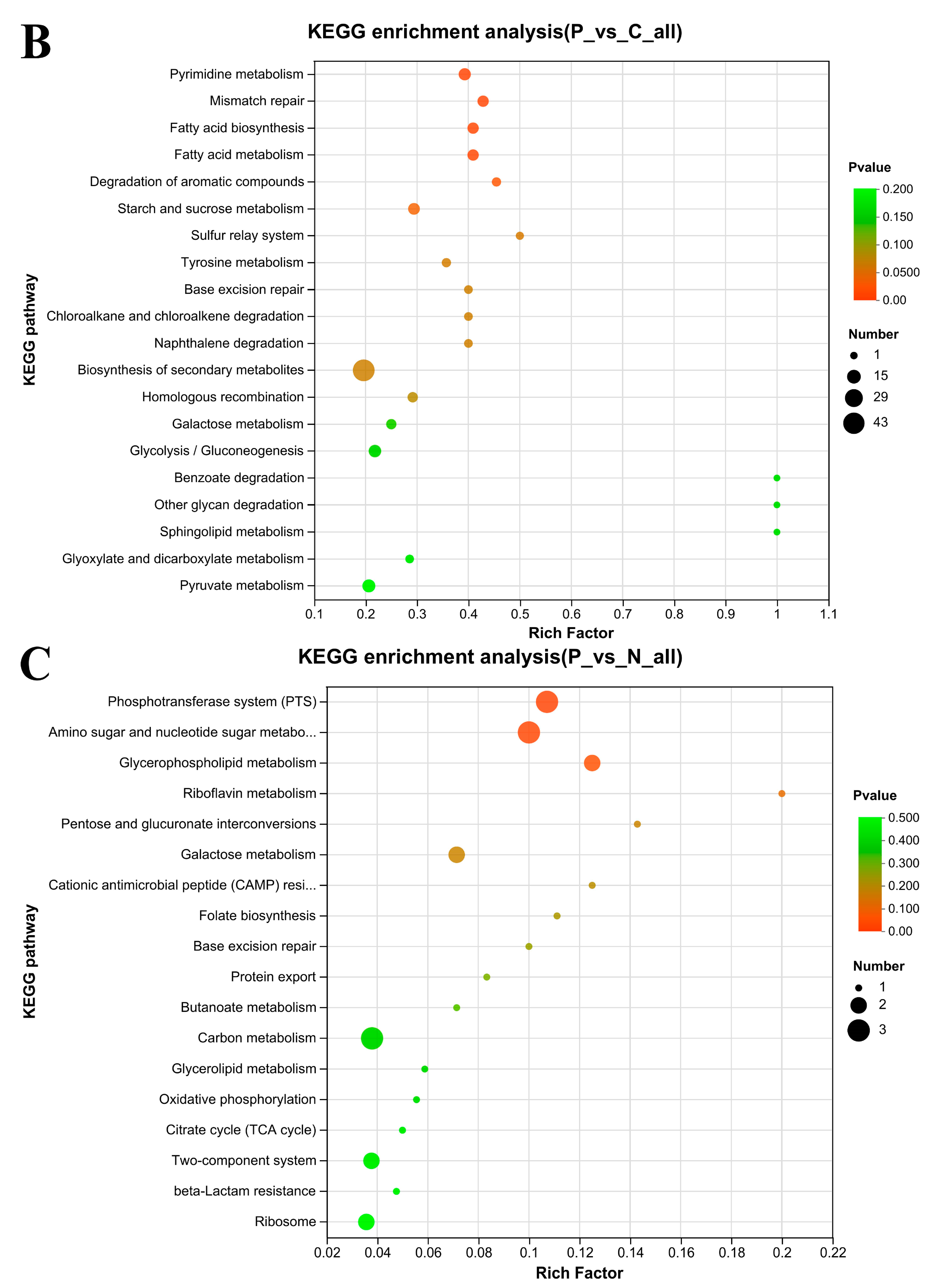
3.7. KEGG Enrichment Analysis
4. Discussion
4.1. Effect of Moonlight Proteins on L. plantarum 67 Adherence and Survival
4.2. Effect of Cell Surface Proteins and Other Proteins on the Adhesion and Survival of L. plantarum 67
4.3. Some KEGG Pathways Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, Z.; Li, X.; Zhang, X.; Du, S.; Jia, M.; Hu, D.; Jia, X.; Cong, B.; Zhang, Y. A new Lactobacillus gasseri strain HMV18 inhibits the growth of pathogenic bacteria. Food Sci. Hum. Wellness 2022, 11, 247–254. [Google Scholar] [CrossRef]
- Wu, H.; Xie, S.; Miao, J.; Li, Y.; Wang, Z.; Wang, M.; Yu, Q. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes 2020, 11, 997–1014. [Google Scholar] [CrossRef] [PubMed]
- Alp, D.; Kuleaşan, H. Adhesion mechanisms of lactic acid bacteria: Conventional and novel approaches for testing. World J. Microbiol. Biotechnol. 2019, 35, 156. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xia, Y.; Cui, J.; Gu, Z.; Song, Y.; Chen, Y.Q.; Chen, H.; Zhang, H.; Chen, W. The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 2014, 16, 15–22. [Google Scholar]
- Jeffery, C. Intracellular proteins moonlighting as bacterial adhesion factors. AIMS Microbiol. 2018, 4, 362–376. [Google Scholar] [CrossRef]
- Jeffery, C.J. Intracellular/surface moonlighting proteins that aid in the attachment of gut microbiota to the host. AIMS Microbiol. 2019, 5, 77. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008, 42, 647–681. [Google Scholar] [CrossRef]
- Cheng, Z.; Yan, X.; Wu, J.; Weng, P.; Wu, Z. Effects of freeze drying in complex lyoprotectants on the survival, and membrane fatty acid composition of Lactobacillus plantarum L1 and Lactobacillus fermentum L2. Cryobiology 2022, 105, 1–9. [Google Scholar] [CrossRef]
- Oluwatosin, S.O.; Tai, S.L.; Fagan-Endres, M.A. Sucrose, maltodextrin and inulin efficacy as cryoprotectant, preservative and prebiotic–towards a freeze dried Lactobacillus plantarum topical probiotic. Biotechnol. Rep. 2022, 33, e00696. [Google Scholar] [CrossRef]
- Chen, D.-W.; Chen, C.-M.; Qu, H.-X.; Ren, C.-Y.; Yan, X.-T.; Huang, Y.-J.; Guan, C.-R.; Zhang, C.-C.; Li, Q.-M.; Gu, R.-X. Screening of Lactobacillus strains that enhance SCFA uptake in intestinal epithelial cells. Eur. Food Res. Technol. 2021, 247, 1049–1060. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, X.; Yang, Y.; Zhang, J.; Zeng, X.; Wu, Z.; Sun, Y.; Pan, D. Prevention of necrotizing enterocolitis through surface layer protein of Lactobacillus acidophilus CICC6074 reducing intestinal epithelial apoptosis. J. Funct. Foods 2018, 47, 91–99. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, X.; Pan, S.; Xu, X.; Wu, T. Effects and Mechanisms of Resveratrol on the Adhesion of Lactobacillus acidophilus NCFM. Probiotics Antimicrob. Proteins 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; E, J.; Ma, R.; Zhang, J.; Yao, C.; Wang, R.; Zhang, Q.; Yang, Y.; Li, J.; Wang, J. The effect of aspartic acid on the freeze-drying survival rate of Lactobacillus plantarum LIP-1 and its inherent mechanism. LWT 2022, 155, 112929. [Google Scholar] [CrossRef]
- Zheng, X.; Fu, N.; Huang, S.; Jeantet, R.; Chen, X.D. Exploring the protective effects of calcium-containing carrier against drying-induced cellular injuries of probiotics using single droplet drying technique. Food Res. Int. 2016, 90, 226–234. [Google Scholar] [CrossRef]
- Lu, W.; Fu, N.; Woo, M.W.; Chen, X.D. Exploring the interactions between Lactobacillus rhamnosus GG and whey protein isolate for preservation of the viability of bacteria through spray drying. Food Funct. 2021, 12, 2995–3008. [Google Scholar] [CrossRef]
- Yang, L.-B.; Guo, G.; Tian, Z.-Q.; Zhou, L.-X.; Zhu, L.-J.; Peng, J.; Sun, C.-Q.; Huang, M.-J. TMT-based quantitative proteomic analysis of the effects of novel antimicrobial peptide AMP-17 against Candida albicans. J. Proteom. 2022, 250, 104385. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Guo, Y.; He, X.; Wang, Y.; Zhao, D.; Ma, Y.; Feng, X.; Zhang, J.; Li, J. TMT-based quantitative proteomics analysis reveals the differential proteins between fresh and frozen-thawed sperm of yak (Bos grunniens). Theriogenology 2023, 200, 60–69. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, X.; Qian, L.; Ai, T.; Xu, Q.; Xiang, W.; Hu, B.; Liu, X.; Wang, J.; Wang, C. Integrated transcriptomic and proteomic analysis of the physiological changes of the liver in domesticated Eurasian perch, Perca fluviatilis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 41, 100957. [Google Scholar] [CrossRef]
- Wen, J.; Niu, X.; Chen, S.; Chen, Z.; Wu, S.; Wang, X.; Yong, Y.; Liu, X.; Yu, Z.; Ma, X. Chitosan oligosaccharide improves the mucosal immunity of small intestine through activating SIgA production in mice: Proteomic analysis. Int. Immunopharmacol. 2022, 109, 108826. [Google Scholar] [CrossRef]
- Hu, X.; Liu, C.; Zhang, H.; Hossen, M.A.; Sameen, D.E.; Dai, J.; Qin, W.; Liu, Y.; Li, S. In vitro digestion of sodium alginate/pectin co-encapsulated Lactobacillus bulgaricus and its application in yogurt bilayer beads. Int. J. Biol. Macromol. 2021, 193, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Bove, P.; Russo, P.; Capozzi, V.; Gallone, A.; Spano, G.; Fiocco, D. Lactobacillus plantarum passage through an oro-gastro-intestinal tract simulator: Carrier matrix effect and transcriptional analysis of genes associated to stress and probiosis. Microbiol. Res. 2013, 168, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Nezhad, M.H.; Knight, M.; Britz, M.L. Evidence of changes in cell surface proteins during growth of Lactobacillus casei under acidic conditions. Food Sci. Biotechnol. 2012, 21, 253–260. [Google Scholar] [CrossRef]
- Muscariello, L.; De Siena, B.; Marasco, R. Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr. Microbiol. 2020, 77, 3831–3841. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef] [PubMed]
- Peighambardoust, S.; Tafti, A.G.; Hesari, J. Application of spray drying for preservation of lactic acid starter cultures: A review. Trends Food Sci. Technol. 2011, 22, 215–224. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Silva, J.; Ho, P.; Teixeira, P.; Malcata, F.X.; Gibbs, P. Relevant factors for the preparation of freeze-dried lactic acid bacteria. Int. Dairy J. 2004, 14, 835–847. [Google Scholar] [CrossRef]
- Jawan, R.; Abbasiliasi, S.; Tan, J.S.; Kapri, M.R.; Mustafa, S.; Halim, M.; Ariff, A.B. Influence of type and concentration of lyoprotectants, storage temperature and storage duration on cell viability and antibacterial activity of freeze-dried lactic acid bacterium, Lactococcus lactis Gh1. Dry. Technol. 2022, 40, 1774–1790. [Google Scholar] [CrossRef]
- Adachi, T.; Kakuta, S.; Aihara, Y.; Kamiya, T.; Watanabe, Y.; Osakabe, N.; Hazato, N.; Miyawaki, A.; Yoshikawa, S.; Usami, T. Visualization of probiotic-mediated Ca2+ signaling in intestinal epithelial cells in vivo. Front. Immunol. 2016, 7, 601. [Google Scholar] [CrossRef]
- Amine, K.M.; Champagne, C.P.; Salmieri, S.; Britten, M.; St-Gelais, D.; Fustier, P.; Lacroix, M. Effect of palmitoylated alginate microencapsulation on viability of Bifidobacterium longum during freeze-drying. LWT-Food Sci. Technol. 2014, 56, 111–117. [Google Scholar] [CrossRef]
- Ji, Y.L.; Pajarillo, E.; Min, J.K.; Chae, J.P.; Kang, D.K. Proteomic and transcriptional analysis of Lactobacillus johnsonii PF01 during bile salt exposure by iTRAQ shotgun proteomics and quantitative RT-PCR. J. Proteome Res. 2013, 12, 432. [Google Scholar]
- Song, S.; Bae, D.-W.; Lim, K.; Griffiths, M.W.; Oh, S. Cold stress improves the ability of Lactobacillus plantarum L67 to survive freezing. Int. J. Food Microbiol. 2014, 191, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Krispin, O.; Allmansberger, R. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 1995, 134, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.P.; Malik, R.K.; Kaur, G. Cell surface proteins play an important role in probiotic activities of Lactobacillus reuteri. Nutrire 2016, 41, 5. [Google Scholar] [CrossRef]
- Post, S.E.; Brito, I.L. Structural insight into protein–protein interactions between intestinal microbiome and host. Curr. Opin. Struct. Biol. 2022, 74, 102354. [Google Scholar] [CrossRef]
- Buck, B.L.; Altermann, E.; Svingerud, T.; Klaenhammer, T.R. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005, 71, 8344–8351. [Google Scholar] [CrossRef]
- Etzold, S.; Kober, O.I.; MacKenzie, D.A.; Tailford, L.E.; Gunning, A.P.; Walshaw, J.; Hemmings, A.M.; Juge, N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 2014, 16, 888–903. [Google Scholar] [CrossRef]
- Yuste, A.; Arosemena, E.L.; Calvo, M.À. Study of the probiotic potential and evaluation of the survival rate of Lactiplantibacillus plantarum lyophilized as a function of cryoprotectant. Sci. Rep. 2021, 11, 19078. [Google Scholar] [CrossRef]
- Romano, N.; Marro, M.; Marsal, M.; Loza-Alvarez, P.; Gomez-Zavaglia, A. Fructose derived oligosaccharides prevent lipid membrane destabilization and DNA conformational alterations during vacuum-drying of Lactobacillus delbrueckii subsp. bulgaricus. Food Res. Int. 2021, 143, 110235. [Google Scholar] [CrossRef]
- Polomska, X.; Wojtatowicz, M.; Zarowska, B.; Szoltysik, M.; Chrzanowska, J. Freeze-drying preservation of yeast adjunct cultures for cheese production. Pol. J. Food Nutr. Sci. 2012, 62, 3. [Google Scholar] [CrossRef]
- Pessione, A.; Bianco, G.L.; Mangiapane, E.; Cirrincione, S.; Pessione, E. Characterization of potentially probiotic lactic acid bacteria isolated from olives: Evaluation of short chain fatty acids production and analysis of the extracellular proteome. Food Res. Int. 2015, 67, 247–254. [Google Scholar] [CrossRef]
- Sánchez García, B.; Urdaci, M.C. Extracellular proteins from Lactobacillus plantarum BMCM12 prevent adhesion of enteropathogens to mucin. Curr. Microbiol. 2012, 64, 592–596. [Google Scholar] [CrossRef]
- Zhai, Z.; Yang, Y.; Wang, H.; Wang, G.; Ren, F.; Li, Z.; Hao, Y. Global transcriptomic analysis of Lactobacillus plantarum CAUH2 in response to hydrogen peroxide stress. Food Microbiol. 2020, 87, 103389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Chen, W.; Wang, M.; Du, G.; Chen, J. A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl. Microbiol. Biotechnol. 2012, 93, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Monedero, V.; Mazé, A.; Boël, G.; Zúñiga, M.; Beaufils, S.; Hartke, A.; Deutscher, J. The phosphotransferase system of Lactobacillus casei: Regulation of carbon metabolism and connection to cold shock response. Microb. Physiol. 2006, 12, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Celebioglu, H.U.; Ejby, M.; Majumder, A.; Købler, C.; Goh, Y.J.; Thorsen, K.; Schmidt, B.; O’Flaherty, S.; Abou Hachem, M.; Lahtinen, S.J. Differential proteome and cellular adhesion analyses of the probiotic bacterium Lactobacillus acidophilus NCFM grown on raffinose—An emerging prebiotic. Proteomics 2016, 16, 1361–1375. [Google Scholar] [CrossRef]
- Degeest, B.; Vuyst, L.D. Correlation of Activities of the Enzymes α-Phosphoglucomutase, UDP-Galactose 4-Epimerase, and UDP-Glucose Pyrophosphorylase with Exopolysaccharide Biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 2000, 66, 3519–3527. [Google Scholar] [CrossRef]
- Perego, M.; Glaser, P.; Minutello, A.; Strauch, M.A.; Leopold, K.; Fischer, W. Incorporation of D-Alanine into Lipoteichoic Acid and Wall Teichoic Acid in Bacillus subtilis: Identification of genes and regulation. J. Biol. Chem. 1995, 270, 15598–15606. [Google Scholar] [CrossRef]
- Lee, J.; Yun, H.M.; Han, G.; Lee, G.J.; Jeon, C.O.; Hyun, S. A bacteria-regulated gut peptide determines host dependence on specific bacteria to support host juvenile development and survival. BMC Biol. 2022, 20, 258. [Google Scholar] [CrossRef]
- De Angelis, M.; Siragusa, S.; Campanella, D.; Di Cagno, R.; Gobbetti, M. Comparative proteomic analysis of biofilm and planktonic cells of Lactobacillus plantarum DB200. Proteomics 2015, 15, 2244–2257. [Google Scholar] [CrossRef] [PubMed]
- Van Wamel, W.J.; van Rossum, G.; Verhoef, J.; Vandenbroucke-Grauls, C.M.; Fluit, A.C. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 1998, 163, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Martín, C.; Fernández-Vega, I.; Suárez, J.E.; Quirós, L.M. Adherence of Lactobacillus salivarius to heLa cells promotes changes in the expression of the genes involved in biosynthesis of their ligands. Front. Immunol. 2020, 10, 3019. [Google Scholar] [CrossRef] [PubMed]


| Strains | Survival Rate after Exposure in Simulated Saliva/(%) | Survival Rate after Exposure in Simulated Saliva and Gastric Juice/(%) | Survival Rate after Exposure in Simulated Saliva, Gastric Juice and Intestinal Juice/(%) | |
|---|---|---|---|---|
| L. plantarum | 67 | 97.19 ± 0.14 eA | 84.29 ± 0.02 bB | 83.23 ± 0.10 aC |
| L160 | 131.11 ± 0.42 aA | 83.75 ± 0.05 cB | 67.63 ± 0.11 bC | |
| L167 | 86.82 ± 0.09 iA | 55.48 ± 0.11 hB | 18.59 ± 0.07 dC | |
| K118 | 95.10 ± 0.58 gA | 29.02 ± 0.02 rB | 3.12 ± 0.00 nC | |
| L169 | 75.18 ± 0.33 mA | 35.54 ± 0.06 mB | 2.81 ± 0.01 oC | |
| L. rhamnosus | L13 | 48.75 ± 0.32 tA | 34.59 ± 0.12 nB | 15.34 ± 0.04 fC |
| O62 | 42.87 ± 0.22 wA | 32.16 ± 0.11 oB | 9.98 ± 0.01 hC | |
| O165 | 96.25 ± 0.11 fA | 30.92 ± 0.05 pB | 7.51 ± 0.01 iC | |
| L63 | 79.91 ± 0.07 lA | 52.66 ± 0.09 iB | 7.34 ± 0.02 jC | |
| L115 | 63.93 ± 0.34 qA | 29.18 ± 0.11 qB | 6.54 ± 0.01 kC | |
| L36 | 87.59 ± 0.19 hA | 6.93 ± 0.02 wB | 1.97 ± 0.00 qC | |
| H113 | 71.95 ± 0.01 oA | 1.44 ± 0.00 zB | 0.77 ± 0.00 tC | |
| e15 | 97.91 ± 0.03 dA | 36.85 ± 0.17 lB | 0.36 ± 0.00 uC | |
| L. fermentum | L7 | 102.33 ± 0.49 cA | 60.24 ± 0.17 gB | 25.09 ± 0.06 cC |
| L73 | 84.65 ± 0.26 jA | 127.73 ± 0.32 aB | 18.26 ± 0.03 eC | |
| O45 | 104.86 ± 0.44 bA | 71.99 ± 0.47 eB | 14.37 ± 0.03gC | |
| L120 | 84.05 ± 0.27 kA | 38.38 ± 0.06 kB | l5.37 ± 0.00 lC | |
| K112 | 49.29 ± 0.09 sA | 15.82 ± 0.06 tB | 3.44 ± 0.01 mC | |
| K161 | 74.39 ± 0.21 nA | 7.35 ± 0.01 vB | 2.14 ± 0.00 pC | |
| R412 | 46.66 ± 0.18 uA | 18.33 ± 0.07 sB | 1.25 ± 0.00 sC | |
| S. thermophilus | F164 | 66.41 ± 0.15 pA | 5.54 ± 0.01 xB | 1.26 ± 0.00 rC |
| Y17 | 45.76 ± 0.35 vA | 2.73 ± 0.01 yB | 0.16 ± 0.00 vC | |
| e114 | 74.41 ± 0.11 nA | 48.24 ± 0.05 jB | 0.13 ± 0.00 wC | |
| K413 | 63.09 ± 0.21 rA | 61.19 ± 0.75 fB | 0.12 ± 0.00 xC | |
| K173 | 41.33 ± 0.40 xB | 77.19 ± 0.16 dA | 0.06 ± 0.00 yC | |
| M613 | 20.80 ± 0.10 yA | 13.06 ± 0.09 uB | 0.04 ± 0.00 zC | |
| Accession | N/C | P/C | P/N | |||
|---|---|---|---|---|---|---|
| FC | p Value | FC | p Value | FC | p Value | |
| Q88WN3 | 0.67 | 0.0031 | - | - | - | - |
| Q88YW7 | - | - | 0.62 | 0.0002 | - | - |
| F9URT0 | 0.61 | 0.0002 | - | - | - | - |
| F9UR61 | 0.52 | 0.0002 | 0.62 | 0.0006 | - | - |
| F9UTV9 | 0.52 | <0.0001 | 0.55 | <0.0001 | - | - |
| F9UTM2 | 0.46 | <0.0001 | 0.66 | 0.0027 | - | - |
| F9UP60 | 0.12 | <0.0001 | 0.21 | 0.0001 | 1.70 | 0.0007 |
| F9ULM1 | 0.46 | 0.0017 | - | - | - | - |
| F9USA9 | 0.50 | <0.0001 | - | - | - | - |
| F9UU92 | 0.62 | 0.0006 | 0.65 | 0.0010 | - | - |
| F9UNI8 | 0.41 | 0.0099 | 0.58 | 0.0457 | - | - |
| F9UU91 | 0.22 | <0.0001 | 0.31 | <0.0001 | - | - |
| F9UU93 | 0.14 | 0.0003 | 0.19 | 0.0004 | - | - |
| F9UUC6 | 0.29 | 0.0002 | 0.64 | 0.0030 | 2.19 | <0.0001 |
| F9ULS6 | 0.55 | 0.0016 | 0.65 | 0.0036 | - | - |
| F9UM21 | 0.47 | 0.0009 | 0.59 | 0.0016 | - | - |
| F9US12 | 0.50 | 0.0026 | 0.66 | 0.0066 | - | - |
| F9US24 | 0.18 | 0.0001 | 0.25 | 0.0001 | - | - |
| F9URE0 | 0.10 | 0.0002 | 0.28 | 0.0005 | 2.76 | <0.0001 |
| F9UUF2 | 0.64 | 0.0033 | - | - | 2.23 | <0.0001 |
| P96349 | 0.58 | 0.0064 | - | - | 1.65 | 0.0011 |
| F9UMI2 | - | - | 1.82 | 0.0015 | 1.81 | 0.0016 |
| Q88VM8 | 0.66 | 0.0034 | - | - | 1.56 | 0.0051 |
| F9UPA0 | 0.34 | <0.0001 | 0.54 | 0.0005 | 1.59 | 0.0058 |
| F9UQ72 | 1.78 | <0.0001 | 1.71 | <0.0001 | - | - |
| Q88UZ8 | 2.11 | 0.0007 | 1.84 | 0.0019 | - | - |
| Q88UZ7 | 1.96 | <0.0001 | 1.82 | <0.0001 | - | - |
| F9USR0 | 1.51 | <0.0001 | - | - | - | - |
| Q88SE8 | - | - | 1.95 | 0.0018 | 1.78 | 0.0022 |
| Q88XY8 | 1.50 | <0.0001 | 1.50 | <0.0001 | - | - |
| F9URS4 | 0.18 | 0.0003 | 0.28 | 0.0004 | 1.50 | 0.0024 |
| F9UQA0 | 0.14 | 0.0002 | 0.18 | 0.0002 | - | - |
| F9UUA0 | 0.18 | <0.0001 | 0.23 | <0.0001 | - | - |
| F9USH2 | 0.52 | 0.0064 | - | - | 1.57 | 0.0295 |
| Q890I8 | 0.23 | <0.0001 | 0.29 | 0.0001 | - | - |
| F9USL7 | 0.41 | 0.0004 | 0.47 | 0.0006 | - | - |
| F9UME2 | 0.30 | 0.0012 | 0.34 | 0.0014 | - | - |
| F9UP14 | 0.11 | <0.0001 | 0.12 | <0.0001 | - | - |
| F9UR18 | 0.60 | 0.0004 | - | - | - | - |
| F9USM7 | 0.36 | 0.0005 | 0.47 | 0.0007 | - | - |
| F9USS1 | 0.61 | 0.0167 | 1.50 | 0.0101 | ||
| F9UN64 | 0.44 | 0.0002 | 0.55 | 0.0006 | - | - |
| F9USV1 | 0.28 | <0.0001 | 0.31 | <0.0001 | - | - |
| F9UTM5 | 0.38 | <0.0001 | 0.36 | <0.0001 | - | - |
| Q88VE9 | 1.75 | 0.0001 | 1.45 | 0.0002 | 0.83 | 0.0044 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Guo, C.; Ren, C.; Xia, Z.; Xu, H.; Qu, H.; Wa, Y.; Guan, C.; Zhang, C.; Qian, J.; et al. Screening of Lactiplantibacillus plantarum 67 with Strong Adhesion to Caco-2 Cells and the Effects of Protective Agents on Its Adhesion Ability during Vacuum Freeze Drying. Foods 2023, 12, 3604. https://doi.org/10.3390/foods12193604
Chen D, Guo C, Ren C, Xia Z, Xu H, Qu H, Wa Y, Guan C, Zhang C, Qian J, et al. Screening of Lactiplantibacillus plantarum 67 with Strong Adhesion to Caco-2 Cells and the Effects of Protective Agents on Its Adhesion Ability during Vacuum Freeze Drying. Foods. 2023; 12(19):3604. https://doi.org/10.3390/foods12193604
Chicago/Turabian StyleChen, Dawei, Congcong Guo, Chenyu Ren, Zihan Xia, Haiyan Xu, Hengxian Qu, Yunchao Wa, Chengran Guan, Chenchen Zhang, Jianya Qian, and et al. 2023. "Screening of Lactiplantibacillus plantarum 67 with Strong Adhesion to Caco-2 Cells and the Effects of Protective Agents on Its Adhesion Ability during Vacuum Freeze Drying" Foods 12, no. 19: 3604. https://doi.org/10.3390/foods12193604
APA StyleChen, D., Guo, C., Ren, C., Xia, Z., Xu, H., Qu, H., Wa, Y., Guan, C., Zhang, C., Qian, J., & Gu, R. (2023). Screening of Lactiplantibacillus plantarum 67 with Strong Adhesion to Caco-2 Cells and the Effects of Protective Agents on Its Adhesion Ability during Vacuum Freeze Drying. Foods, 12(19), 3604. https://doi.org/10.3390/foods12193604






